DOI:10.32604/biocell.2021.09629

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.09629 |  www.techscience.com/journal/biocell |
| Article |
Newly identified genetic variant rs2294693 in UNC5CL gene is associated with decreased risk of esophageal carcinoma in the J&K Population–India
1Human Genetics Research Group, Shri Mata Vaishno Devi University, Katra, Jammu and Kashmir, 182320, India
2Surgical Oncologist, SMVD Narayana Super Specialty Hospital, Katra, Jammu and Kashmir, 182320, India
*Address correspondence to: Ruchi Shah, ruchimicro81@gmail.com; Swarkar Sharma, swarkar.sharma@smvdu.ac.in
Received: 10 January 2020; Accepted: 14 May 2020
Abstract: Esophageal cancer is the second most common type of cancer after lung carcinoma in the state of Jammu and Kashmir (J&K). The understanding of genetics in Esophageal cancer development is poor in the state. Genome wide association studies (GWAS) has proved to be unsurpassed tool in identification of new loci associated with different cancers. GWAS in Chinese population has identified SNP rs2294693 present in UNC5CL (UNC-5 Family C-Terminal like) to be associated with non-cardia gastric cancer. We performed a case control association study and genotyped the SNP rs2294693 using Taqman allele discrimination assay in 566 individuals (166 esophageal cancer patients and 400 controls) belonging to the J&K population. A statistically significant protective association with allelic odds ratio of 0.73 (0.56–0.94 at 95% CI) and p value = 0.016 was observed. This is the first study in relation to esophageal cancer in the Jammu and Kashmir population, so far it has been studied in association with gastric carcinoma in the Chinese population only. The results indicate that the polymorphism rs2294693 is associated with esophageal cancer susceptibility and the mutant (T) allele might be a protective factor for esophageal cancer among Jammu and Kashmir population. Further the functional characterization of the variation is also warranted.
Keywords: Esophageal cancer; Single nucleotide polymorphism; Unc-5 Family C-Terminal (UNC5CL); Candidate gene
Esophageal cancer is the eighth most common cancer worldwide and the sixth leading cause of death from cancer throughout the world (Zhang et al., 2012; Yousefi et al., 2018). It has been observed that about 5,72,034 new cases have been reported in 2018 and each year accounts for about 90% of the 456,000 ESCC (esophageal squamous cell carcinoma) cases (Abnet et al., 2018). The mortality rate of esophageal cancer closely follows the geographical patterns for incidence (Kachala, 2010). Esophageal cancer has been divided into two major subtypes: Esophageal Squamous Cell Carcinoma (ESCC) and Esophageal Adeno Carcinoma (EAC) (Blot and McLaughlin, 1999). ESCC is most prevalent in developing countries like India and Iran, while EAC is common in developed countries like the USA (Mir and Dar, 2009; Barbhuiya et al., 2018). About 90% of the esophageal cancers are ESCC, 5% are EAC, and 5% are other rare forms of esophageal cancer in Kashmir valley (Mir and Dar, 2009). A number of risk factors are associated with esophageal carcinoma. Alcohol consumption, hot drinks, and smoking are being primarily associated with ESCC, and obesity, smoking, and acid reflux are associated with EAC (Hazelton et al., 2015).
Esophageal cancer is the second-largest prevailing cancer in Jammu and Kashmir, India, after lung cancer (Dikshit et al., 2012), and it accounts for 60% of all cancers along with gastric cancer in the region (Shah et al., 2020). The population of Jammu and Kashmir (J&K) is highly diverse in terms of ethnicity and culture (Sharma et al., 2018). The population of Jammu and Kashmir is ethnically distinct and have special dietary habits and weather. Due to these climatic conditions of Kashmir valley, the population usually depend on preserved foods like smoked pickle, sundried vegetables, and these food products contain carcinogens like N-nitrosodimethylamine (NDMA), methyl nitrosourea (NMU), N-nitrosopyrrolidine (NPYR), that could lead to adverse effects in various organs leading to favorable condition for cancers (Chikan et al., 2012). Studies have shown that both cases, as well as controls in the population of J&K, are exposed to the same type of dietary habits. These studies also suggest that there is a chance of a higher rate of esophageal cancer in the population that is exposed to common dietary risk factors than those without such exposures (Murtaza et al., 2006). There has been a rapid increase in ESCC in Kashmir according to the cancer registry of SKIMS (Sher-E-Kashmir Institute of Medical Sciences) from years 2001–2003. Incidence of ESCC is low in Jammu and Ladakh compared to Kashmir valley (Khuroo et al., 1992; Iqbal et al., 2015). Alterations in the genetic makeup of an individual play an important role in esophageal carcinogenesis (Xing et al., 2003). Genome-wide studies have been widely used to identify the novel common variants associated with different cancers. GWAS (Genomic Wide Association Studies) in the Asian population identified variant rs2294693 of UNC5CL associated with gastric non-cardia cancer risk (Hu et al., 2016). This genetic variant in UNC5CL falls in a genomic region that is in close proximity with two SNPs, rs10484761 and rs2494938, which are a hotspot for upper gastrointestinal cancers and has been associated with esophageal squamous cell carcinoma and non-cardia gastric cancer in Chinese populations (Hu et al., 2016). In the current study, we tried to explore the genetic variant in the population of J&K (Jammu and Kashmir) to find if it is associated with the increased or decreased risk of ESCC. It should be noted that both the frequency of SNPs and the extent of LD varies significantly between different populations (Erichsen and Chanock, 2004). In order to determine the clinical implication of the variant, it is essential to replicate the newly identified variants in other ethnicities (Jing et al., 2014) to ascertain and strengthen the findings of GWAS.
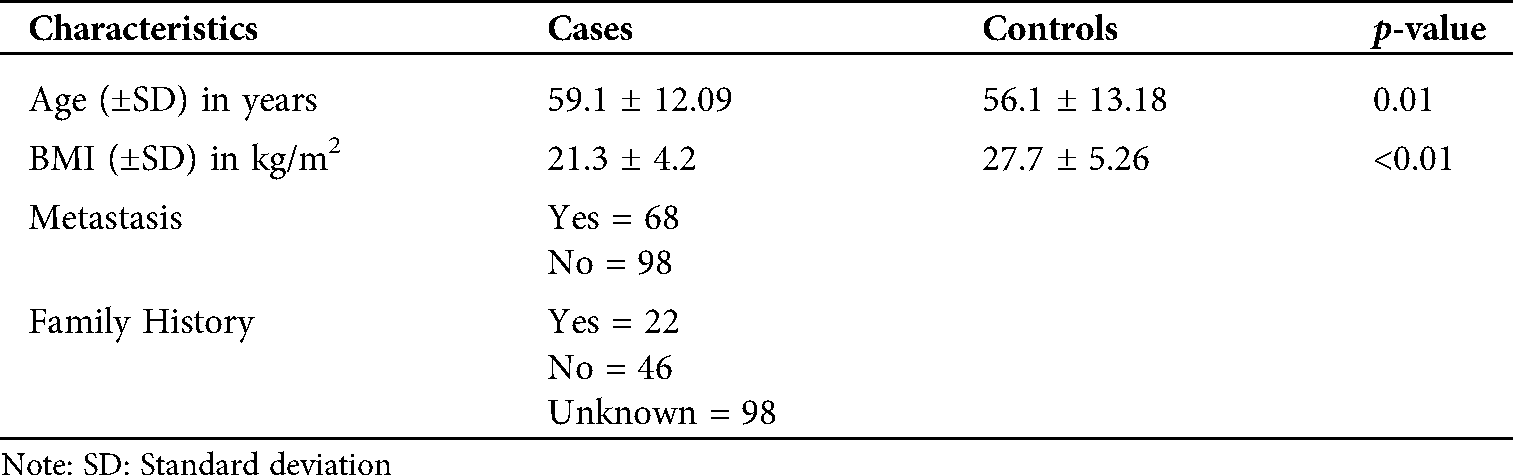
A total of 566 individuals (166 cancerous and 400 healthy controls) were recruited for the study after approval from the Institutional Ethical Review Board (IERB)–Shri Mata Vaishno Devi University (SMVDU) under notification number (SMVDU/IERB/16/43) dated 12/05/2016. The cancer cases were recruited only after the histopathological confirmation. All the patients recruited were neither on chemotherapy nor radiotherapy. All the control samples recruited had no prior family history of ESCC or related malignancy. A 2 mL-venous blood sample was collected, and written informed consent was obtained from all the participants. Details of both cases and controls have been coded and no information which reveals the identity of the subjects has been divulged. The details of cases and controls are given in Tab. S1.
The genomic DNA was isolated by phenol–chloroform method given by Ghatak et al. (2013) and Flexi gene® DNA Kit. The DNA was quantified using the Eppendorf’s Bio-spectrophotometerTM at wavelengths of 260 nm and 280 nm. The amount of DNA was calculated using the following formula: DNA ng/mL = OD at 260 nm × 50X dilution factor. The ratio at 260/280 was taken as criteria to check the purity of DNA.
The TaqMan allele discrimination assay was adopted to perform the genotyping using real-time PCR (Agilent Stratagene M × 3005p, Wald Brown, Germany) following the procedure given by Bhat et al. (2019) (21). TaqMan ®SNP Genotyping assay labelled with FAM and VIC (ThermoFisher Scientific). Following is the primer sequence of the probes used in the study: TGGTATATTGGAAATTGAGACAGCA(A/G) CTTCTCTGCATCAAGGAGACTCTTA; and UNG Mastermix II (Applied Biosystems, ThermoFisher Scientific, Foster City, CA, USA) were used. Following thermal cycling conditions were adopted; 10 min at 95°C, 40 cycles of 95°C for 15 s, and 60°C for 1 min. In each 96-well, 3 NTC’s (non-template controls) were used for quality control. The genotype data were analyzed by Agilent MX3005p Post PCR detection software. As a quality control measure, 40 cases and 40 controls were randomly selected and validated by genotyping, no mismatch was found.
The Chi-square (χ2) analysis was performed to test the genotypic frequency distribution for the Hardy–Weinberg equilibrium. Logistic regression analysis was used to estimate the odds ratio (OR), at 95% confidence interval (CI) and respective level of significance as a p-value. Covariates for adjustment in this study were age, gender, and BMI (Body mass index). The statistical analyses was performed using the Statistical Package for the Social Sciences (SPSS) version 23.
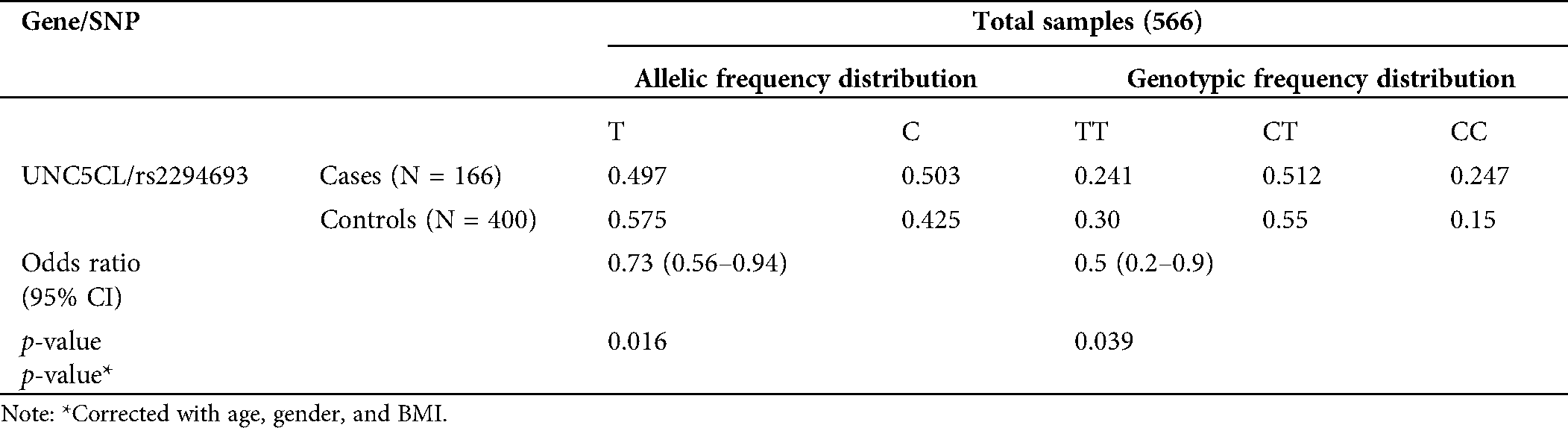
The case-control association study included 166 cases and 400 controls (N = 566). The genotypic and allelic frequency distribution for the studied population is given in Tab. 1. The frequency distribution of the population was in accordance with the Hardy-Weinberg equilibrium (p = 0.10). The T allele of the SNP (single nucleotide polymorphism) rs2294693 was significantly associated with the reduced risk of esophageal carcinoma in the Jammu and Kashmir population, having an allelic odds ratio (OR) 0.73 (95% CI, 0.56–0.94, p = 0.016) (Tab. 1). XT (TT + TC) genotypes under the dominant model when compared against CC genotype yielded corrected OR 0.5 (0.34–0.84, at 95% CI, p = 0.006). After correction with confounding factors like age, gender, and BMI, the observed OR observed was 0.5 (0.2–0.9, at 95% CI, p = 0.039).
Table 1: Allele frequency distribution and risk associated with variant rs2294693 of UNC5CL in the studied population group from the Jammu and Kashmir, India
UNC5CL is also called ZU5 or ZUD (Stelzer et al., 2018). It has pro-inflammatory activity and specific expression in the epithelium, which demonstrates its association with mucosal diseases like Inflammatory Bowel Disease (IBD), and it needs to be explored in future genetic association studies (Heinz et al., 2012). In the current study, the interaction of the UNC5CL gene was also predicted using the Genemania tool of BioGRID version 3.4 (Stark et al., 2006). UNC5CL has been found to be physically interacting with NF-k Band REL gene (Fig. S1). NF-κB is a protein-coding gene and transcription regulator (Stelzer et al., 2018). NF-κB has an important role in the initiation, development, and metastasis of human cancers (Xia et al., 2014). REL is a proto-oncogene and plays a vital role in cell differentiation and lymphopoiesis. REL/NFkB pathway regulates immunity, apoptosis, and cell proliferation. Many studies have shown a correlation between REL/NF-κB factors, gene expression, and their role in malignancy (Kojima et al., 2004). It has been indicated that functional inactivation of NF-κB in many tumor cell types induce apoptosis and alienate oncogenicity (Rayet and Gelinas, 1999). The interactive functional role of UNC5CL, NF-κB, and REL in the pathogenesis of cancer needs further elucidation.
In a combined Asian population (Beijing, Henan, and Korea), a GWAS study found that the variant rs2294693 of UNC5CL showed risk with gastric non-cardia cancer (p = 2.50 × 10−8) and nominal risk for cardia cancer (p = 1.47 × 10−2) (Hu et al., 2016). In the above study, T is the reference allele, and C is the risk allele. The result showed that EAF (Effect Allele Frequency) in cases is 0.277, and EAF in controls is 0.226 with OR = 1.14 (1.09–1.20, p < 0.05). The present study shows that the variant rs2294693 of UNC5CL gene is showing protection against esophageal carcinoma in the population of J&K population. The frequency of allele T in controls (0.575) is higher than cases (0.497), which may be a protective factor for esophageal cancer in the studied population. These results may be due to differences in ethnicity, lifestyle, and disease prevalence, as well as possible limitations due to the relatively small sample size.
The above-investigated pattern of association can occur when the investigated variant is correlated, through interactive effects or linkage disequilibrium (LD), with a causal variant (Lin et al., 2007). Single nucleotide polymorphism (SNPs) varies in frequencies within ethnic groups (Moorjani et al., 2013; Sharma et al., 2017). It is pertinent to evaluate the variants, which are in strong LD with the studied SNP, in order to find the contributory variant associated with disease. We also evaluated the LD of the variant rs2294693 of the UNC5CL gene using the Ensemble database (www.ensemble.org). The variant rs2294693 is an intronic SNP in UNC5CL, but it is in close proximity with 5’UTR variant rs9369260 of TSPO gene (Translocator protein gene) (Fig. S2). TSPO has been associated with cell proliferation, apoptosis, and mitochondrial function. It has been investigated that the expression of TSPO protein is associated with tumor malignancy (Bhoola et al., 2018). TSPO encodes PBR (peripheral type benzodiazepine receptor) protein, which has been linked to regulation of apoptosis and immune response; because of these functions, it has been associated with a number of diseases and cancers. It has been investigated that expression level of PBR is increased in a number of malignancies, which also include esophageal cancer. PBR is considered an independent prognostic factor in ESCC (Chen et al., 2017). We can hypothesize that as the rs2294693 in UNC5CL and rs9369260 in TSPO gene are in close proximity with each other, there must be a mechanism of interaction between the two and a pathway that links these two genes with ESCC.
This variant has not been studied so far in any Indian population. Our study is the first one to explore this variant in association with ESCC in J&K. Evaluation of this SNP in different Indian ethnic groups will present a better insight towards its significance, and future studies should address whether these differences in association may be related to the local incidence of cancer or not. Though the power of the study is above 90% on the basis of the incidence of ESCC in the J&K region, it is very important to replicate the study in the large cohort for a conclusive statement.
Drug response is just as complex as disease genetics, resulting not only from underlying genotypic variation at many loci, but also from variation at the level of gene expression, post-translational modification of proteins, drug dose, drug interactions, diet, and other non-genetic factors. Therefore, individual genes will be associated with effects on drug response. To have no data from the population severely undermines the importance of research that needs to be undertaken in these specific domains. This present study investigated the association of UNC5CL gene polymorphisms with esophageal cancer among an Indian subpopulation. The mutant allele T of the genetic variant rs2294693 of UNC5CL gene is a protective factor for the development of esophageal cancer in the studied population. Evaluation of this SNP in different Indian ethnic groups will present a better insight towards the significance, and future studies should address whether these differences in association direction may be related to the local incidence of cancer or not. These results are the first step towards the genetic elucidations of cancers in the region. This Region in the past has suffered heavily due to the turmoil related to political conditions which led to the shifting of focus from diseases and looking at the recent data, these diseases have now taken alarming proportions. It is worth to mention that the region studied is the lung cancer capital of India, where smoking is at the highest. The esophagus being the doorway for lung cancer is the primary site and hotspot for the malignancy. The investigated information can be used to create diagnostic and prognostic markers. Cancer controls programs can be successful across the globe only if the cancer menace is uprooted at the regional level also. SNP study helps the medical practitioners to provide the treatment on the basis of the assessment of risk. As already informed in a previous revision, the focus on these SNPs will help in establishing the biomarkers for the esophageal cancers in the region.
Acknowledgement: RS and SS acknowledges Women Scientist Scheme (WOS-A), Department of Science and Technology, Grant No. (SR/WOS-A/LS1067/2015) Government of India for fellowship
Availability of Data and Materials: Data generated and analyzed during study is not available publicly but can be made available from the corresponding author upon reasonable request.
Authors Contribution: RS and SS have planned the study, VS, IS, HS and ER edited the manuscript critically. SV, AB, GR helped in sampling and compilation of tables and figures. SK provided the samples. RS and SS also acknowledge Dr. Nazir Ahmed Dar, Department of Biochemistry, University of Kashmir, for providing samples from his repository.
Ethical Approval: The study was approved by the Institutional Ethics Review board (IERB) of Shri Mata Vaishno Devi University (SMVDU) vide IERB Serial No. SMVDU/IERB/16/41. The informed written consent was taken into account from each participant and all the parameters were recorded in pre-designed Performa. In this study all experimental research work was performed according to guidelines issued by Institutional Ethics Review board committee.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abnet CC, Arnold M, Wei WQ. (2018). Epidemiology of esophageal squamous cell carcinoma. Gastroenterology 154: 360–373. DOI 10.1053/j.gastro.2017.08.023. [Google Scholar] [CrossRef]
Barbhuiya MA, Kashyap MK, Puttamallesh VN, Kumar RV, Wu X, Pandey A, Gowda H. (2018). Identification of spleen tyrosine kinase as a potential therapeutic target for esophageal squamous cell carcinoma using reverse phase protein arrays. Oncotarget 9: 18422–18434. DOI 10.18632/oncotarget.24853. [Google Scholar] [CrossRef]
Bhat A, Shah R, Bhat GR, Verma S, Sharma V, Sharma I, Pandita M, Bakshi D, Sharma B, Suri J, Kumar R. (2019). Association of ARID5B and IKZF1 variants with leukemia from Northern India. Genetic Testing and Molecular Biomarkers 23: 176–179. DOI 10.1089/gtmb.2018.0283. [Google Scholar] [CrossRef]
Bhoola NH, Mbita Z, Hull R, Dlamini Z. (2018). Translocator Protein (TSPO) as a potential biomarker in human cancers. International Journal of Molecular Sciences 19: 2176. DOI 10.3390/ijms19082176. [Google Scholar] [CrossRef]
Blot WJ, McLaughlin JK. (1999). The changing epidemiology of esophageal cancer. Seminars in Oncology 26: 2. [Google Scholar]
Chen YF, Xie JD, Jiang YC, Chen DT, Pan JH, Chen YH, Yuan YF, Wen ZS, Zeng WA. (2017). The prognostic value of peripheral benzodiazepine receptor in patients with esophageal squamous cell carcinoma. Journal of Cancer 8: 3343–3355. DOI 10.7150/jca.20739. [Google Scholar] [CrossRef]
Chikan NA, Shabir N, Shaffi S, Mir MR, Patel TN. (2012). N-nitrosodimethylamine in the Kashmiri diet and possible roles in the high incidence of gastrointestinal cancers. Asian Pacific Journal of Cancer Prevention 13: 1077–1079. DOI 10.7314/APJCP.2012.13.3.1077. [Google Scholar] [CrossRef]
Dikshit R, Gupta PC, Ramasundarahettige C, Gajalakshmi V, Aleksandrowicz L, Badwe R, Kumar R, Roy S, Suraweera W, Bray F, Mallath M, Singh PK, Sinha DN, Shet AS, Gelband H, Jha P. (2012). Cancer mortality in India: A nationally representative survey. The Lancet 379: 1807–1816. DOI 10.1016/S0140-6736(12)60358-4. [Google Scholar] [CrossRef]
Erichsen H, Chanock S. (2004). SNPs in cancer research and treatment. British Journal of Cancer 90: 747–751. DOI 10.1038/sj.bjc.6601574. [Google Scholar] [CrossRef]
Ghatak S, Muthukumaran RB, Nachimuthu SK. (2013). A simple method of genomic DNA extraction from human samples for PCR-RFLP analysis. Journal of Biomolecular Techniques 24: 224. [Google Scholar]
Hazelton WD, Curtius K, Inadomi JM, Vaughan TL, Meza R, Rubenstein JH, Hur C, Luebeck EG. (2015). The role of gastroesophageal reflux and other factors during progression to esophageal adenocarcinoma. Cancer Epidemiology Biomarkers & Prevention 24: 1012–1023. DOI 10.1158/1055-9965.EPI-15-0323-T. [Google Scholar] [CrossRef]
Heinz LX, Rebsamen M, Rossi DC, Staehli F, Schroder K, Quadroni M, Gross O, Schneider P, Tschopp J. (2012). The death domain-containing protein Unc5CL is a novel MyD88-independent activator of the pro-inflammatory IRAK signaling cascade. Cell Death & Differentiation 19: 722–731. DOI 10.1038/cdd.2011.147. [Google Scholar] [CrossRef]
Hu N, Wang Z, Song X, Wei L, Kim BS, Freedman ND, Baek J, Burdette L, Chang J, Chung C, Dawsey SM, Ding T, Gao YT, Giffen C, Han Y, Hong M, Huang J, Kim HS, Koh WP, Liao LM, Mao YM, Qiao YL, Shu XO, Tan W, Wang C, Wu C, Wu MJ, Xiang YB, Yeager M, Yook JH, Yuan JM, Zhang P, Zhao XK, Zheng W, Song K, Wang LD, Lin D, Chanock SJ, Goldstein AM, Taylor PR, Abnet CC. (2016). Genome-wide association study of gastric adenocarcinoma in Asia: A comparison of associations between cardia and non-cardia tumours. Gut 65: 1611–1618. DOI 10.1136/gutjnl-2015-309340. [Google Scholar] [CrossRef]
Iqbal B, Shah IA, Bhat GA, Bhat AB, Rafiq R, Nabi S, Malekhzadeh R, Abnet CC, Boffetta P, Jenab M, Dar NA. (2015). Impediments in foreign collaboration and conducting a high throughput molecular epidemiology research in India, an assessment from a feasibility study. SpringerPlus 4: 287. DOI 10.1186/s40064-015-1046-z. [Google Scholar] [CrossRef]
Jing L, Su L, Ring BZ. (2014). Ethnic background and genetic variation in the evaluation of cancer risk: A systematic review. PLoS One 9: e97522. DOI 10.1371/journal.pone.0097522. [Google Scholar] [CrossRef]
Kachala R. (2010). Systematic review: Epidemiology of oesophageal cancer in SubSaharan Africa. Malawi Medical Journal 22: 65–70. DOI 10.4314/mmj.v22i3.62190. [Google Scholar] [CrossRef]
Khuroo M, Zargar S, Mahajan R, Banday M. (1992). High incidence of oesophageal and gastric cancer in Kashmir in a population with special personal and dietary habits. Gut 33: 11–15. DOI 10.1136/gut.33.1.11. [Google Scholar] [CrossRef]
Kojima M, Morisaki T, Sasaki N, Nakano K, Mibu R, Tanaka M, Katano M. (2004). Increased nuclear factor-kB activation in human colorectal carcinoma and its correlation with tumor progression. Anticancer Research 24: 675–682. [Google Scholar]
Lin PI, Vance JM, Pericak-Vance MA, Martin ER. (2007). No gene is an island: The flip-flop phenomenon. American Journal of Human Genetics 80: 531–538. DOI 10.1086/512133. [Google Scholar] [CrossRef]
Mir MM, Dar NA. (2009). Esophageal cancer in Kashmir (IndiaAn enigma for researchers. International Journal of Health Sciences 3: 71. [Google Scholar]
Moorjani P, Thangaraj K, Patterson N, Lipson M, Loh PR, Govindaraj P, Berger B, Reich D, Singh L. (2013). Genetic evidence for recent population mixture in India. American Journal of Human Genetics 93: 422–438. DOI 10.1016/j.ajhg.2013.07.006. [Google Scholar] [CrossRef]
Murtaza I, Mushtaq D, Margoob MA, Dutt A, Wani NA, Ahmad I, Lal Bhat M. (2006). A study on p53 gene alterations in esophageal squamous cell carcinoma and their correlation to common dietary risk factors among population of the Kashmir valley. World Journal of Gastroenterology 12: 4033. DOI 10.3748/wjg.v12.i25.4033. [Google Scholar] [CrossRef]
Rayet B, Gelinas C. (1999). Aberrant rel/nfkb genes and activity in human cancer. Oncogene 18: 6938–6947. DOI 10.1038/sj.onc.1203221. [Google Scholar] [CrossRef]
Shah R, Khaitan PG, Pandita TK, Rafiq A, Abrol D, Suri J, Kaul S, Kumar R, Sharma S. (2020). Gastric cancer in Jammu and Kashmir, India: A review of genetic perspectives. Journal of Cancer Research and Therapeutics, DOI 10.4103/jcrt.JCRT_12_19. [Google Scholar] [CrossRef]
Sharma I, Sharma V, Khan A, Kumar P, Rai E, Bamezai RNK, Vilar M, Sharma S. (2018). Ancient human migrations to and through Jammu Kashmir-India were not of males exclusively. Scientific Reports 8: 851. DOI 10.1038/s41598-017-18893-8. [Google Scholar] [CrossRef]
Sharma V, Sharma I, Sethi I, Mahajan A, Singh G, Angural A, Bhanwer AJS, Dhar MK, Singh V, Rai E, Sharma S. (2017). Replication of newly identified type 2 diabetes susceptible loci in Northwest Indian population. Diabetes Research and Clinical Practice 126: 160–163. DOI 10.1016/j.diabres.2017.02.013. [Google Scholar] [CrossRef]
Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. (2006). BioGRID: A general repository for interaction datasets. Nucleic Acids Research 34: D535–D539. DOI 10.1093/nar/gkj109. [Google Scholar] [CrossRef]
Stelzer G, Rosen R, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Iny Stein T, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. (2018). The GeneCards Suite: From gene data mining to disease genome sequence analysis. Current Protocols in Bioinformatics 54: 1.30.1–1.30.33. [Google Scholar]
Xia Y, Shen S, Verma IM. (2014). NF-κB, an active player in human cancers. Cancer Immunology Research 2: 823–830. DOI 10.1158/2326-6066.CIR-14-0112. [Google Scholar] [CrossRef]
Xing D, Tan W, Lin D. (2003). Genetic polymorphisms and susceptibility to esophageal cancer among Chinese population. Oncology Reports 10: 1615–1623. [Google Scholar]
Yousefi MS, Sharifi-Esfahani M, Pourgholam-Amiji N, Afshar M, Sadeghi-Gandomani H, Otroshi O, Salehiniya H. (2018). Esophageal cancer in the world: Incidence, mortality and risk factors. Biomedical Research and Therapy 5: 2504–2517. DOI 10.15419/bmrat.v5i7.460. [Google Scholar] [CrossRef]
Zhang HZ, Jin GF, Shen HB. (2012). Epidemiologic differences in esophageal cancer between Asian and Western populations. Chinese Journal of Cancer 31: 281–286. DOI 10.5732/cjc.011.10390. [Google Scholar] [CrossRef]
Supplementary Files
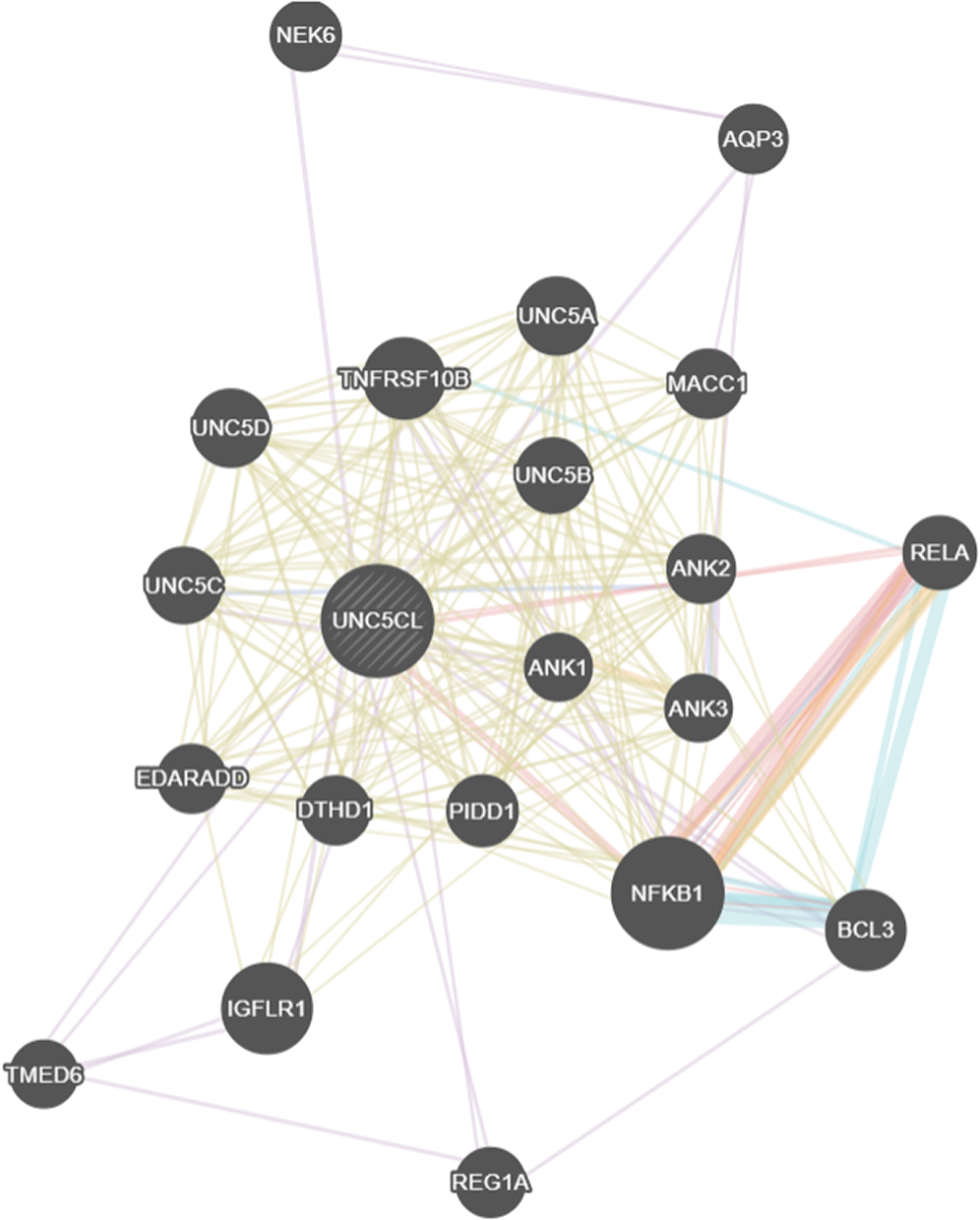
SUPPLEMENTARY FIGURE S1: The Genemania network interaction of UNC5CL with NF-k Band REL.
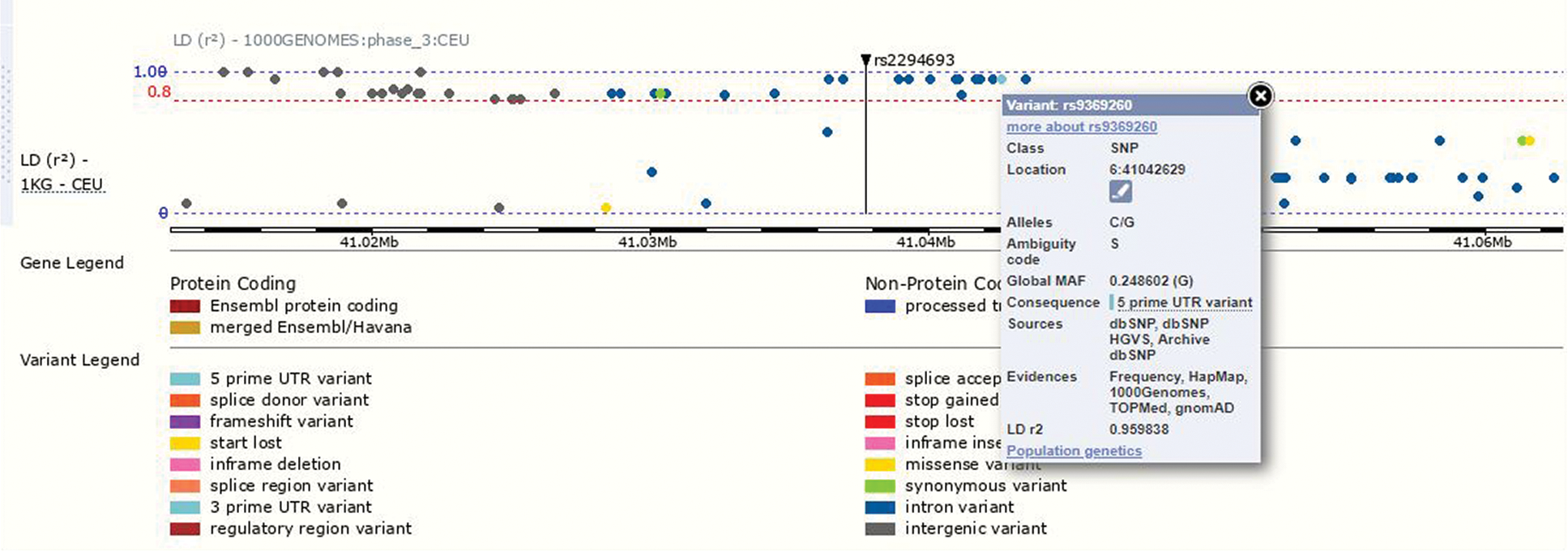
SUPPLEMENTARY FIGURE S2: The variant rs2294693 of UNC5CL shows close proximity with 5’UTR variant rs9369260 of the TSPO2 gene.
Supplementary Table 1: Details and clinical features of the cases and controls of J&K Population, India
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |