DOI:10.32604/biocell.2021.012519

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.012519 |  www.techscience.com/journal/biocell |
| Article |
Hypoxia-associated circular RNA RPPH1 modulates triple-negative breast cancer cell growth via the miR-1296-5p/TRIM14 axis
Department of Breast Surgery, Affiliated Tumor Hospital of Xinjiang Medical University, Urumqi, 830011, China
*Address correspondence to: Qian Zhao, nva9lca@163.com
Received: 02 July 2020; Accepted: 26 October 2020
Abstract: Hypoxia affects the advancement, metastasis, and metabolism of breast cancer (BC). The circular RNA ribonuclease P RNA component H1 (circRPPH1) (has_circ_0000515) is implicated in tumor progression. Nevertheless, the regulatory mechanism related to circRPPH1 in hypoxia-mediated triple-negative breast cancer (TNBC) progression is indistinct. The expression levels of circRPPH1, miR-1296-5p, tripartite motif-containing 14 (TRIM14) mRNA in tissue samples and cells were examined through quantitative real-time polymerase chain reaction (qRT-PCR). Cell viability, migration, and invasion were determined with Cell Counting Kit-8 (CCK-8) or transwell assays. The levels of glucose consumption and lactate production were assessed via the Glucose Assay Kit or Lactate Assay Kit. The protein levels of TRIM14, Glucose Transporter GLUT1 (GLUT1), and lactic dehydrogenase A (LDHA) were detected by western blot analysis. The targeting relationship between circRPPH1 or TRIM14 and miR-1296-5p was verified with dual-luciferase reporter assay. The role of circRPPH1 was confirmed via xenograft assay. We verified that circRPPH1 and TRIM14 expression were increased while miR-1296-5p expression was decreased in BC tissues and hypoxia-cultured TNBC cells. Functionally, circRPPH1 silencing reversed the promoting effect of hypoxia on viability, migration, invasion, and glycolysis of TNBC cells. CircRPPH1 knockdown repressed decreased TNBC cell growth in vivo. Mechanistically, circRPPH1 sponged miR-1296-5p to modulate TRIM14 expression. Also, miR-1296-5p silencing restored circRPPH1 inhibition-mediated influence on the viability, migration, invasion, and glycolysis of hypoxia-treated TNBC cells. TRIM14 elevation overturned the inhibitory impact of miR-1296-5p mimic on viability, migration, invasion, and glycolysis of hypoxia-cultured TNBC cells. In conclusion, hypoxia-induced circRPPH1 fostered TNBC progression through regulation of the miR-1296-5p/TRIM14 axis, indicating that circRPPH1 was a promising target for TNBC treatment.
Keywords: TNBC; circRPPH1; miR-1296-5p; TRIM14; Hypoxia
Breast cancer (BC) is the most common cancer in women and the leading cause of cancer-related deaths worldwide, and its most malignant subtype is triple-negative breast cancer (TNBC) (Torre et al., 2015, Carey et al., 2010). Hypoxia is a vital characteristic of solid tumors, which results in a more aggressive phenotype by elevating the potential for metastasis and proliferation (Brahimi-Horn et al., 2007; Harris, 2002). Hypoxia is involved in regulating a variety of cellular response pathways, including metabolism, proliferation, angiogenesis, and DNA damage (Bristow and Hill, 2008). Moreover, hypoxia-induced glycolysis can maintain the viability of cancer cells and accelerate cancer cell growth (Huang and Zong, 2017). Hence, it is essential to explore new mechanisms of TNBC progression under hypoxic conditions to develop new targets for BC treatment.
Circular RNAs (circRNAs) are a type of covalently closed RNA transcripts, which are highly conserved and stable (Rybak-Wolf et al., 2015). It has been demonstrated that some circRNAs can regulate the transcription and translation of proteins and peptides through sponging microRNAs (miRNAs), interacting with RNA-pol II, or binding to RNA-binding proteins (Li et al., 2018). Strikingly, circRNAs have become vital regulators in a range of human diseases and cancers (Han et al., 2018). For instance, circRNA circ_403658 accelerated bladder cancer advancement via regulation of LDHA under hypoxia (Wei et al., 2019). Moreover, hypoxia-induced circRNA circDENND2A constrained miR-625-5p expression, which promoted glioma progression (Su et al., 2019). Circular RNA ribonuclease P RNA component H1 (circRPPH1), also termed as has_circ_0000515, was proved to be involved in cervical cancer progression (Tang et al., 2019). However, the role of circRPPH1 in hypoxia-mediated TNBC progression is unclear.
MiRNAs are a class of small molecular RNAs that primarily regulate gene expression at post-transcriptional levels (He and Hannon, 2004). They have been shown to modulate cell viability, differentiation, proliferation, invasion, and migration in various cancers (Sassen et al., 2008). For example, reduced miR-491-5p expression contributed to gastric cancer progression via modulation of SNAIL and FGFR4 expression (Yu et al., 2018). MiRNA-1296-5p (miR-1296-5p) has been proved to be related to tumor progression, such as osteosarcoma (Wang et al., 2020), gastric cancer (Shan et al., 2017), and BC (Chen et al., 2017). However, the regulatory mechanisms of miR-1296-5p in TNBC progression have not been fully elucidated.
The tripartite motif (TRIM) family proteins are composed of 3 zinc-binding regions: coiled-coil, B-box, and RING finger domains, which were associated with multiple cellular and biological functions, such as development, oncogenesis, differentiation, and innate immunity (Hatakeyama, 2017; Khan et al., 2019). Studies indicated that the tripartite motif-containing 14 (TRIM14) acted as an oncogene in various tumors, such as colorectal cancer (Jin et al., 2018), osteosarcoma (Xu et al., 2017), and glioblastoma (Feng et al., 2019). Also, TRIM14 had been reported to play a tumor repressive role in non-small cell lung cancer (Hai et al., 2017). Besides, the forcing expression of TRIM14 was connected with BC advancement (Hu et al., 2019). Nevertheless, the role of TRIM14 in hypoxia-mediated TNBC progression is unclear.
In this study, we verified circRPPH1 was upregulated in BC tissues and hypoxia-treated TNBC cells. Also, circRPPH1 knockdown reduced hypoxia-mediated TNBC progression via regulating the miR-1296-5p/TRIM14 axis. The research provided a novel mechanism to understand the progress of TNBC.
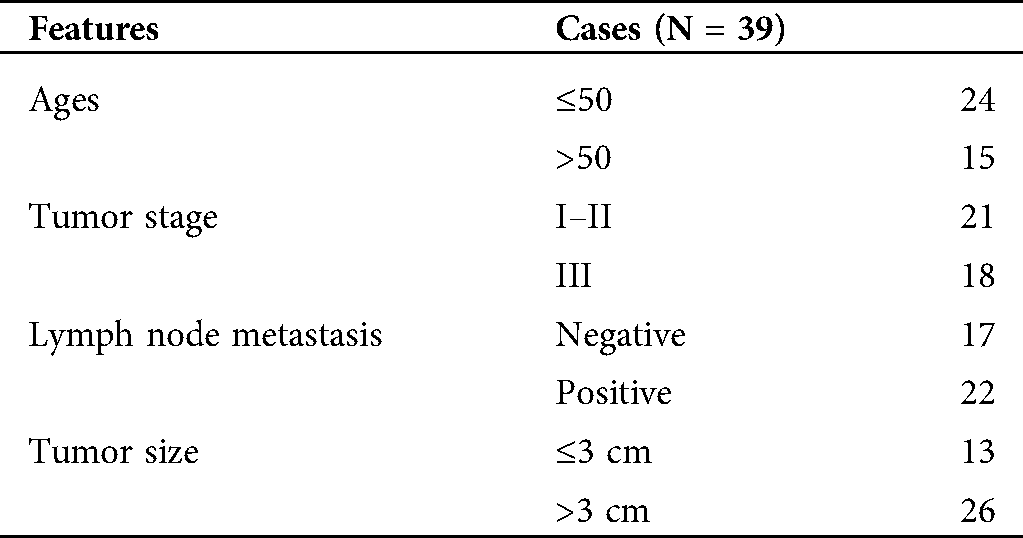
The research was authorized and supervised by the ethics committee of Affiliated Tumor Hospital of Xinjiang Medical University and was performed in accordance with the ethical guidelines of Helsinki. BC tissues and adjacent normal tissues were obtained from 39 BC patients who underwent surgery at Affiliated Tumor Hospital of Xinjiang Medical University. All tumor tissues and adjacent normal tissues were confirmed by experienced pathologists. None of the patients who participated in the study had received chemotherapy or radiotherapy before surgery. The clinical staging of BC patients was based on the 7th edition of the American Cancer Council staging system. All participants had provided the written informed consent. The Clinicopathological features of BC patients were presented in Tab. 1.
Table 1: The clinicopathological features of BC patients
In brief, BC tissues and adjacent normal tissues were fixed in formaldehyde (4%) (Sigma, Sigma, St Louis, MO, USA), embedded in paraffin, and cut into 4-μm sections. Subsequently, the sections were treated with an immunohistochemical labeling kit (MaxVision Biotechnology, Fuzhou, China) and labeled with a primary antibody against Ki67 (ab15580, 1:200, Abcam, Cambridge, MA, USA). The picture was obtained with an inverted microscope (Olympus, Tokyo, Japan).
Human TNBC cells MDA-MB-231 and MDA-MB-468, as well as human non-tumor mammary gland cells MCF-10A, were bought from American Type Culture Collection (Rockville, MD, USA). All cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Hyclone, Logan, UT, USA) supplemented with fetal bovine serum (10%, FBS, Hyclone), penicillin (100 U/mL, Sigma), and streptomycin (100 g/mL, Sigma) in an incubator with 5% CO2 at 37°C.
For hypoxia treatment, cells were cultured in an Invivo2 400 hypoxic workstation (Ruskin Technologies, Bridgend, UK) with 94% N2, 5% CO2, and 1% O2 for different times (0, 3, 6, 12, 24, or 48 h) (Wu and Yotnda, 2011).
Small interference RNA targeting circRPPH1 (si-circRPPH1: 5’-CCGGAGCTTGGAACAGACT-3’) and matching control (si-NC: 5’-UCUCCGAACGUGUCACGUTT-3’), miR-1296-5p mimics (miR-1296-5p) and corresponding control (miR-NC), and miR-1296-5p inhibitors (anti-miR-1296-5p) and negative control (anti-miR-NC) were synthesized by Ribobio (Guangzhou, China). The overexpression vectors for TRIM14 were obtained via cloning the sequence of TRIM14 into the pcDNA3.1 vector (vector) (Life Technologies, Carlsbad, CA, USA). When 70–90% confluence was reached, the specified vectors or oligonucleotides were transfected into MDA-MB-231 and MDA-MB-468 cells with Lipofectamine 3000 reagent (Life Technologies). The cells were cultured in hypoxic conditions at different times after transfection (Chen et al., 2009).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA (tissue specimens and cells) was extracted with TRIzol reagent (Sigma). For the RNase R digestion, total RNA of MDA-MB-231 and MDA-MB-468 cells were treated with RNase R (3 U/μg, Epicentre Technologies, Madison, WI, USA) at 37°C for 15 min. For complementary DNA production, total RNA was reversely transcribed by a High-Capacity complementary DNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) or MicroRNA Reverse Transcription Kit (Applied Biosystems). QRT-PCR was executed with the SYBR Green PCR Master Mix (Applied Biosystems) in the CFX96 Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA). The expression levels of circRPPH1, RPPH1, miR-326, miR-1908-5p, miR-1036-5p, miR-296-5p, miR-1296-5p, miR-328-3p, miR-3612, miR-663a, and TRIM14 were figured via the 2−ΔΔCt method, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 small nuclear RNA (snRNA) was used as the internal control. Primer sequences were displayed as below: circRPPH1 (Forward: 5’-GGTCAGACTGGGCAGGAGAT-3’, Reverse: 5’-GAGTGACAGGACGCACTCAG-3’), RPPH1 (Forward: 5’-GCCGGARCTTGGAACAGA-3’, Reverse: 5’-ACCTCACCTCAGCCATTGAACT-3’), TRIM14 (Forward: 5’-GGATTTGTGTCTCCGTTCTG-3’, Reverse: 5’-TCTGTCTGCCTGGTATTCTG-3’), GAPDH (Forward: 5’-GAAGGTGAAGGTCGGAGTC-3’, Reverse: 5’-GAAGATGGTGATGGGATTTC-3’), miR-1296-5p (Forward: 5’-GTTAGGGCCCTGGCTCC-3’, Reverse: 5’-CAGTGCGTGTCGTGGAGT-3’), miR-326 (Forward: 5’-GGCGCCCAGAUAAUGCG-3’, Reverse: 5’-CGTGCAGGGTCCGAGGTC-3’), miR-1908-5p (Forward: 5’-ACACTCCAGCTGGGGCGGCGGGGACG-3’, Reverse: 5’-CTCAACTGGTGTCGTGGA-3’), miR-1306-5p (Forward: 5’-GGCAGAGGAGGGCTGTTC-3’, Reverse: 5’-GTGCGTGTCGTGGAGTCG-3’), miR-296-5p (Forward: 5’-GTATCCAGTGCAGGGTCCGA-3’, Reverse: 5’-CGACGAGGGCCCCCCCT-3’), miR-328-3p (Forward: 5’-TGCGGCTGGCCCTCTCTGCCC-3’, Reverse: 5’-CCAGTGCAGGGTCCGAGGT-3’), miR-3612 (Forward: 5’-AGGCATCTTGAGAAAT-3’, Reverse: 5’-GCGAGCACAGAATTAATACGAC-3’), miR-663a (Forward: 5’-AGGCGGGGCGCCGCGGGACCGC-3’, Reverse: 5’-GCGAGCACAGAATTAATACGAC-3’), as well as U6 snRNA (Forward: 5’-GCTTCGGCAGCACATATACTAAAAT-3’, Reverse: 5’-CGCTTCACGAATTTGCGTGTCAT-3’).
Cell Counting Kit-8 (CCK-8) assay
MDA-MB-231 and MDA-MB-468 cells were transfected and cultured in normoxia or hypoxia at different times. Then, CCK-8 solution (10 μL, Beyotime, Shanghai, China) was added and incubated for 2 h. The absorbance at 450 nm was measured with a microplate reader (Bio-Rad).
For migration assay, the transfected MDA-MB-231 and MDA-MB-468 cells (100 μL, 1 × 105 cells/mL) in RPMI-1640 medium without FBS were added to the upper of the transwell chamber (Costar, Cambridge, MA, USA). And the RPMI-1640 medium with FBS (10%) was added to the lower chamber. The cells were cultured in a hypoxia chamber in normoxia or hypoxia. Subsequently, the cells on the lower surface were fixed with glutaraldehyde (5%) and then stained with 0.1% crystal violet (0.1%; Sigma). The migration and invasion assays had the same procedures, but the invasion assay used a transwell chamber with Matrigel (BD Biosciences), and the number of cells inoculated was 10 times that of the migration assay. The number of migrated and invasive cells was assessed by an inverted microscope (Olympus) at 100 × amplification.
Transfected MDA-MB-231 and MDA-MB-468 cells (1 × 105 cells/well) were cultured overnight. Next, the cells were cultured in normoxia or hypoxia for 48 h. The levels of glucose consumption and lactate production were assessed via the Glucose Assay Kit (Sigma) or Lactate Assay Kit (Sigma). The relative levels of glucose consumption and lactate production in the treatment group were normalized to the normoxic group.
Total protein was extracted with lysis buffer (Beyotime) with protease inhibitor (Beyotime). Western blot analysis was executed, referring to a previous study (Ren et al., 2019). Total protein was separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride (PVDF) membranes (Beyotime). The Bull Serum Albumin (BSA, 5%) was utilized to block the PVDF membranes. The immunoblot was visualized through the enhanced chemiluminescence solution (Beyotime). The primary antibodies were displayed as blows: anti-Glucose Transporter GLUT1 (GLUT1) (ab15309, 1:500, Abcam), anti-lactic dehydrogenase A (LDHA) (ab135366, 1:1000, Abcam), anti-TRIM14 (ab185349, 1:800, Abcam), and anti-GAPDH (ab9484, 1:1000, Abcam). Goat anti-rabbit IgG (ab205718, 1:2000, Abcam) was used as a secondary antibody, and GAPDH was used as a loading control.
The biotinylated-circRPPH1 probe or Oligo probe were obtained from Sigma. The RNA pull-down assay was conducted according to a previous study (Wang et al., 2016). Briefly, the biotinylated-circRPPH1 probe was incubated with C-1 magnetic beads (Life Technologies) for the generation of the probe-coated beads. Following this, the probe-coated beads were incubated with sonicated MDA-MB-231 and MDA-MB-468 cells. The bound RNA was purified through TRIzol reagent (Sigma). The levels of circRPPH1, miR-326, miR-1908-5p, miR-1036-5p, miR-296-5p, miR-1296-5p, miR-328-3p, miR-3612, and miR-663a were evaluated with qRT-PCR.
Dual-luciferase reporter assay
The targets of circRPPH1 were predicted by starBase and circBank databases. The binding sites between miR-1296-5p and TRIM14 were predicted by the starBase database. The sequences of wild type (WT) circRPPH1 and 3’UTR of TRIM14 and their mutant (MUT) sequence were synthesized and inserted into the pmirGLO luciferase vectors (GeneCreat, Wuhan, China). The luciferase reporter vectors were co-transfected into MDA-MB-231 and MDA-MB-468 with miR-NC or miR-1296-5p, respectively. The luciferase activities were determined with the luciferase reporter assay kit (Promega).
The protocols of xenograft assay were authorized by the ethics committee of Affiliated Tumor Hospital of Xinjiang Medical University and were conducted according to the guidelines for the care and use of laboratory animals (GB/T 35892-2018). The lentiviral vectors with sh-circRPPH1 or matching control (sh-NC) were established by Genepharma (Shanghai, China). The MDA-MB-468 cells (1 × 107) transfected with lentiviral-mediated sh-circRPPH1 or sh-NC were suspended in 100 μL phosphate-buffered saline. Next, the transfected MDA-MB-468 cells were subcutaneously injected into the right flank of 10 BALB/c nude mice (5 weeks old, Experimental Animal Center, Shanghai, China), 5 mice in each group. Tumors volume was calculated according to the following equation: Volume = (length × width2)/2. Tumor volume was measured every 5 days with a caliper. After 30 days, all mice were killed for subsequent analysis.
The normal distribution was assessed through a Kolmogorov-Smirnov test. SPSS 20.0 software (IBM Corporation, Armonk, NY, USA) was utilized for statistical analysis. Data were expressed as mean ± standard deviation, which was acquired from at least 3 replicate experiments. The variance homogeneity was determined by the F-test. Differences were deemed significant if p < 0.05. Student’s t-test or one-way variance analysis was utilized for the comparison of the differences between two or among more groups.
CircRPPH1 was upregulated in BC tissues and hypoxic TNBC cells
Previous research (microarray data: GSE101123) exhibited that circRPPH1 was highly expressed in BC tissues (Fig. 1A). CircRPPH1 (hsa_circ_0000515) is derived from the RPPH1 gene (exon1), whose spliced mature sequence length is 229 bp (Fig. 1B). To verify the expression pattern of circRPPH1 in BC, we first examined the expression of circRPPH1 in 39 paired BC tissues and adjacent normal tissues. QRT-PCR presented that circRPPH1 expression was observably elevated in BC tissues compared with the adjacent normal tissues (Fig. 1C). According to immunohistochemistry assay, more Ki67-positive cells were observed in BC tissues when relative to adjacent normal tissues (Suppl. Fig. 3). Consistently, circRPPH1 expression was increased in TNBC cells (MDA-MB-231 and MDA-MB-468) relative to the MCF-10A cells (Fig. 1D). In addition, the loop structure of circRPPH1 was further verified by RNase R digestion. As exhibited in Figs. 1E and 1F, circRPPH1 was resistant to RNase R digestion compared with the liner RPPH1 gene. Also, qRT-PCR demonstrated that circRPPH1 expression was gradually elevated in MDA-MB-231 and MDA-MB-468 cells with the extension of hypoxia time, and the cells treated with hypoxia for 48 h were selected for subsequent analysis (Figs. 1G and 1H). These results manifested that circRPPH1 might be involved in hypoxia-mediated TNBC progression.
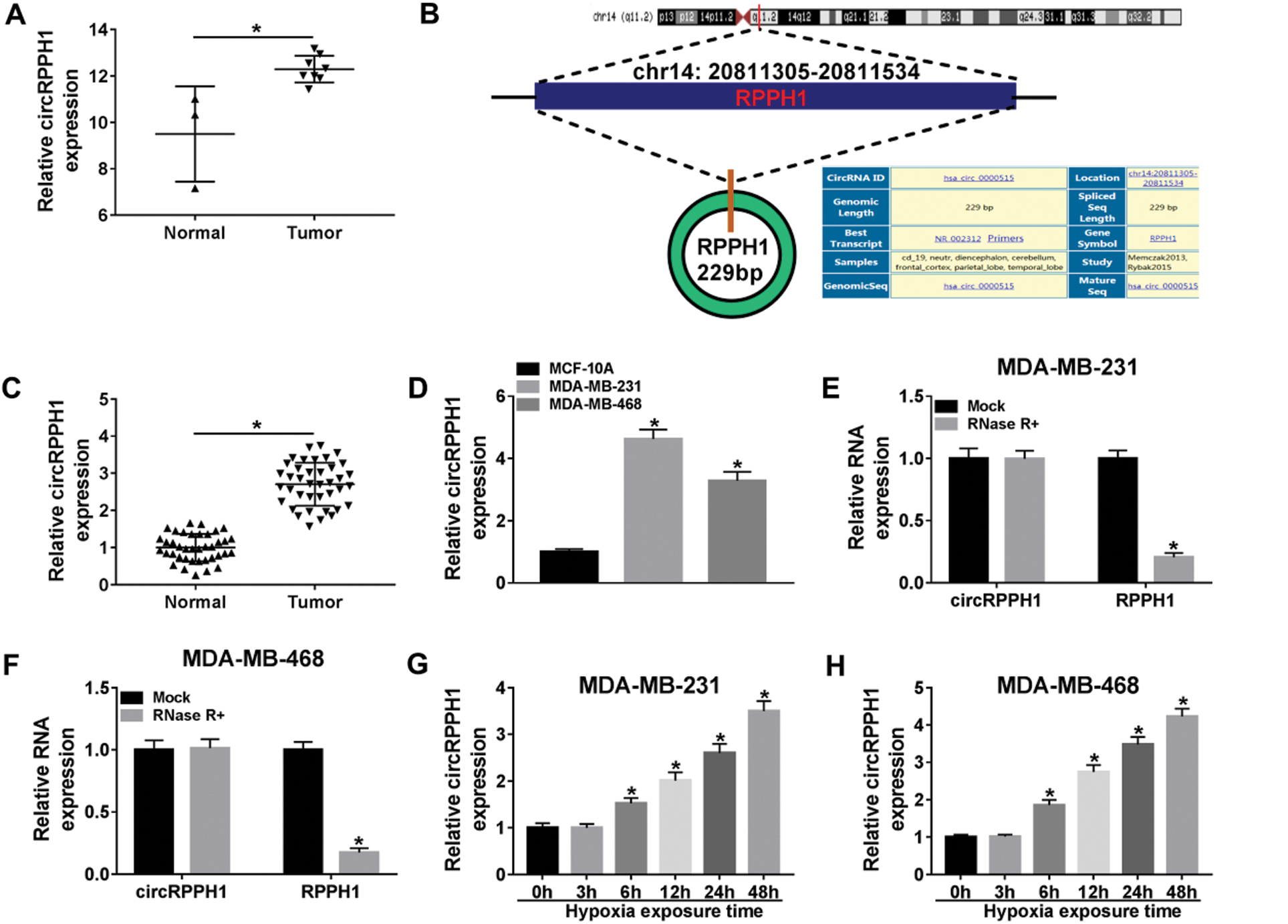
Figure 1: Expression levels of circRPPH1 in BC tissues and hypoxia-cultured TNBC cells.
CircRPPH1 knockdown reversed the promoting effect of hypoxia on cell viability, migration, invasion, and glycolysis in TNBC cells
In consideration of the upregulation of circRPPH1 in BC tissues and hypoxia-cultured TNBC cells, we explored the role of circRPPH1 in hypoxia-mediated TNBC progression via loss-of-function experiments. QRT-PCR manifested that the elevation of circRPPH1 in hypoxia-cultured MDA-MB-231 and MDA-MB-468 cells was restored after si-circRPPH1 introduction (Figs. 2A and 2B). CCK-8 assay revealed that the viability of MDA-MB-231 and MDA-MB-468 cells was enhanced after hypoxia treatment, while this influence was restored by circRPPH1 inhibition (Figs. 2C and 2D). Transwell assay demonstrated that the number of migration and invasion of MDA-MB-231 and MDA-MB-468 cells was elevated after hypoxic treatment, but this trend was reversed by circRPPH1 knockdown (Figs. 2E and 2F). Moreover, silenced circRPPH1 expression overturned the elevation of glucose consumption and lactate production in hypoxia-cultured MDA-MB-231 and MDA-MB-468 cells (Figs. 2G–2J). Furthermore, the inhibition of circRPPH1 restored the increase of GLUT1 and LDHA protein levels in hypoxia-treated MDA-MB-231 and MDA-MB-468 cells (Figs. 2K and 2L). Taken together, these findings suggested that circRPPH1 knockdown could overturn hypoxia-mediated the malignant behaviors of BC cells.
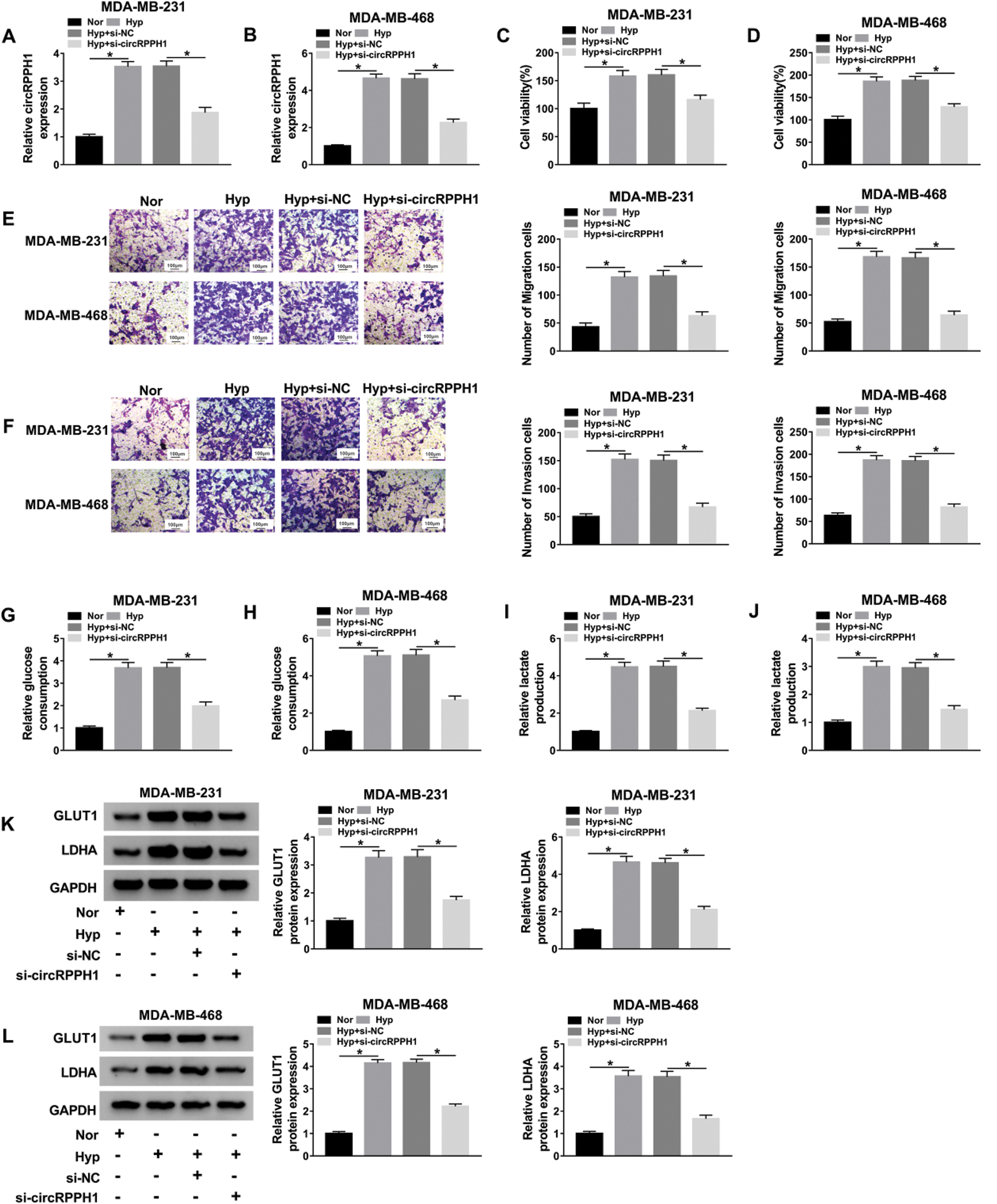
Figure 2: Effects of circRPPH1 silencing on the malignant behaviors of hypoxia-cultured TNBC cells.
CircRPPH1 functioned as a sponge for miR-1296-5p
To explore the underlying molecular mechanism of circRPPH1 in hypoxia-mediated TNBC progression, we used a 3’ terminal-biotinylated-circRPPH1 probe for the verification of the miRNAs that could interact with circRPPH1. As presented in Figs. 3A and 3B, the circRPPH1 probe could pull down circRPPH1 in MDA-MB-231 and MDA-MB-468 cells, and circRPPH1 overexpression enhanced the pull-down efficiency (Figs. 3A and 3B). Both the circBank and starBase databases exhibited that 8 miRNAs (miR-326, miR-1908-5p, miR-1036-5p, miR-296-5p, miR-1296-5p, miR-328-3p, miR-3612, and miR-663a) possessed the complementary sites for circRPPH1 (Fig. 3C). QRT-PCR displayed that miR-296-5p and miR-1296-5p were abundantly pulled down by circRPPH1 probe in both MDA-MB-231 and MDA-MB-468 cells, and miR-1296-5p was higher compared to miR-296-5p (Figs. 3D and 3E). The potential binding sites between circRPPH1 and miR-1296-5p were presented in Fig. 3F. Also, dual-luciferase reporter assay indicated that miR-1296-5p overexpression reduced the luciferase intensity of the luciferase vectors with circRPPH1 WT in MDA-MB-231 and MDA-MB-468 cells in comparison to the control group, but the luciferase activity of the luciferase vectors with circRPPH1 MUT did not change (Figs. 3G and 3H). Furthermore, miR-1296-5p expression was elevated in circRPPH1-silenced MDA-MB-231 and MDA-MB-468 cells (Figs. 3I and 3J). We observed that miR-1296-5p was downregulated in BC tissues and cells (Figs. 3K and 3L). Additionally, miR-1296-5p expression was gradually decreased in MDA-MB-231 and MDA-MB-468 cells with the increase of hypoxia time (Figs. 3M and 3N). Together, these data proved that circRPPH1 acted as a sponge for miR-1296-5p in TNBC cells.
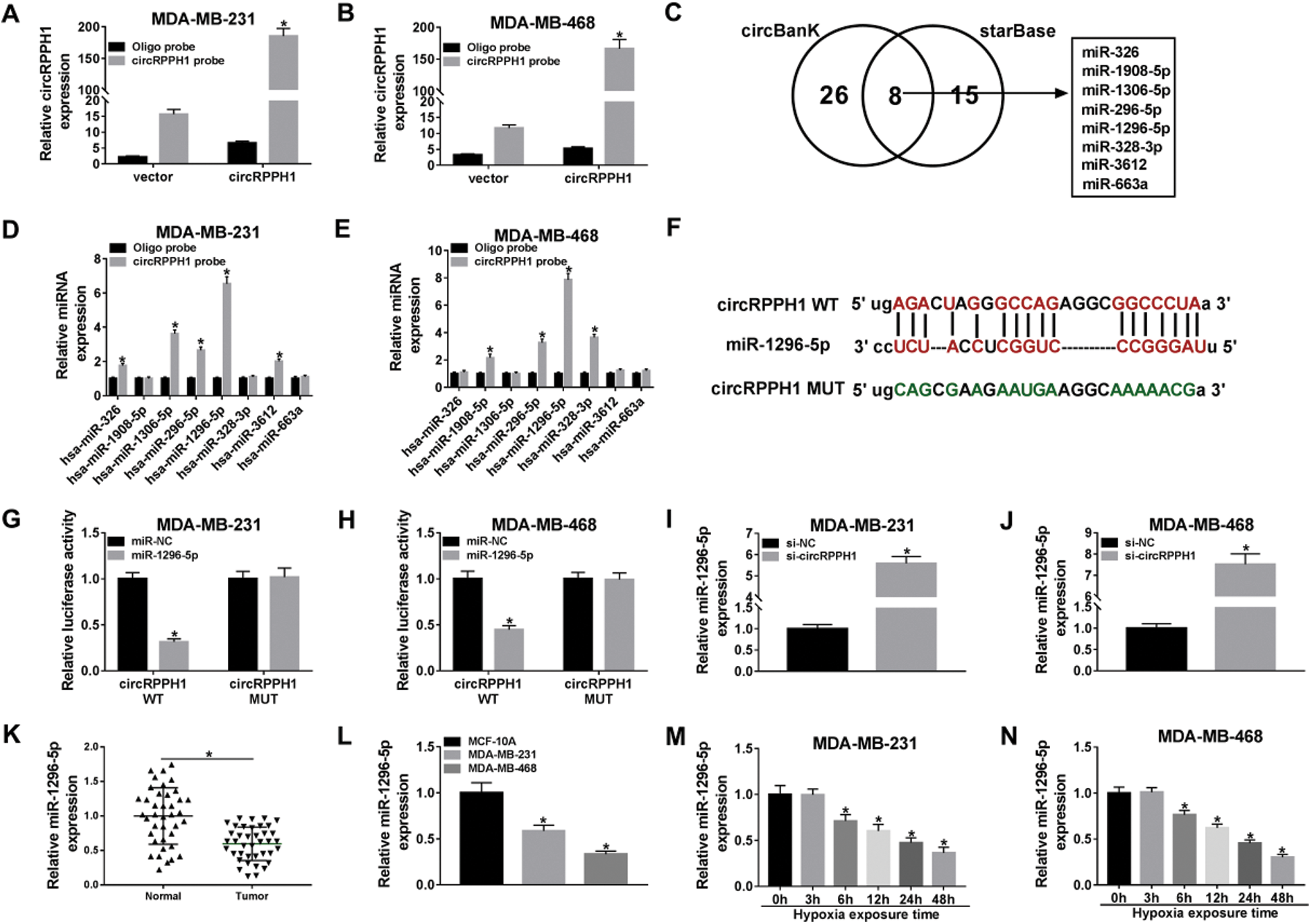
Figure 3: MiR-1296-5p acted as a target for circRPPH1.
MiR-1296-5p silencing overturned circRPPH1 knockdown-mediated impacts on the viability, migration, invasion, and glycolysis of hypoxia-treated TNBC cells
Next, we further verified whether circRPPH1 exerted its function via miR-1296-5p in hypoxia-mediated TNBC progression. We found that circRPPH1 inhibition elevated miR-1296-5p expression in hypoxia-cultured MDA-MB-231 and MDA-MB-468 cells, but this tendency was restored after anti-miR-1296-5p transfection (Figs. 4A and 4B). CCK-8 assay indicated that miR-1296-5p silencing abolished the inhibition of viability of hypoxia-cultured MDA-MB-231 and MDA-MB-468 cells mediated by the inhibition of circRPPH1 expression (Figs. 4C and 4D). Transwell assay revealed that miR-1296-5p inhibitor overturned the suppressive impact of circRPPH1 silencing on migration and invasion of hypoxia-cultured MDA-MB-231 and MDA-MB-468 cells (Figs. 4E–4H; Suppl. Figs. 1A–1D). The pictures for migration and invasion were exhibited in Suppl. Fig. 1. The silence of circRPPH1 reduced glucose consumption and lactate production in hypoxia-treated MDA-MB-231 and MDA-MB-468 cells, while this effect was recovered by miR-1296-5p silencing (Figs. 4I–4L). Furthermore, the suppressive influence of circRPPH1 knockdown on the levels of GLUT1 and LDHA protein of hypoxia-cultured MDA-MB-231 and MDA-MB-468 cells were restored by the miR-1296-5p inhibitor (Figs. 4M and 4N). These results demonstrated that circRPPH1 modulated the malignant behaviors of hypoxia-cultured TNBC cells through miR-1296-5p.
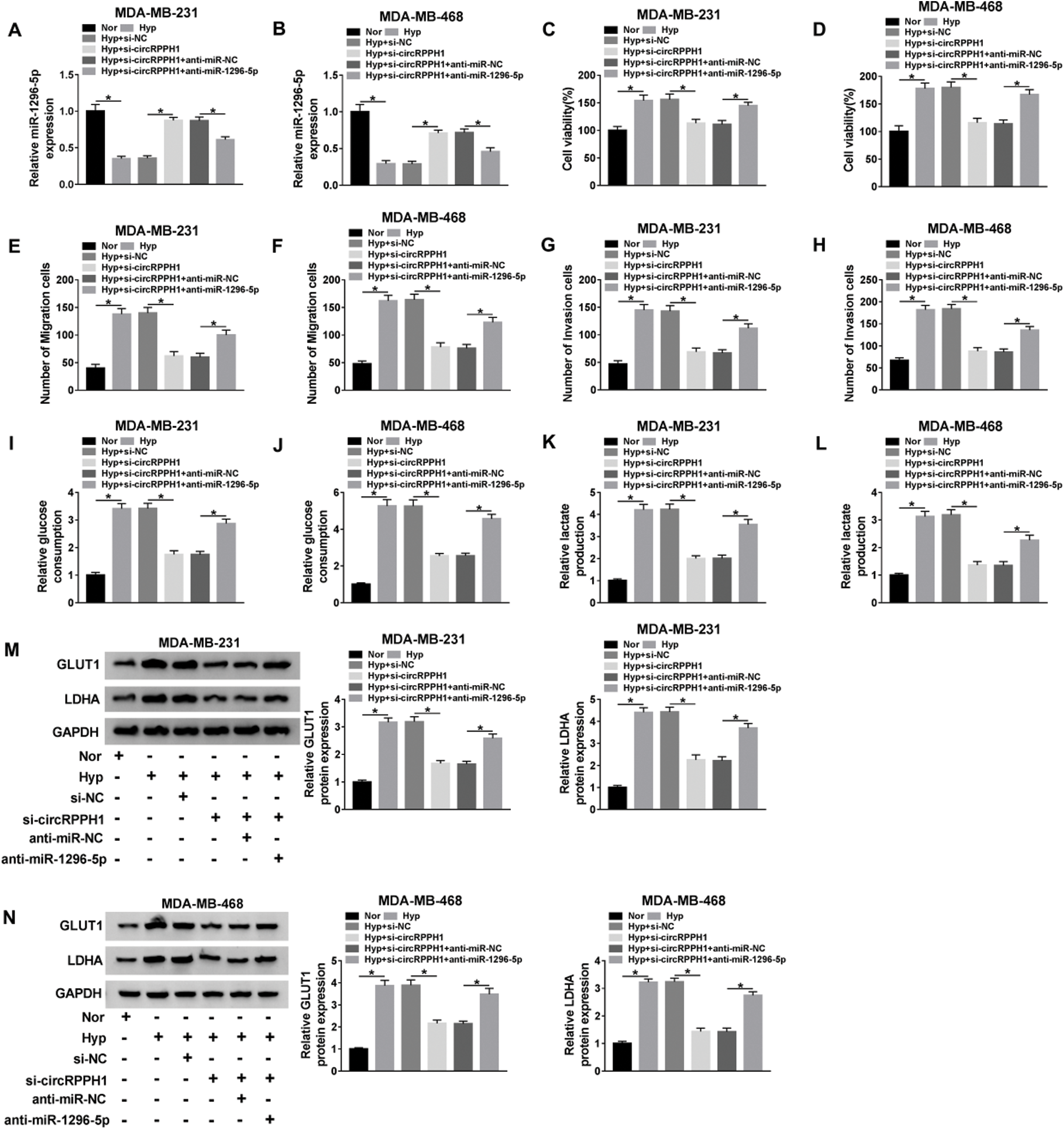
Figure 4: Hypoxia-induced circRPPH1 played its role through miR-1296-5p.
TRIM14 was identified as a downstream target of miR-1296-5p
Considering the above results, we found that TRIM14 might be a target of miR-1296-5p (Fig. 5A). Furthermore, the luciferase activity of the luciferase vectors with TRIM14 3’UTR WT was decreased by the miR-1296-5p mimic in MDA-MB-231 and MDA-MB-468 cells, while there was no overt change in the luciferase vectors with TRIM14 3’UTR MUT (Figs. 5B and 5C). Also, miR-1296-5p mimic decreased the level of TRIM14 protein in MDA-MB-231 and MDA-MB-468 cells (Figs. 5D and 5E). Moreover, TRIM14 protein level was reduced in circRPPH1-silenced MDA-MB-231 and MDA-MB-468 cells, while this decrease was reversed after miR-1296-5p silencing (Figs. 5F and 5G). We also observed that the level of TRIM14 protein was increased in BC tissues and cells (Figs. 5H and 5I). In addition, TRIM14 protein level was gradually elevated in MDA-MB-231 and MDA-MB-468 cells with the enhancement of hypoxia time (Figs. 5J and 5K). Collectively, these data indicated that TRIM14 acted as a target of miR-1296-5p in TNBC cells.

Figure 5: TRIM14 was targeted by miR-1296-5p in BC cells.
TRIM14 elevation restored miR-1296-5p mimic-mediated effects on the viability, migration, invasion, and glycolysis of hypoxia-treated TNBC cells
Given that miR-1296-5p targeted TRIM14 in TNBC cells, we explored whether miR-1296-5p regulated hypoxia-mediated TNBC progression via TRIM14. Western bot analysis presented that miR-1296-5p overexpression reduced the level of TRIM14 protein in hypoxia-cultured MDA-MB-231 and MDA-MB-468, while this tendency was abolished after TRIM14 overexpression (Figs. 6A and 6B). Moreover, the inhibitory influence of miR-1296-5p mimic on viability, migration, and invasion of hypoxia-cultured MDA-MB-231 and MDA-MB-468 cells was abolished by elevating TRIM14 expression (Figs. 6C–6F; Suppl. Figs. 2A and 2B). The pictures for migration and invasion were exhibited in Suppl. Fig. 2. Furthermore, TRIM14 elevation overturned the decrease of glucose consumption and lactate production in miR-1296-5p-elevated MDA-MB-231 and MDA-MB-468 cells under hypoxia treatment (Figs. 6G–6J). In addition, the downregulation of GLUT1 and LDHA in miR-1296-5p-increased MDA-MB-231 and MDA-MB-468 cells were reserved by TRIM14 overexpression under hypoxia treatment (Figs. 6K and 6L). Therefore, these data manifested that miR-1296-5p modulated the malignant behaviors of hypoxia-cultured TNBC cells through TRIM14.

Figure 6: MiR-1296-5p regulated the malignant behaviors of hypoxia-treated TNBC cells through TRIM14.
CircRPPH1 knockdown impeded tumor growth in vivo
Subsequently, we further verified the role of circRPPH1 in TNBC progression by xenograft assay. The results exhibited that circRPPH1 silencing reduced tumor volume and weight relative to the control group (Figs. 7A and 7B). Furthermore, circRPPH1 expression was reduced, and miR-1296-5p expression was elevated in xenograft tumors in the sh-circRPPH1 group compared to the sh-NC group (Figs. 7C and 7D). Also, circRPPH1 silencing decreased the level of TRIM14 protein in xenograft tumors (Fig. 7E). These data demonstrated that circRPPH1 silencing reduced TNBC growth in vivo.
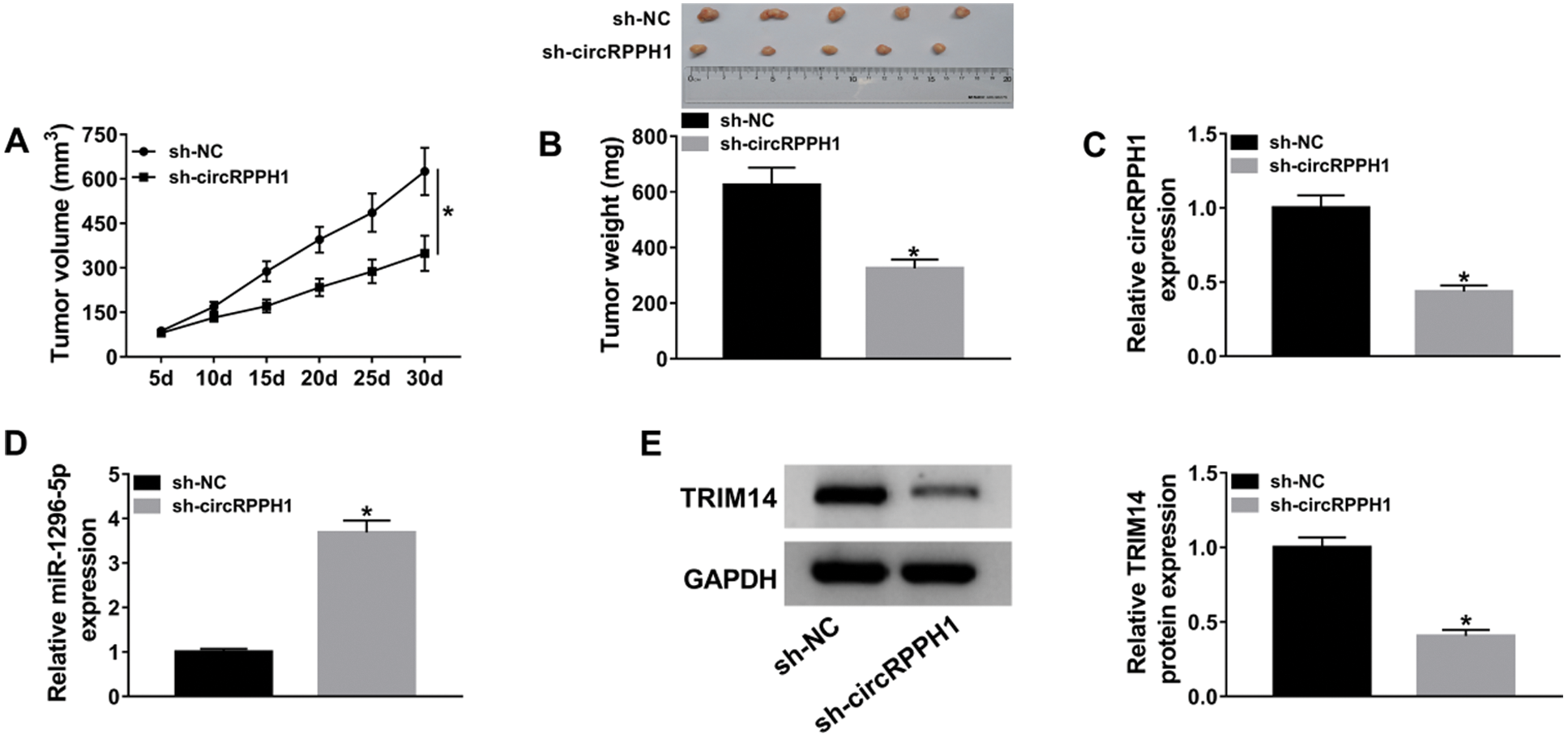
Figure 7: CircRPPH1 knockdown impeded TNBC growth in vivo.
Currently, metastatic BC patients have a relatively poor response to available therapies, and approximately 90% of BC patients die due to metastases (Gilkes and Semenza, 2013). TNBC has a high histological grade and a poor prognosis compared with other BC subtypes (Foulkes et al., 2010). The oxygen content in tumor tissue is an important determinant of metastasis, and tumor hypoxia has been identified as a poor prognostic indicator for patients not related to histopathological parameters (Gong et al., 2018; Vaupel, 2009). Some circRNAs have been proved to be related to the hypoxic state of tumors (Boeckel et al., 2015). For example, circRNA circ_403658 was induced by hypoxia, which contributed to cancer cell malignant behaviors through activating LDHA in bladder cancer (Wei et al., 2019). Ou et al. (2019) indicated that circRNA cric_0000977 regulated HIF1A-mediated pancreatic cancer cell immune escape through modulating HIF1A and ADAM10 via targeting miR-153 (Ou et al., 2019). In BC, HIF1α-associated circDENND4C accelerated cell proliferation under hypoxia treatment (Liang et al., 2017), and circDENND4C silencing blocked cell invasion, migration, and glycolysis through sponging miR-200b/c in a hypoxic environment (Ren et al., 2019). In this study, circRPPH1 expression was increased in hypoxia-cultured TNBC cells, and circRPPH1 knockdown impeded viability, migration, invasion, and glycolysis of hypoxia-cultured TNBC cells in vitro and suppressed tumor growth in vivo. Tang et al. demonstrated that circRPPH1 acted as an oncogene in cervical cancer, and circRPPH1 facilitated cervical cancer advancement via elevating ELK1 expression (Tang et al., 2019). Therefore, these results manifested that hypoxia-induced circRPPH1 played a promoting role in TNBC progression.
Accumulated evidence manifested that circRNAs can function as miRNAs sponges in cancers (Ren et al., 2019; Su et al., 2019; Tang et al., 2019). Herein, we confirmed that miR-1296-5p acted as a target for circRPPH1. Moreover, miR-1296-5p downregulation abolished circRPPH1 inhibition-mediated effects on the malignant behaviors of TNBC cells under hypoxic conditions. These data indicated that circRPPH1 regulated the malignancy of hypoxia-induced TNBC cells by sponging miR-1296-5p. MiR-1296-5p had been uncovered to exert a tumor-suppressive role in gastric cancer via targeting EGFR, CDK6, or ERBB2 (Jia et al., 2019; Shan et al., 2017). Another report manifested that miR-1296-5p constrained the invasion, migration, and proliferation of osteosarcoma via repression of NOTCH2 expression (Wang et al., 2020). Also, miR-1296-5p could curb cell proliferation via targeting ERBB2 in ERBB2-positive BC (Chen et al., 2017). Hence, these data revealed that miR-1296-5p played a tumor-suppressive role in TNBC, and hypoxia-induced circRPPH1 modulated the malignant behaviors of TNBC cells sponging miR-1296-5p.
Additionally, we found that TRIM14 was a target of miR-1296-5p. A previous study unmasked that TRIM14 facilitated cell invasion and proliferation through activating the AKT pathway in osteosarcoma (Xu et al., 2017). One report pointed out that TRIM14 could be repressed by miR-15b in oral tongue squamous cell cancer, thereby decreasing cell resistance to cisplatin and cancer-initiating cell phenotypes (Wang et al., 2017). Moreover, TRIM14 knockdown accelerated apoptosis and repressed proliferation of BC cells (Hu et al., 2019). Herein, circRPPH1 adsorbed miR-1296-5p to regulate TRIM14 expression, and TRIM14 overexpression restored miR-1296-5p upregulation-mediated effects on the malignant behaviors of TNBC cells under a hypoxic environment. Therefore, we inferred that hypoxia-induced circRPPH1 regulated the malignant behaviors of TNBC cells via modulating TRIM14 expression by sponging miR-1296-5p.
In conclusion, hypoxia-induced circRPPH1 played a carcinogenic role in TNBC progression. Moreover, circRPPH1 knockdown repressed TNBC advancement via the miR-1296-5p/TRIM14 axis. This research provided a novel mechanism to understand B TNBC advancement under hypoxia and manifested that circRPPH1 was a hopeful target for BC treatment.
Availability of Data and Materials: The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions: DL designed and supervised the study. DJ conducted the experiments and drafted the manuscript. JF collected and analyzed the data. QZ contributed to the methodology and analyzed the data. MZ edited the manuscript. All authors read and approved the final manuscript.
Funding Statement: The project was funded by the Xinjiang Uygur Autonomous Region Natural Science Foundation (No. 2017D01C379).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Boeckel JN, Jaé N, Heumüller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S. (2015). Identification and characterization of hypoxia-regulated endothelial circular RNA. Circulation Research 117: 884–890. DOI 10.1161/CIRCRESAHA.115.306319. [Google Scholar] [CrossRef]
Brahimi-Horn MC, Chiche J, Pouysségur J. (2007). Hypoxia and cancer. Journal of Molecular Medicine 85: 1301–1307. DOI 10.1007/s00109-007-0281-3. [Google Scholar] [CrossRef]
Bristow RG, Hill RP. (2008). Hypoxia, DNA repair and genetic instability. Nature Reviews Cancer 8: 180–192. DOI 10.1038/nrc2344. [Google Scholar] [CrossRef]
Chen G, He M, Yin Y, Yan T, Cheng W, Huang Z, Zhang L, Zhang H, Liu P, Zhu W, Zhu Y. (2017). miR-1296-5p decreases ERBB2 expression to inhibit the cell proliferation in ERBB2-positive breast cancer. Cancer Cell International 17: 95. DOI 10.1186/s12935-017-0466-y. [Google Scholar] [CrossRef]
Chen L, Feng P, Li S, Long D, Cheng J, Lu Y, Zhou D. (2009). Effect of hypoxia-inducible factor-1α silencing on the sensitivity of human brain glioma cells to doxorubicin and etoposide. Neurochemical Research 34: 984–990. DOI 10.1007/s11064-008-9864-9. [Google Scholar] [CrossRef]
Carey L, Winer E, Viale G, Cameron D, Gianni L. (2010). Triple-negative breast cancer: Disease entity or title of convenience? Nature Reviews Clinical Oncology 7: 683–692. DOI 10.1038/nrclinonc.2010.154. [Google Scholar] [CrossRef]
Feng S, Cai X, Li Y, Jian X, Zhang L, Li B. (2019). Tripartite motif-containing 14 (TRIM14) promotes epithelial-mesenchymal transition via ZEB2 in glioblastoma cells. Journal of Experimental & Clinical Cancer Research 38: 57. DOI 10.1186/s13046-019-1070-x. [Google Scholar] [CrossRef]
Foulkes WD, Smith IE, Reis-Filho JS. (2010). Triple-negative breast cancer. New England Journal of Medicine 363: 1938–1948. DOI 10.1056/NEJMra1001389. [Google Scholar] [CrossRef]
Gilkes DM, Semenza GL. (2013). Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncology 9: 1623–1636. DOI 10.2217/fon.13.92. [Google Scholar] [CrossRef]
Gong IY, Fox NS, Huang V, Boutros PC. (2018). Prediction of early breast cancer patient survival using ensembles of hypoxia signatures. PLoS One 13: e0204123. DOI 10.1371/journal.pone.0204123. [Google Scholar] [CrossRef]
Hai J, Zhu CQ, Wang T, Organ SL, Shepherd FA, Tsao MS. (2017). TRIM14 is a putative tumor suppressor and regulator of innate immune response in non-small cell lung cancer. Scientific Reports 7: 39692. DOI 10.1038/srep39692. [Google Scholar] [CrossRef]
Han B, Chao J, Yao H. (2018). Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacology & Therapeutics 187: 31–44. DOI 10.1016/j.pharmthera.2018.01.010. [Google Scholar] [CrossRef]
Harris AL. (2002). Hypoxia—a key regulatory factor in tumour growth. Nature Reviews Cancer 2: 38–47. DOI 10.1038/nrc704. [Google Scholar] [CrossRef]
Hatakeyama S. (2017). TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends in Biochemical Sciences 42: 297–311. DOI 10.1016/j.tibs.2017.01.002. [Google Scholar] [CrossRef]
He L, Hannon GJ. (2004). MicroRNAs: Small RNAs with a big role in gene regulation. Nature Reviews Genetics 5: 522–531. DOI 10.1038/nrg1379. [Google Scholar] [CrossRef]
Hu G, Pen W, Wang M. (2019). TRIM14 promotes breast cancer cell proliferation by inhibiting apoptosis. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics 27: 439–447. DOI 10.3727/096504018X15214994641786. [Google Scholar] [CrossRef]
Huang R, Zong X. (2017). Aberrant cancer metabolism in epithelial–mesenchymal transition and cancer metastasis: Mechanisms in cancer progression. Critical Reviews in Oncology/Hematology 115: 13–22. DOI 10.1016/j.critrevonc.2017.04.005. [Google Scholar] [CrossRef]
Jia Y, Zhao LM, Bai HY, Zhang C, Dai SL, Lv HL, Shan BE. (2019). The tumor-suppressive function of miR-1296-5p by targeting EGFR and CDK6 in gastric cancer. Bioscience Reports 39: BSR20181556. DOI 10.1042/BSR20181556. [Google Scholar] [CrossRef]
Jin Z, Li H, Hong X, Ying G, Lu X, Zhuang L, Wu S. (2018). TRIM14 promotes colorectal cancer cell migration and invasion through the SPHK1/STAT3 pathway. Cancer Cell International 18: 202. DOI 10.1186/s12935-018-0701-1. [Google Scholar] [CrossRef]
Khan R, Khan A, Ali A, Idrees M. (2019). The interplay between viruses and TRIM family proteins. Reviews in Medical Virology 29: e2028. DOI 10.1002/rmv.2028. [Google Scholar] [CrossRef]
Li X, Yang L, Chen LL. (2018). The biogenesis, functions, and challenges of circular RNAs. Molecular Cell 71: 428–442. DOI 10.1016/j.molcel.2018.06.034. [Google Scholar] [CrossRef]
Liang G, Liu Z, Tan L, Su AN, Jiang WG, Gong C. (2017). HIF1α-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Research 37: 4337–4343. [Google Scholar]
Ou ZL, Luo Z, Wei W, Liang S, Gao TL, Lu YB. (2019). Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: Role of circ_0000977/miR-153 axis. RNA Biology 16: 1592–1603. DOI 10.1080/15476286.2019.1649585. [Google Scholar] [CrossRef]
Ren S, Liu J, Feng Y, Li Z, He L, Li L, Cao X, Wang Z, Zhang Y. (2019). Knockdown of circDENND4C inhibits glycolysis, migration and invasion by up-regulating miR-200b/c in breast cancer under hypoxia. Journal of Experimental & Clinical Cancer Research 38: 388. DOI 10.1186/s13046-019-1398-2. [Google Scholar] [CrossRef]
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Molecular Cell 58: 870–885. DOI 10.1016/j.molcel.2015.03.027. [Google Scholar] [CrossRef]
Sassen S, Miska EA, Caldas C. (2008). MicroRNA—Implications for cancer. Virchows Archiv 452: 1–10. DOI 10.1007/s00428-007-0532-2. [Google Scholar] [CrossRef]
Shan X, Wen W, Zhu D, Yan T, Cheng W, Huang Z, Zhang L, Zhang H, Wang T, Zhu W, Zhu Y, Zhu J. (2017). miR 1296-5p inhibits the migration and invasion of gastric cancer cells by repressing ERBB2 expression. PLoS One 12: e0170298. DOI 10.1371/journal.pone.0170298. [Google Scholar] [CrossRef]
Su H, Zou D, Sun Y, Dai Y. (2019). Hypoxia-associated circDENND2A promotes glioma aggressiveness by sponging miR-625-5p. Cellular & Molecular Biology Letters 24: 24. DOI 10.1186/s11658-019-0149-x. [Google Scholar] [CrossRef]
Tang Q, Chen Z, Zhao L. (2019). Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging 11: 9982–9999. [Google Scholar]
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. (2015). Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians 65: 87–108. DOI 10.3322/caac.21262. [Google Scholar] [CrossRef]
Vaupel P. (2009). Prognostic potential of the pre-therapeutic tumor oxygenation status. Advances in Experimental Medicine and Biology 645: 241–246. [Google Scholar]
Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF. (2016). A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. European Heart Journal 37: 2602–2611. DOI 10.1093/eurheartj/ehv713. [Google Scholar] [CrossRef]
Wang L, Hu K, Chao Y, Wang X. (2020). MicroRNA-1296-5p suppresses the proliferation, migration, and invasion of human osteosarcoma cells by targeting NOTCH2. Journal of Cellular Biochemistry 121: 2038–2046. DOI 10.1002/jcb.29438. [Google Scholar] [CrossRef]
Wang X, Guo H, Yao B, Helms J. (2017). miR-15b inhibits cancer-initiating cell phenotypes and chemoresistance of cisplatin by targeting TRIM14 in oral tongue squamous cell cancer. Oncology Reports 37: 2720–2726. DOI 10.3892/or.2017.5532. [Google Scholar] [CrossRef]
Wei Y, Zhang Y, Meng Q, Cui L, Xu C. (2019). Hypoxia-induced circular RNA has_circRNA_403658 promotes bladder cancer cell growth through activation of LDHA. American Journal of Translational Research 11: 6838–6849. [Google Scholar]
Wu D, Yotnda P. (2011). Induction and testing of hypoxia in cell culture. JoVE: Journal of Visualized Experiments 12: 2899. [Google Scholar]
Xu G, Guo Y, Xu D, Wang Y, Shen Y, Wang F, Lv Y, Song F, Jiang D, Zhang Y, Lou Y, Meng Y, Yang Y, Kang Y. (2017). TRIM14 regulates cell proliferation and invasion in osteosarcoma via promotion of the AKT signaling pathway. Scientific Reports 7: 42411. DOI 10.1038/srep42411. [Google Scholar] [CrossRef]
Yu T, Wang LN, Li W, Zuo QF, Li MM, Zou QM, Xiao B. (2018). Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Science 109: 1393–1403. DOI 10.1111/cas.13583. [Google Scholar] [CrossRef]
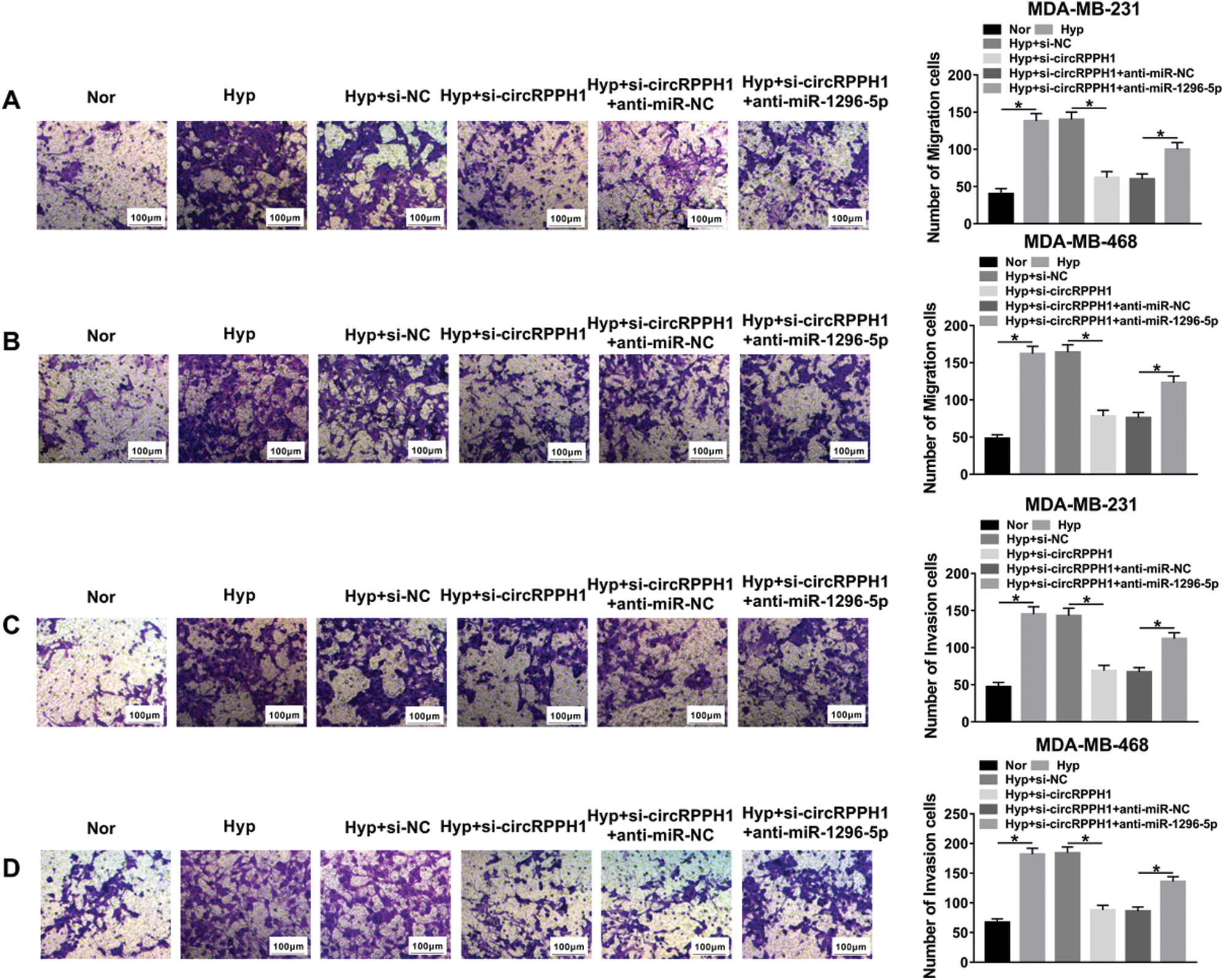
SupplEMENTARY Figure 1: CircRPPH1 regulated migration and invasion of hypoxia-treated TNBC cells by miR-1296-5p.

SUPPLEMENTARY Figure 2: MiR-1296-5p regulated migration and invasion of hypoxia-treated TNBC cells by TRIM14.
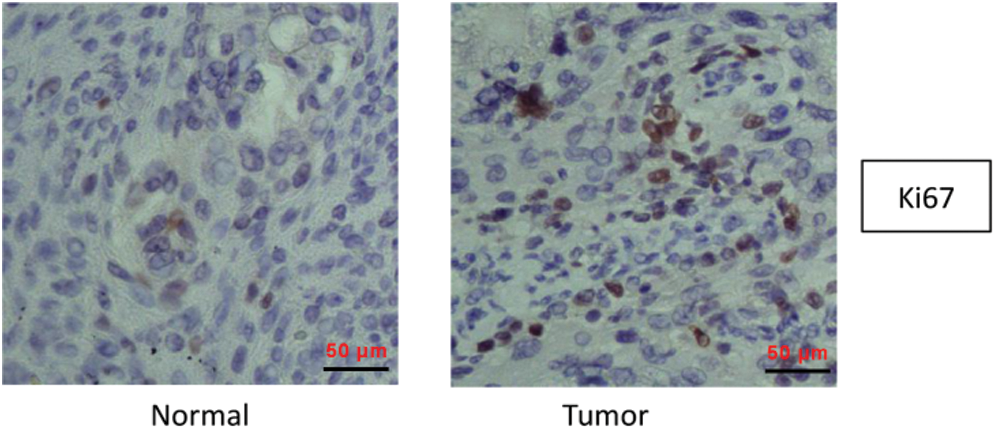
SUPPLEMENTARY Figure 3: Immunohistochemistry assay was used to detect Ki67 expression in BC tissues and adjacent normal tissues.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |