DOI:10.32604/biocell.2021.014193

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.014193 |  www.techscience.com/journal/biocell |
| Article |
Elevated nuclear phospho-eIF4E body levels are associated with tumor progression and poor prognosis for acute myeloid leukemia
1Department of Hematology, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, 310006, China
2College of Life Sciences, China Jiliang University, Hangzhou, 310018, China
*Address correspondence to: Hong Zhou, rainbow0223@sina.com
Received: 08 September 2020; Accepted: 21 December 2020
Abstract: Uncontrolled proliferation is a hallmark of cancer cells, yet the molecular mechanisms that contribute to this proliferation are unclear. Therapeutic treatment of cancer is suboptimal in many cases, with no accurate index by which to evaluate the success of treatment or patient prognosis. In this study, we explored the protein levels of nuclear phospho-eIF4E in acute myeloid leukemia (AML) cell lines and primary leukemia samples by Western blot and immunofluorescence and as well analyzed transcriptomes by RNA-seq. We found nuclear phospho-eIF4E, an exporter of oncogenic mRNAs, to be abundant in AML. Further, nuclear phospho-eIF4E abundance was significantly associated with tumor burden as well as the response of AML patients to chemotherapy. The results demonstrate “massive clustering and export of oncogenic mRNAs to the translation machinery” by highly abundant RNA-nuclear phospho-eIF4E bodies. This is an efficient mechanism that may drive the proliferation of cancer cells. Herein, nuclear phospho-eIF4E bodies were identified as potential markers of AML, which may be useful for prognosis and as targets for cancer therapy.
Keywords: Acute myeloid leukemia; Nuclear phospho-eIF4E; Poor prognosis
Acute myeloid leukemia (AML) is a heterogeneous group of malignant disorders characterized by dysregulated proliferation of hematopoietic stem cells and myeloid progenitors. Based on statistics released by the National Central Cancer Registry (NCCR), in 2015, there were approximately 4,292,000 new cancer cases diagnosed in China (an average of 12,000 new cases per day). Of these, 75,300 new leukemia cases were diagnosed with an overall mortality rate of 53.4% (Chen et al., 2016; Smith et al., 2004). The annual incidence of AML in China is 1.62/100,000, with 80% adult acute leukemia (Chen et al., 2016). AML morbidity and mortality are on the rise and are increasing yearly.
Although various chemotherapeutic regimens have been introduced for AML, patient outcomes have not markedly improved, with significant numbers of patients relapsing and dying soon after treatment (Lonetti et al., 2019). It is necessary to improve the screening and evaluation of relevant biomarkers that can be used to monitor patient treatment responses and disease progression. Further, individualized patient treatment will significantly improve outcomes and will ensure appropriate drug use. Therefore, the identification of new AML biomarkers for therapeutic evaluation and patient prognosis is of significant scientific and clinical importance.
Eukaryotic translation initiation factor 4E (eIF4E) is a potent oncogene that is frequently elevated in approximately 30% of human cancers including, blast phase chronic myeloid leukemia (CML-BC), Hodgkin and non-Hodgkin lymphomas, and M4 and M5 subtypes of AML (Borden and Culjkovic-Kraljacic, 2010; Smith et al., 2004) (Assouline et al., 2009; Jin et al., 2013). Physiologically, within the cytoplasm, eIF4E acts to rate-limit translation initiation, whereas in the nucleus, eIF4E forms nuclear bodies that promote cytoplasmic export of a subset of growth-promoting mRNAs (Borden and Culjkovic-Kraljacic, 2010). eIF4E must be phosphorylated at Ser209 to promote tumor development (Culjkovic et al., 2006; Robichaud et al., 2015). Phosphorylation of nuclear eIF4E appears to be important in the control of mRNA transport and the cancer-transforming properties of eIF4E. Elevated eIF4E levels are correlated with a poor prognosis.
As an eIF4E inhibitor, ribavirin has been used for the treatment of AML in phase II clinical trials. However, most of the treated AML patients failed to achieve long-term disease-free status and ultimately relapsed (Assouline et al., 2009; Smith et al., 2004). However, a small molecule inhibitor of the MAPK-activated protein kinase MNK1 reduces phosphorylation of eIF4E and effectively hampers the progress of leukemia (Lim et al., 2013). These results suggest that phospho-eIF4E (p-eIF4E), not eIF4E, is key to the progression of leukemia. We have previously shown that the traditional Chinese medicine homoharringtonine (HHT) selectively reduces levels of p-eIF4E, especially nuclear p-eIF4E, as well as its downstream effector Mcl-1, but not total eIF4E (Gu et al., 2015). HHT potently inhibits the growth of a distinct subset of AML cells and primary leukemia cells that contain high levels of nuclear p-eIF4E (Gu et al., 2015). These results suggest that nuclear p-eIF4E may be an ideal therapeutic target for AML. Therefore, we assessed the distribution, physiological function, clinical expression, and the effect of treatment on nuclear p-eIF4E in AML.
AML cell lines were purchased from ATCC. Primary AML cells and normal bone marrow cells, cord blood, peripheral blood were isolated from AML patients or healthy volunteers with immunodensity cell separation platforms. All experiments were approved by the ethics committee of the Affiliated Hang Zhou First People’s Hospital, Zhejiang University School of Medicine. AML cell lines were cultured in RPMI-1640 supplemented with 10% fetal calf serum (FCS) at 37°C in a 95% air, 5% CO2 humidified incubator.
AML cell lines, primary AML cells, normal bone marrow cells, cord blood cells, peripheral blood cells were fixed with 3.7% paraformaldehyde in PBS for 20 min on slides at room temperature (RT), then blocked and permeabilized with PBST containing 10% FBST (Fetal Bovine Serum and Tween 20) for 30 min. Cells were stained with primary antibodies overnight at 4°C, and then with FITC- or rhodamine-conjugated secondary antibodies for 60 min at RT. After three washes with PBS, the slides were mounted in Vectashield with 4’,6-diamidino-2-phenylindole (DAPI). Fluorescence was observed with a Zeiss Confocal Laser Scanning Microscope. The antibodies used for immunofluorescence staining were reactive with p-eIF4E from Abcam (Cambridge, MA).
Immuno-purification of nuclear phospho-eIF4E bodies from leukemia cells
Nuclear p-eIF4E bodies were purified from primary AML cells. Cells (2 × 108) were harvested by centrifugation at 2,000 rpm for 5 min at 4°C, washed with PBS three times, resuspended in PBS containing protease inhibitors, and transferred to a 7 mL Dounce tissue homogenizer for Dounce homogenization. The suspension was collected and centrifuged at 1,000 rpm for 5 min at 4°C. The supernatant was transferred to a fresh tube and centrifuged at 6,000 rpm for 5 min at 4°C. The pellet (containing nuclear p-eIF4E bodies) was resuspended in 1 mL of PBS containing protease inhibitors and pretreated with 5 μg of normal rabbit IgG for 1 h and then resuspended with 50 μL protein A/G beads for 30 min at 4°C. The suspension was centrifuged at 1,000 rpm for 5 min at 4°C, the supernatants transferred to a fresh tube, incubated with 2 μg of anti-p-eIF4E antibody overnight at 4°C, and incubated with 50 μL of protein A/G beads for 2 h at 4°C. After washing six times with PBS by centrifugation at 1,000 rpm for 5 min at 4°C, 0.5 mL of elution buffer (0.1M glycine-HCl, pH 2.8) was added to elute nuclear p-eIF4E bodies from protein A/G beads for 10 min at RT. The quality of purified nuclear p-eIF4E bodies was examined by immunofluorescence staining with a confocal laser scanning microscope.
Cells were washed twice with PBS (pH 7.2) and total cellular protein extracted with radioimmuno-precipitation assay buffer (RIPA). Protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide gels) and then transferred to nitrocellulose membranes, blocked with 5% nonfat milk in tris-buffered saline with Tween 20 (TBST), and incubated with primary antibodies overnight at 4°C. After three washes with TBST, membranes were probed with a horseradish peroxidase-conjugated secondary antibody for 1 h at RT, and signals were detected by chemiluminescence. The antibodies used for Western blot analysis were reactive with; Histone-2, BIN1, EPS15, SMARCA2, TPT1, eIF4E, and GAPDH from Cell Signaling Technology (Beverly, MA). Others from Abcam (Cambridge, MA).
Separation of nuclear phospho-eIF4E bodies in the cell nuclei
Myeloid leukemia cells (AML cell lines, primary cells, and normal cells from peripheral blood) were collected, then lysed with 0.5 mL of buffer A (10 mM Tris pH 7.5, 10 mM NaCl, 3 mM MgCl2, 1 mM PMSF, 0.05% NP40) for 5 min and centrifuged at 700 rpm for 5 min at 4°C. The supernatant was removed, and the nuclei were washed with 1.5 mL of Buffer B (10 mM Tris pH 7.5,10 mM NaCl, 3 mM MgCl2, 1 mM PMSF) at 700 rpm for 5 min at 4°C. The separation effect of nuclear phospho-eIF4E bodies in cell nuclei was examined with immunofluorescence staining under a confocal laser scanning microscope.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from KG-1 cells and nuclear phospho-eIF4E bodies using Trizol reagent (Life Technologies) according to the manufacturer’s instructions. Np9, cyclin D1, HOXA9, and Meis1 mRNAs were amplified using the following specific primers.
Np9: forward, 5’-CTCCACGGAGATGTCTGCA-3’
Reverse, 5’-CCCACATTTCCCCCTTTTC-3’
Cyclin D1: forward, 5’-CGATGCCAACCTCCTCAACGAC-3’
Reverse, 5’-CCAGCATCCAGGTGGCGACG-3’
HOXA9: forward, 5’-ATGGCCACCACTGGGGCCCTG-3’
Reverse, 5’-CTCGTCTTTTGCTCGGTCTTT-3’
Meis 1: forward, 5’-CTAACACACCCTTACCCTTCTG-3’
Reverse, 5’-TCTATCATGGGCTGCACTATTC-3’
β-actin: forward, 5’- GGTCATCACCATTGGCAATG -3’
Reverse, 5’- TCCATGCCCAGGAAGGAA-3’
All results are presented as the mean ± standard deviation. The two groups were compared using the unpaired Student’s t-test, and multiple comparisons were conducted using one-way analysis of variance, followed by Tukey’s post hoc test in GraphPad Prism 7 (GraphPad Software, Inc. San Diego, USA). Correlation analysis was conducted using the Pearson correlation coefficient. p < 0.05 was considered to indicate a statistically significant difference.
p-eIF4E is widely distributed in the cytoplasm and nucleus of AML cells. Although up to 68% of cellular eIF4E is within the nucleus (Robichaud et al., 2015), the percentage of nuclear p-eIF4E is less clear. We evaluated the levels of nuclear p-eIF4E in seven AML cell lines, cells from 14 AML patients (from bone marrow), and normal bone marrow cells, cord blood cells, peripheral blood cells from 12 healthy donors (Tab. 1).We detected higher levels (≥40%) of p-eIF4E in all AML cell lines, and in the majority (64.29%, 9/14) of cells from AML patients, as judged by Western blot analysis (Figs. 1a–1c). The immunofluorescent analysis confirmed p-eIF4E to be within the nucleus of AML cell lines (33.1–76.7%) and in cells of AML patients (29.7–79.2%), but not in normal bone marrow cells, cord blood cells, peripheral blood cells (Figs. 1d–1f).
Table 1: Results of nuclear p-eIF4E by western blot in AML cell lines, bone marrow of AML patients and normal hematopoietic cells from PB, BM and cord blood
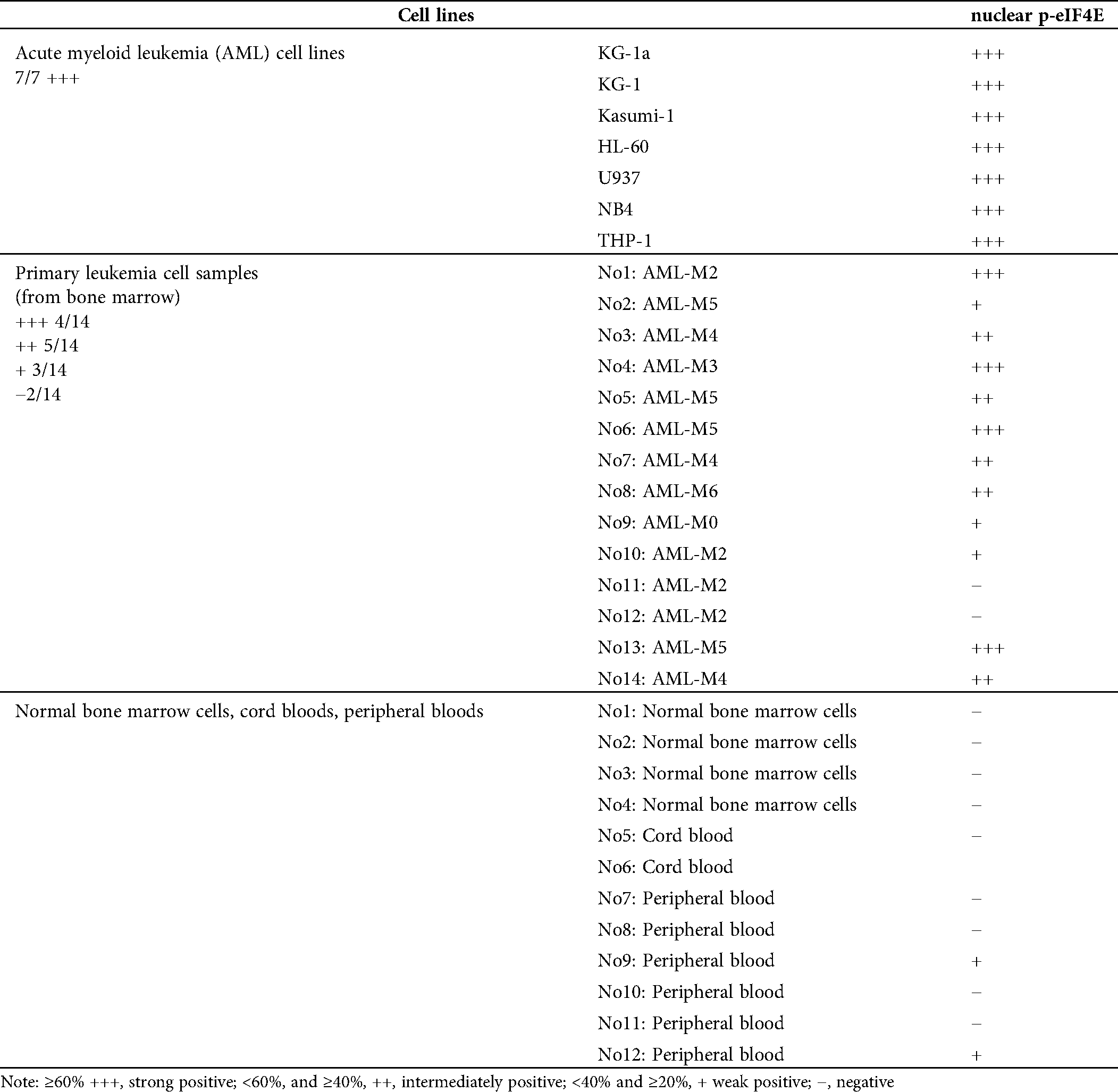
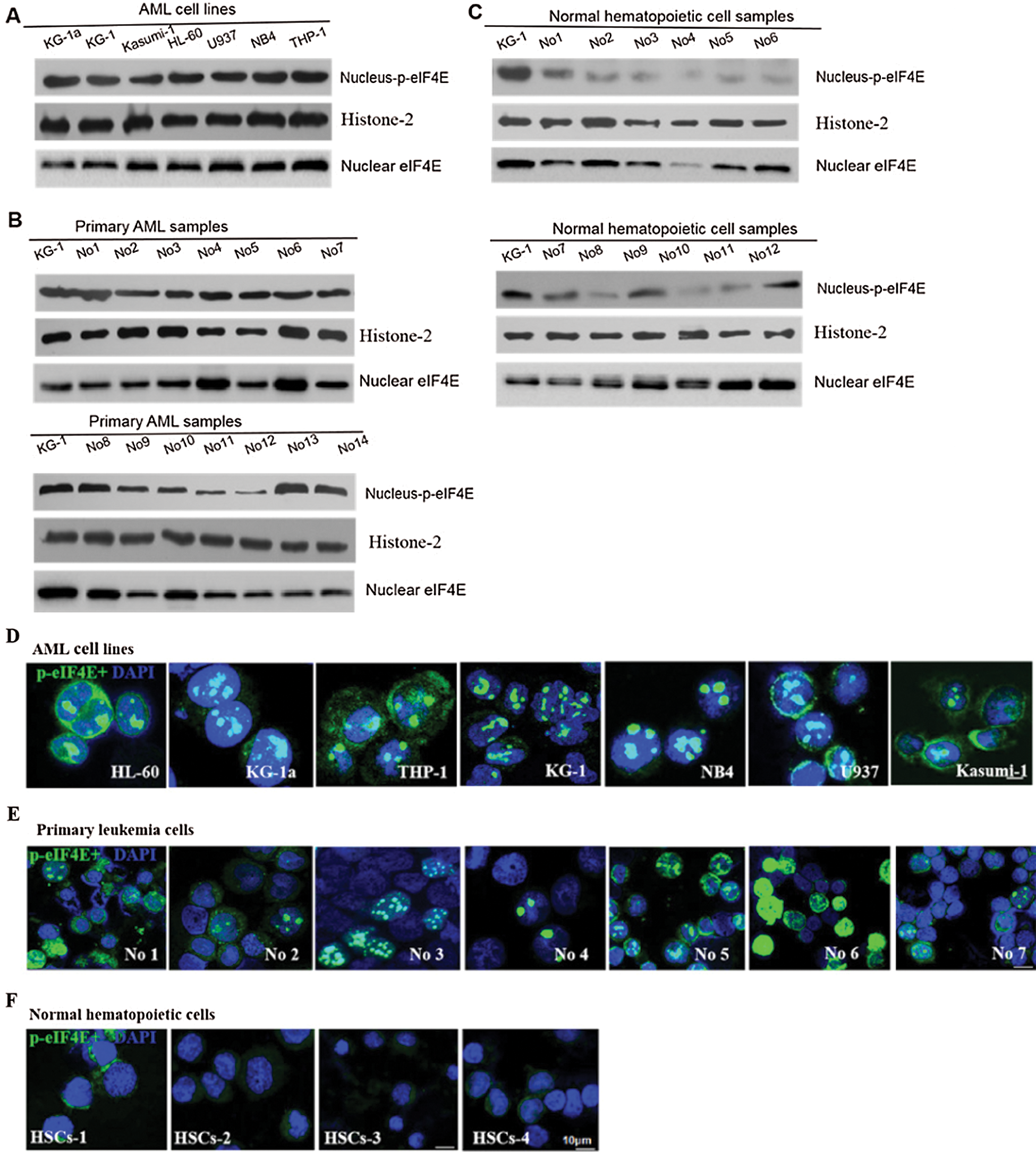
Figure 1: Nuclear distribution of p-eIF4E in AML cells.
Nuclear p-eIF4E is required for cell proliferation
Nuclear p-eIF4E is predominantly found in proliferating cells. Correlations were sought between nuclear p-eIF4E levels and cell proliferation, which is known to increase levels of SUMO1- p-eIF4E in a dose-dependent manner (Xu et al., 2010). A similar dose-dependent increase in nuclear p-eIF4E levels was also observed in KG-1 cells (Figs. 2a and 2b). Conversely, FBS starvation depleted nuclear p-eIF4E and induced cell growth arrest in KG-1 cells. We analyzed the effect of nuclear p-eIF4E inhibition on leukemia cell proliferation. We treated KG-1 cells with the small molecule inhibitor, CGP57380, for 72 h, after which the cells were analyzed for cell activity and nuclear p-eIF4E levels. We found that nuclear p-eIF4E levels were decreased in CGP57380 treated cells, which correlated with reduced leukemia cell proliferation (Figs. 2c–2e).
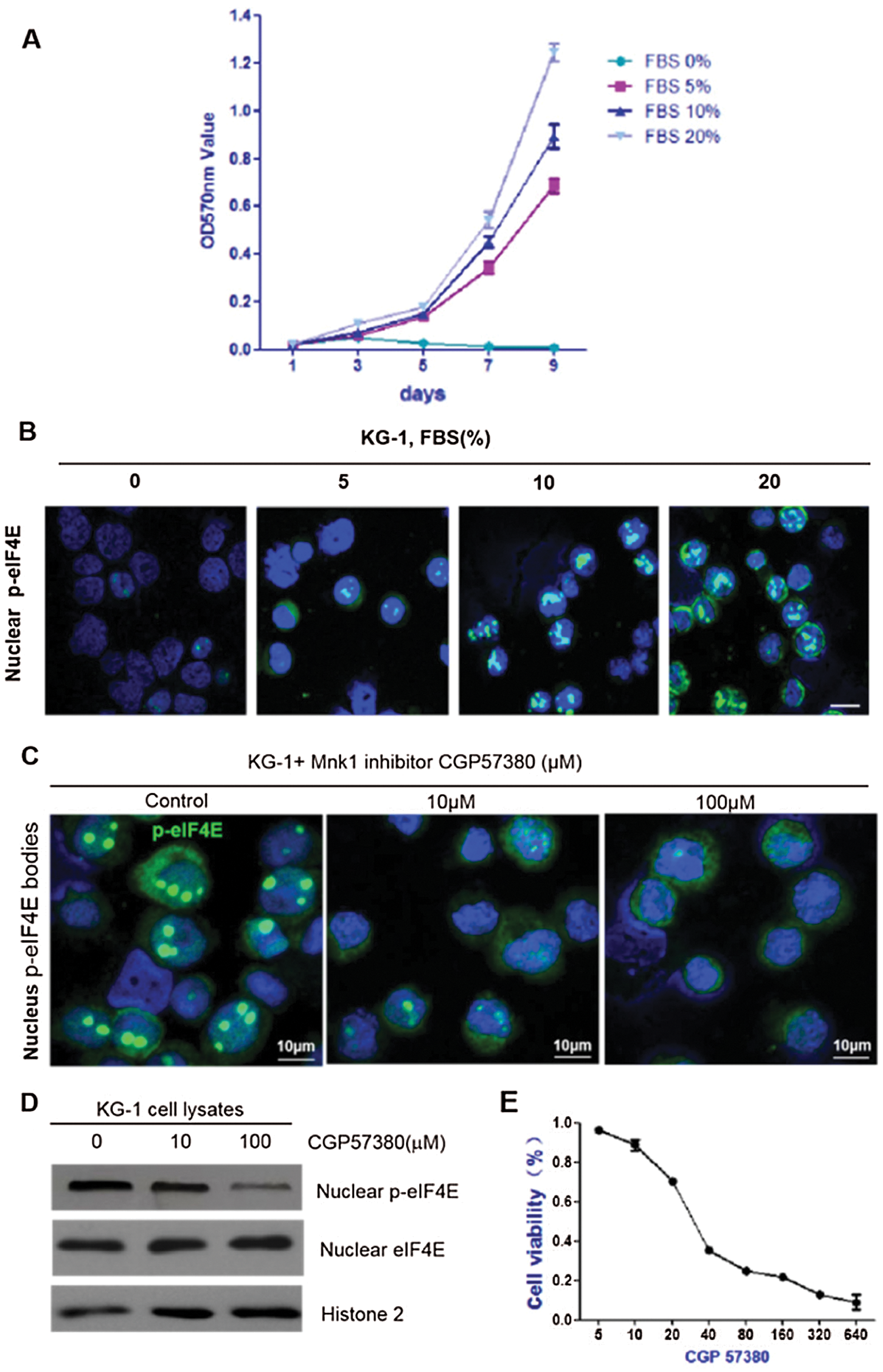
Figure 2: Nuclear p-eIF4E is required for cell proliferation.
We next analyzed the transcriptome of two clinical AML samples by RNA-seq, one with high and one with low expression of nuclear p-eIF4E, as well as one normal control sample (Fig. 3a). We found that most of the gene expressions of the two AML samples were higher than the normal control, so we directly analyzed the differences between the high- and low-expression AML samples. For these two samples, 35 genes were identified that are known to be associated with cellular proliferation, including EPS15 (16.04 vs. 1), SMARCA2 (16.39 vs. 1), and BIN1 (16.81 vs. 1) (Fig. 3b). EPS15 is a ubiquitous protein involved in cell growth regulation, mitogenic signal regulation, and the control of cellular proliferation. Further, enhanced expression of EPS15 homology domain 1 is associated with a poor prognosis for some cancers (Meng et al., 2015). A global transcription activator, SNF2L2, a protein encoded by SMARCA2 in humans, is required for transcriptional activation of genes normally repressed by chromatin. BIN1 encodes several isoforms of a nucleocytoplasmic adaptor protein, one of which was initially identified as an MYC-interacting protein with tumor suppressor activity (Wang et al., 2017). Our Western blot analysis found that the levels of these three proteins were greater in patients with elevated levels of p-eIF4E than in patients with lower levels of p-eIF4E (Fig. 3c).
We also found differentially expressed genes associated with leukemia stem cells, especially TPT1 (15.43 vs. 1) (Fig. 3d). TPT1 (translationally-controlled 1) is known to participate in various cellular activities, including protein synthesis, cell growth, and cell survival, which is highly expressed in tumor cells. In addition, TPT1 has been identified as a direct target of the tumor suppressor, TP53/p53 (Bae et al., 2017), which plays an important role in maintaining the self-renewal and proliferative ability of stem cells. Western blot analysis confirmed that levels of TPT1 were higher in patients with elevated levels of p-eIF4E than in patients with lower levels of p-eIF4E (Fig. 3e).
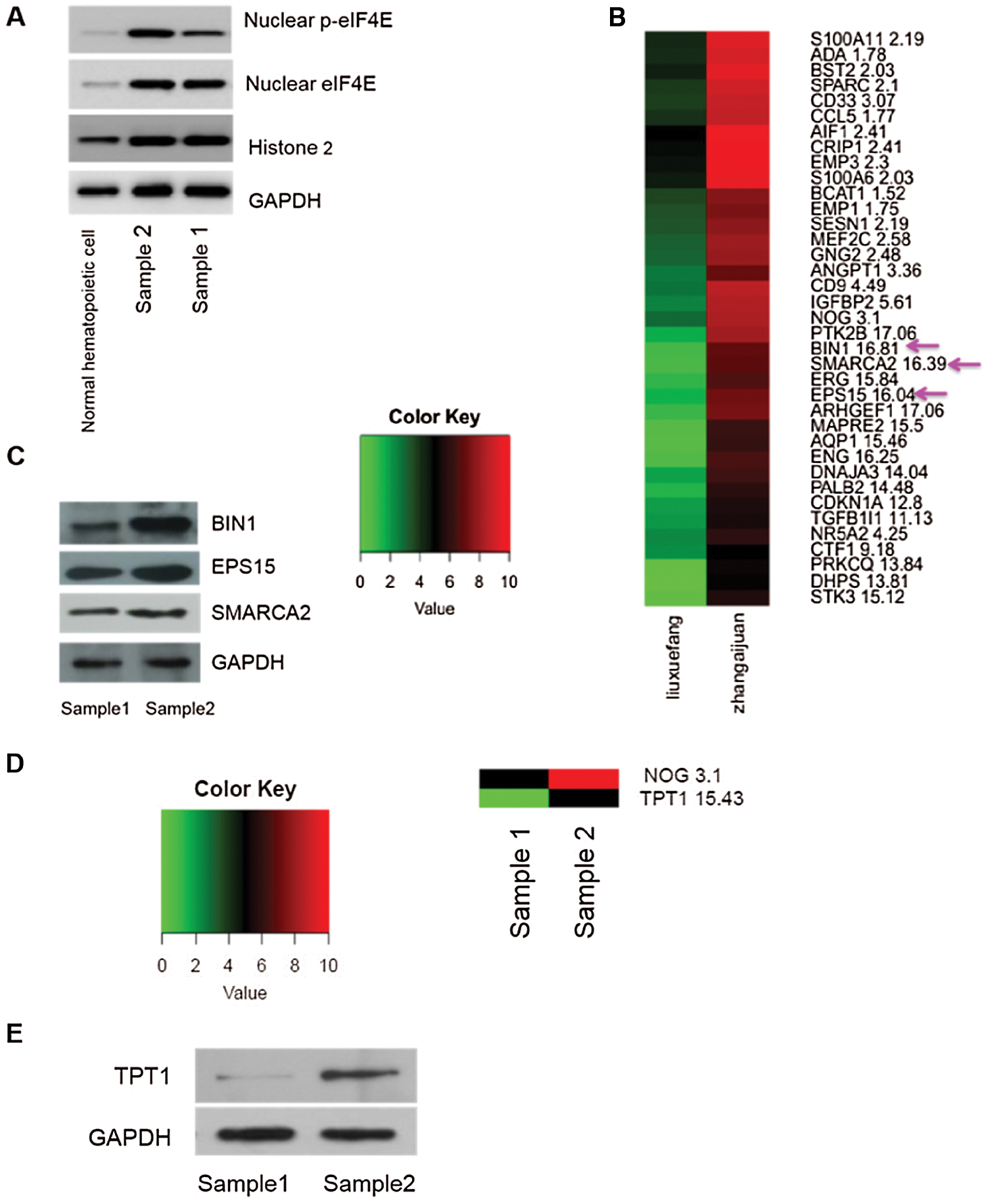
Figure 3: Greater nuclear p-eIF4E is associated with cell proliferation.
Nuclear p-eIF4E participates in cancer cell clustering and export of nuclear oncogenic mRNAs
eIF4E phosphorylation is associated with the translation of a small subset of c-Myc, Cyclin D1, and HoxA9 mRNAs, which have critical roles in cell survival (Zhou et al., 2016). There is evidence that mRNA export of nuclear p-eIF4E is linked to its oncogenic transformative capacity (Zhou et al., 2016). To verify this observation, an mRNA nuclear profile of KG-1 leukemia cell p-eIF4E bodies was performed. A total of 27,410 mRNAs were identified in nuclear p-eIF4E bodies from leukemia cells that could be grouped into 20 clusters based on their biological activities, including ribosome biogenesis in eukaryotes, proteasome function, spliceosome function, DNA replication, base excision repair, protein export, aminoacyl-tRNA biosynthesis, and RNA transport (Fig. 4a). Most importantly, 1,448 of these mRNAs are involved in cellular proliferation and apoptosis. These results clearly indicate that the nuclear p-eIF4E bodies in cancer cells contain large amounts of mRNAs.
Co-expression of various oncogenic mRNAs plays an important role in the uncontrolled proliferation of cancer cells. For example, co-expression of oncogenic transcription factors HOXA9 and Meis1 is sufficient to transform primary bone marrow cells and to induce leukemia (Wang et al., 2005). To determine whether oncogenic mRNAs are enriched in nuclear p-eIF4E bodies, we extracted mRNAs from the purified nuclear p-eIF4E bodies of KG-1 cells and measured the level of representative oncogenic mRNAs associated with uncontrolled proliferation of cancer cells, including viral Np9 (Chen et al., 2013; Denne et al., 2007), cyclin-D1 (Ju et al., 2014; Katz et al., 2014), HOXA9 (Brumatti et al., 2013; Li et al., 2012; Sun et al., 2013), and Meis1 (Bisaillon et al., 2011; Orlovsky et al., 2011). RT-PCR analysis showed that the levels of Np9, cyclin-D1, HOXA9, and Meis1 were 6.31-, 6.57-, 6.19-, and 10.83-fold higher in p-eIF4E bodies than in KG-1 cells (Fig. 4b). This result indicates that these oncogenic mRNAs are highly enriched within p-eIF4E bodies of leukemia cells. The enrichment of diverse oncogenic mRNAs within nuclear p-eIF4E bodies may be required for subsequent exporting of these oncogenic mRNAs from the nucleus to the cytoplasm.
To examine whether p-eIF4E was essential for mRNA export, we next determined whether the level of nuclear p-eIF4E bodies was correlated with cytoplasmic RNA levels in leukemia cells by acridine orange staining. We observed that leukemia cells with abundant p-eIF4E bodies exhibited strong RNA staining (arrows), whereas those cells without p-eIF4E bodies, such as mitotic cells (asterisks), were weakly stained for RNA (Fig. 4c). These results imply that p-eIF4E participates in both clustering and export of nuclear oncogenic mRNAs.
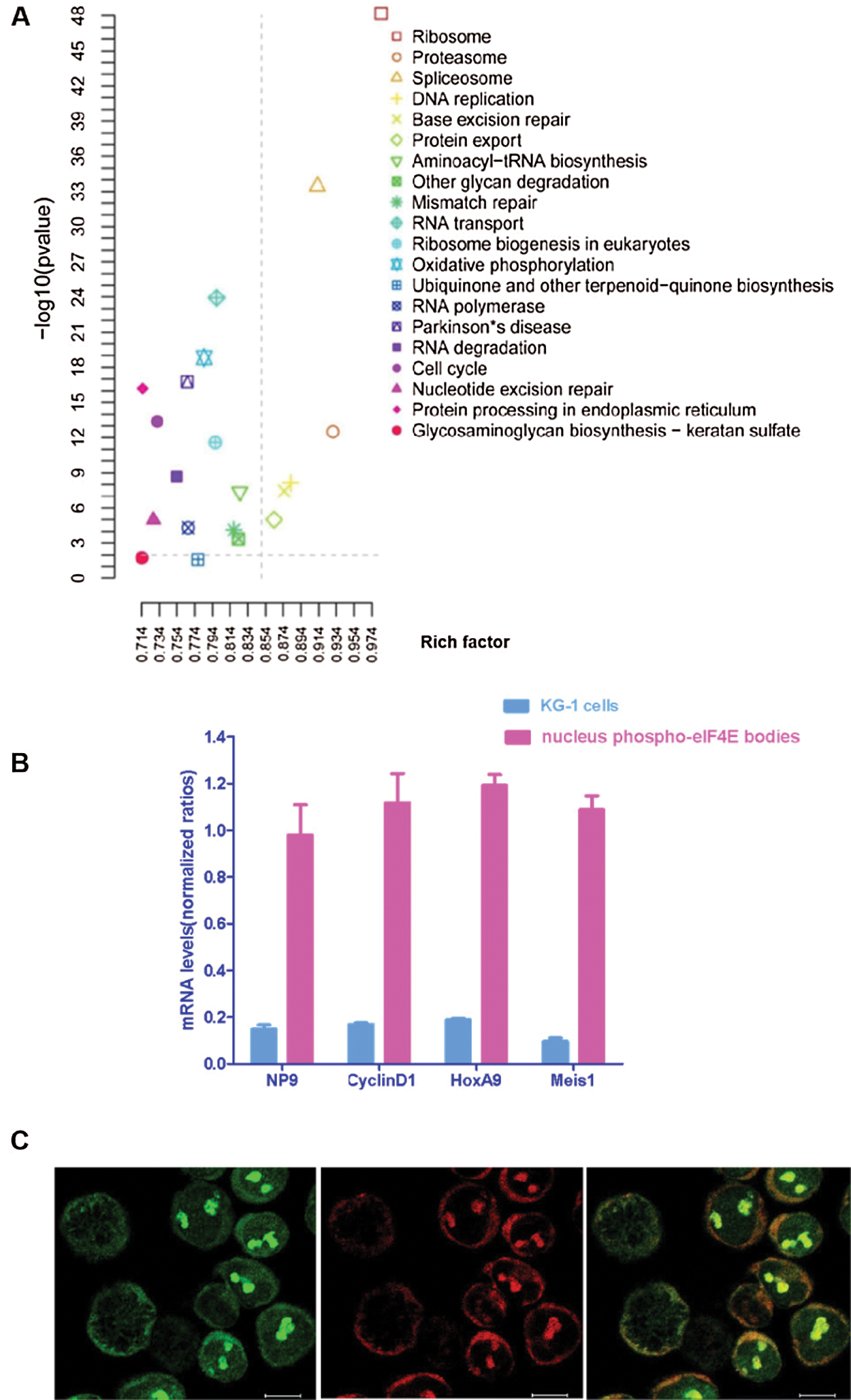
Figure 4: Nuclear p-eIF4E bodies contain a variety of mRNAs and participate in the enrichment and trafficking of oncogenic RNAs.
Nuclear p-eIF4E is correlated with tumor progression and poor patient prognosis
eIF4E regulates cell proliferation, and its dysregulation induces tumorigenesis (Bitterman and Polunovsky, 2015). eIF4E phosphorylation is also associated with cancer progression, with p-eIF4E important in carcinogenesis by the promotion of post-transcriptional regulation of cancer-related genes. Hence, p-eIF4E may be a marker for poor prognosis (Furic et al., 2010). We collected samples from 53 primary AML patients (these 53 primary AML patients all treatment with IA or HAA or DA, according to NCCN Clinical Practice Guidelines in Oncology: Acute Myeloid Leukemia (2020)) and analyzed nuclear p-eIF4E levels for correlation with clinical outcomes (Tab. 2). We observed that nuclear p-eIF4E levels were higher in hyperplastic bone marrow than in hypoplastic bone marrow, suggesting that the levels of nuclear p-eIF4E correlate with the malignant proliferative potential of bone marrow in leukemia patients, p < 0.001 (Fig. 5a). Importantly, we found that patients with highly abundant nuclear p-eIF4E exhibited poorer outcomes (at the end of the second course of treatment, the treatment response was evaluated as CR (complete response), PR (partial response), NR (non-response), PR and NR were defined as poor outcome) than patients with less abundant nuclear p-eIF4E, p < 0.001 (Fig. 5b). These results suggest that higher nuclear p-eIF4E levels are not only required for the uncontrolled proliferation of cancer cells but are also an indicator of a poor leukemia prognosis. We also found that human AML cells exhibited a close relationship between malignancy and nuclear p-eIF4E over-expression, R2 = 0.8859 (Fig. 5c). These results indicate that nuclear p-eIF4E abundance is not only positively correlated with the proliferative potential and tumor burden of cancer cells, but also serves as an indicator of a poor response to chemotherapy.
Table 2: Clinical information of Primary AML patients
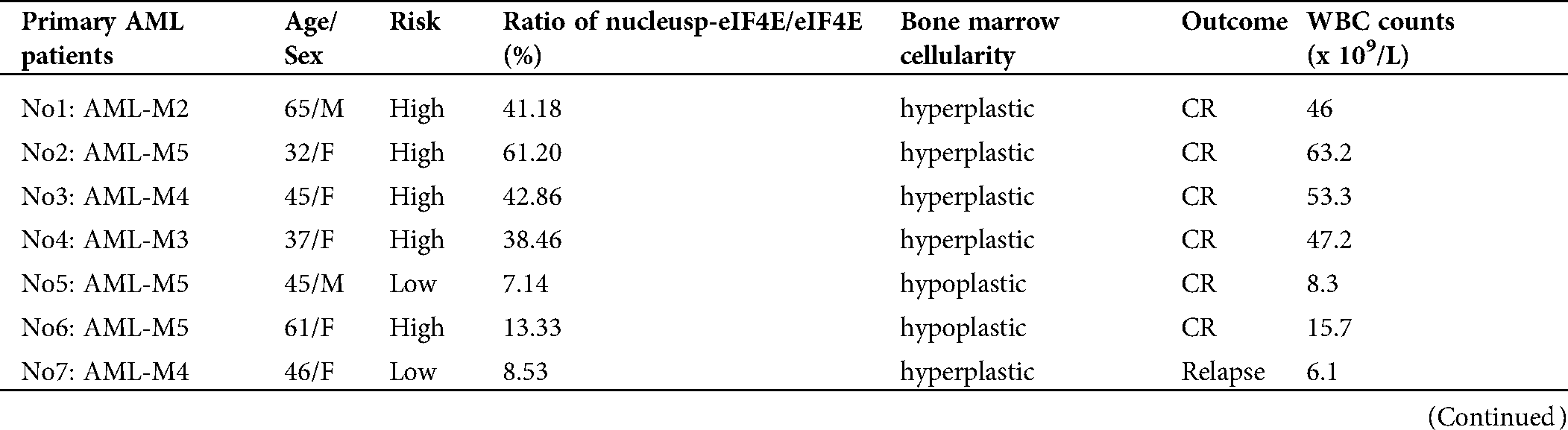
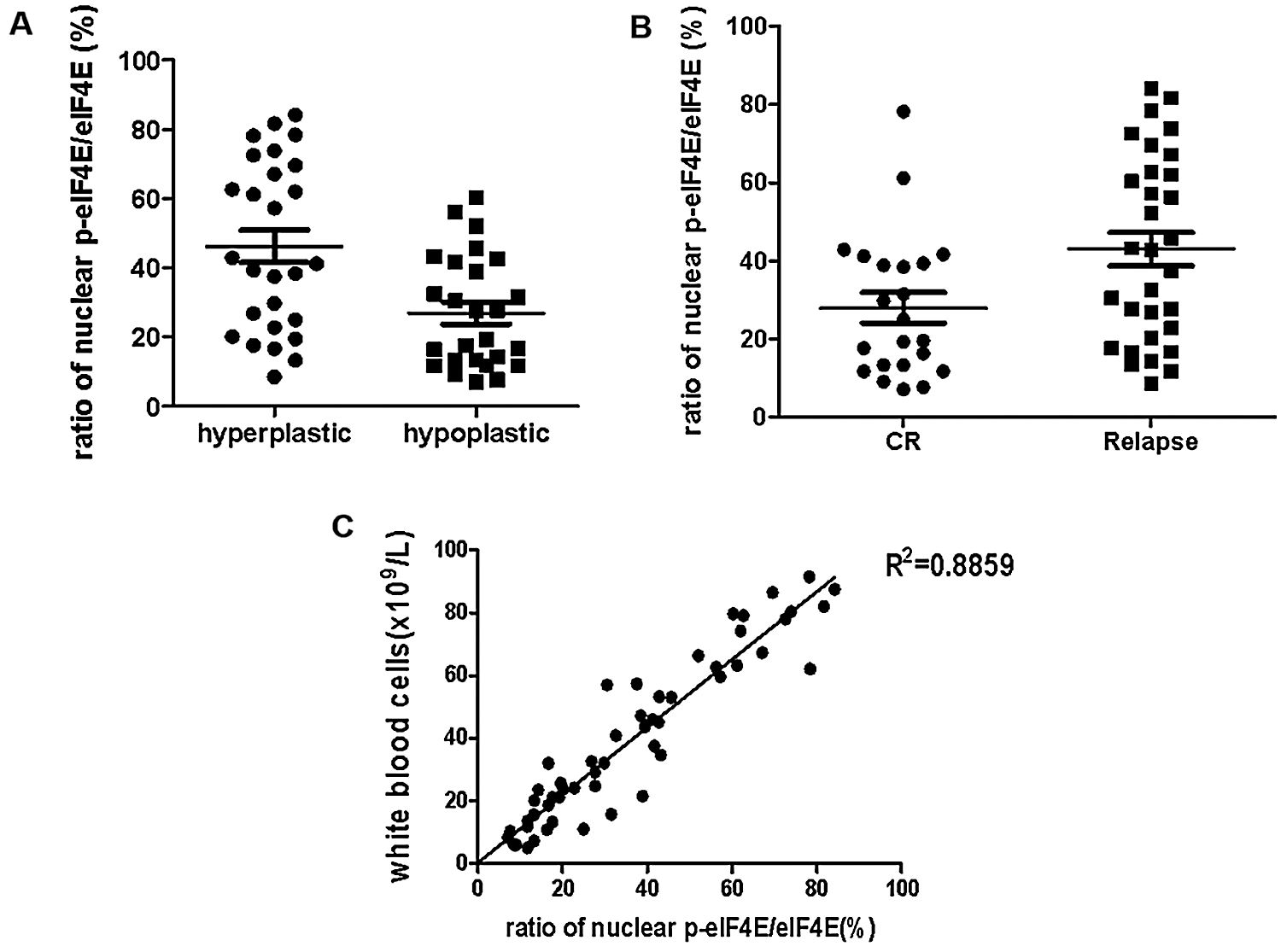
Figure 5: Nuclear p-eIF4E body abundance is correlated with cell proliferation, tumor burden, and outcomes in human leukemia.
eIF4E is associated with nuclear bodies within the nucleus (Cohen et al., 2001; Strudwick and Borden, 2002). Furthermore, the non-phosphorylated form of eIF4E is insufficient for malignant transformation and resists tumorigenesis in a model of prostate cancer (Furic et al., 2010). In contrast, there is a positive correlation between increased eIF4E phosphorylation and cellular proliferation (Flynn and Proud, 1996; Gingras et al., 1999), which suggests phosphorylation of eIF4E to be critical to tumorigenesis (Furic et al., 2010).
The biochemical function of p-eIF4E is unknown, despite its known involvement in several human disorders. Our findings establish eIF4E phosphorylation as a critical event in tumorigenesis. We have identified nuclear p-eIF4E to be associated with cancer cell proliferation. Although nuclear p-eIF4E is found in normal proliferating blood cells, its abundance is much lower than that in cancer cells. Importantly, we found that nuclear p-eIF4E participates in the clustering and export of oncogenic nuclear mRNAs essential to cell proliferation. These findings raise the possibility that chemical compounds that prevent phosphorylation of nuclear eIF4E may be potential anticancer drugs.
Consistent with these results, we demonstrated a variety of oncogenic mRNAs, which promote the proliferation of cancer cells, to be markedly clustered within cancer cell nuclear p-eIF4E bodies. These included viral Np9 (Chen et al., 2013; Denne et al., 2007), cyclin-D1 (Ju et al., 2014; Katz et al., 2014), HOXA9 (Brumatti et al., 2013; Li et al., 2012; Sun et al., 2013), and Meis1 (Bisaillon et al., 2011; Orlovsky et al., 2011). Of particular note, the abundance of nuclear p-eIF4E bodies was correlated with the levels of HOXA9 and Meis1, whose co-expression is critical for the transformation of primary bone marrow cells and leukemogenesis (Wang et al., 2005). Further, we found that abundant nuclear p-eIF4E bodies are present in approximately two-thirds of leukemia patients. Most importantly, the abundance of nuclear p-eIF4E bodies was positively associated with tumor burden and poorer clinical outcomes. These findings suggest that nuclear p-eIF4E bodies play a critical role in clustering and export of nuclear oncogenic mRNAs as well as subsequent cell proliferation.
Although a definitive link between the uncontrolled proliferation of cancer cells and elevated oncogenic mRNAs has been well established in cancer (Khavari and Rinn, 2007; Nieminen et al., 2013; Tang et al., 2009), little is known about the factors that facilitate the export of oncogenic mRNAs from the nucleus to the cytoplasm. Herein, we have identified nuclear p-eIF4E bodies to be critical nuclear organelles that regulate the clustering and export of nuclear oncogenic mRNAs. Proteomic studies have demonstrated nearly half of nuclear p-eIF4E body proteins to be involved in nuclear RNA trafficking, including sorting, assembling, transport, stability, and even metabolism of RNAs. These studies are consistent with a close correlation between nuclear p-eIF4E body abundance and oncogenic mRNA levels in cancer cells and normal blood cells. Although the mechanistic basis for the anti-oncogenic activity of p-eIF4E is unclear, we recently reported that small-molecule induction of phospho-eIF4E sumoylation results in degradation of phosphorylated serine residue 209 (Gu et al., 2015). SUMO1-p-eIF4E, a critical component of nuclear p-eIF4E bodies, has been shown to be essential for the activation of mRNA translation (Xu et al., 2010). These results indicate that mRNA trafficking from the nucleus to the cytoplasm is an important function of nuclear p-eIF4E bodies.
We propose a model by which nuclear p-eIF4E bodies regulate the uncontrolled proliferation of cancer cells. In the nuclei of cancer cells, a variety of amplified oncogenic mRNAs cluster within nuclear p-eIF4E bodies, which then facilitate the export of oncogenic mRNAs. Oncogenic mRNAs are then translated into oncogenic proteins that result in the over-proliferation of cancer cells.
In summary, “clustering and excessive export of amplified oncogenic mRNAs to the translation machinery of the cytoplasm” by abundant nuclear p-eIF4E bodies is a novel regulatory mechanism for over-proliferation by cancer cells. Further studies will be performed to characterize the function of proteins and mRNAs of nuclear p-eIF4E bodies. Such a characterization will contribute to an understanding of the uncontrolled proliferation of cancer cells and raise the possibility that compounds capable of inhibiting eIF4E phosphorylation may act as anticancer drugs.
Availability of Data and Materials: The datasets used during the present study are available from the corresponding author upon reasonable request.
Author Contribution: Hong Zhou conceived and designed the experiments and drafted the manuscript. Xiaofeng Jia and Fan Yang participated in the design of the study and performed the statistical analysis. Hong Zhou, Xiaofeng Jia, and Fan Yang performed the experiments. All authors read and approved the final paper.
Ethics Approval: The present study was approved by the Ethics Committee of Affiliated Hangzhou First People’s Hospital, No. 146-01. From March, 2018 to December, 2019. Written informed consent was obtained from every subject.
Funding Statement: This work was supported in part by the National Natural Science Foundation of China (Grant No. 81600129), the Science and Technology Project of Hangzhou (Grant No. 2016Z01), the Science and Technology Project of Hangzhou (Grant No. 2017A11) and Natural Science Foundation of Zhejiang Province (Grant No. LY21H080001).
Conflicts of Interest: The authors declare that they have no conflict of interests.
Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, Caplan S, Leber B, Roy DC, Miller WH Jr, Borden KL (2009). Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AMLA proof-of-principle clinical trial with ribavirin. Blood 114: 257–260. [Google Scholar]
Bae SY, Byun S, Bae SH, Min DS, Woo HA, Lee K. (2017). TPT1 (tumor protein, translationally-controlled 1) negatively regulates autophagy through the BECN1 interactome and an MTORC1-mediated pathway. Autophagy 13: 820–833. DOI 10.1080/15548627.2017.1287650. [Google Scholar] [CrossRef]
Bisaillon R, Wilhelm BT, Krosl J, Sauvageau G. (2011). C-terminal domain of MEIS1 converts PKNOX1 (PREP1) into a HOXA9-collaborating oncoprotein. Blood 118: 4682–4689. DOI 10.1182/blood-2011-05-354076. [Google Scholar] [CrossRef]
Bitterman PB, Polunovsky VA. (2015). eIF4E-mediated translational control of cancer incidence. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 1849: 774–780. DOI 10.1016/j.bbagrm.2014.09.007. [Google Scholar] [CrossRef]
Borden KL, Culjkovic-Kraljacic B. (2010). Ribavirin as an anti-cancer therapy: Acute myeloid leukemia and beyond. Leukemia & Lymphoma 51: 1805–1815. DOI 10.3109/10428194.2010.496506. [Google Scholar] [CrossRef]
Brumatti G, Salmanidis M, Kok CH, Bilardi RA, Sandow JJ, Silke N, Mason K, Visser J, Jabbour AM, Glaser SP, Okamoto T, Bouillet P, D’Andrea RJ, Ekert PG. (2013). HoxA9 regulated Bcl-2 expression mediates survival of myeloid progenitors and the severity of HoxA9-dependent leukemia. Oncotarget 4: 1933–1947. DOI 10.18632/oncotarget.1306. [Google Scholar] [CrossRef]
Chen T, Meng Z, Gan Y, Wang X, Xu F, Gu Y, Xu X, Tang J, Zhou H, Zhang X, Gan X, Van Ness C, Xu G, Huang L, Zhang X, Fang Y, Wu J, Zheng S, Jin J, Huang W, Xu R. (2013). The viral oncogene Np9 acts as a critical molecular switch for co-activating β-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia 27: 1469–1478. DOI 10.1038/leu.2013.8. [Google Scholar] [CrossRef]
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. (2016). Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians 66: 115–132. DOI 10.3322/caac.21338. [Google Scholar] [CrossRef]
Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL. (2001). PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO Journal 20: 4547–4559. DOI 10.1093/emboj/20.16.4547. [Google Scholar] [CrossRef]
Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. (2006). eIF4E is a central node of an RNA regulon that governs cellular proliferation. Journal of Cell Biology 175: 415–426. DOI 10.1083/jcb.200607020. [Google Scholar] [CrossRef]
Denne M, Sauter M, Armbruester V, Licht JD, Roemer K, Mueller-Lantzsch N. (2007). Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. Journal of Virology 81: 5607–5616. DOI 10.1128/JVI.02771-06. [Google Scholar] [CrossRef]
Flynn A, Proud CG. (1996). Insulin and phorbol ester stimulate initiation factor eIF-4E phosphorylation by distinct pathways in Chinese hamster ovary cells overexpressing the insulin receptor. European Journal of Biochemistry 236: 40–47. DOI 10.1111/j.1432-1033.1996.00040.x. [Google Scholar] [CrossRef]
Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, Pandolfi PP, Saad F, Sonenberg N. (2010). eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proceedings of the National Academy of Sciences of the United States of America 107: 14134–14139. DOI 10.1073/pnas.1005320107. [Google Scholar] [CrossRef]
Gingras AC, Raught B, Sonenberg N. (1999). eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annual Review of Biochemistry 68: 913–963. DOI 10.1146/annurev.biochem.68.1.913. [Google Scholar] [CrossRef]
Gu Y, Zhou H, Gan Y, Zhang J, Chen J, Gan X, Li H, Zheng W, Meng Z, Ma X, Wang X, Xu X, Xu G, Lu X, Liang Y, Zhang X, Lu X, Huang W, Xu R. (2015). Small-molecule induction of phospho-eIF4E sumoylation and degradation via targeting its phosphorylated serine 209 residue. Oncotarget 6: 15111–15121. DOI 10.18632/oncotarget.3615. [Google Scholar] [CrossRef]
Jin J, Wang JX, Chen FF, Wu DP, Hu J, Zhou JF, Hu JD, Wang JM, Li JY, Huang XJ, Ma J, Ji CY, Xu XP, Yu K, Ren HY, Zhou YH, Tong Y, Lou YJ, Ni WM, Tong HY, Wang HF, Mi YC, Du X, Chen BA, Shen Y, Chen Z, Chen SJ. (2013). Homoharringtonine-based induction regimens for patients with de-novo acute myeloid leukaemia: A multicentre, open-label, randomised, controlled phase 3 trial. Lancet Oncology 14: 599–608. DOI 10.1016/S1470-2045(13)70152-9. [Google Scholar] [CrossRef]
Ju X, Casimiro MC, Gormley M, Meng H, Jiao X, Katiyar S, Crosariol M, Chen K, Wang M, Quong AA, Lisanti MP, Ertel A, Pestell RG. (2014). Identification of a cyclin D1 network in prostate cancer that antagonizes epithelial-mesenchymal restraint. Cancer Research 74: 508–519. DOI 10.1158/0008-5472.CAN-13-1313. [Google Scholar] [CrossRef]
Katz SG, Labelle JL, Meng H, Valeriano RP, Fisher JK, Sun H, Rodig SJ, Kleinstein SH, Walensky LD. (2014). Mantle cell lymphoma in cyclin D1 transgenic mice with Bim-deficient B cells. Blood 123: 884–893. [Google Scholar]
Khavari TA, Rinn J. (2007). Ras/Erk MAPK signaling in epidermal homeostasis and neoplasia. Cell Cycle 6: 2928–2931. DOI 10.4161/cc.6.23.4998. [Google Scholar] [CrossRef]
Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, Jiang X, He C, Hyjek E, Zhang J, Zhang Z, Elkahloun A, Cao D, Shen C, Wunderlich M, Wang Y, Neilly MB, Jin J, Wei M, Lu J, Valk P, Delwel R, Lowenberg B, Le Beau MM, Vardiman J, Mulloy JC, Zeleznik-Le NJ, Liu PP, Zhang J, Chen J. (2012). miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nature Communications 3: 688. DOI 10.1038/ncomms1681. [Google Scholar] [CrossRef]
Lim S, Saw TY, Zhang M, Janes MR, Nacro K, Hill J, Lim AQ, Chang CT, Fruman DA, Rizzieri DA, Tan SY, Fan H, Chuah CT, Ong ST. (2013). Targeting of the MNK-eIF4E axis in blast crisis chronic myeloid leukemia inhibits leukemia stem cell function. Proceedings of the National Academy of Sciences of the United States of America 110: E2298–E2307. DOI 10.1073/pnas.1301838110. [Google Scholar] [CrossRef]
Lonetti A, Iacobucci I, Masetti R. (2019). Successes and challenges for diagnosis and therapy of acute leukemia. Journal of Oncology 2019: 3408318. DOI 10.1155/2019/3408318. [Google Scholar] [CrossRef]
Meng Q, Sun W, Li M, Zhao Y, Chen X, Sun L, Cai L. (2015). Increased expression of Eps15 Homology Domain 1 is associated with poor prognosis in resected small cell lung cancer. Journal of Cancer 6: 990–995. DOI 10.7150/jca.11650. [Google Scholar] [CrossRef]
Nieminen AI, Eskelinen VM, Haikala HM, Tervonen TA, Yan Y, Partanen JI, Klefström J. (2013). Myc-induced AMPK-phospho p53 pathway activates Bak to sensitize mitochondrial apoptosis. Proceedings of the National Academy of Sciences of the United States of America 110: E1839–E1848. DOI 10.1073/pnas.1208530110. [Google Scholar] [CrossRef]
Orlovsky K, Kalinkovich A, Rozovskaia T, Shezen E, Itkin T, Alder H, Ozer HG, Carramusa L, Avigdor A, Volinia S, Buchberg A, Mazo A, Kollet O, Largman C, Croce CM, Nakamura T, Lapidot T, Canaani E. (2011). Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation. Proceedings of the National Academy of Sciences of the United States of America 108: 7956–7961. DOI 10.1073/pnas.1103154108. [Google Scholar] [CrossRef]
Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, Larsson O, Muller WJ, Miller WH, Sonenberg N. (2015). Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene 34: 2032–2042. DOI 10.1038/onc.2014.146. [Google Scholar] [CrossRef]
Smith M, Barnett M, Bassan R, Gatta G, Tondini C, Kern W. (2004). Adult acute myeloid leukaemia. Critical Reviews in Oncology/Hematology 50: 197–222. DOI 10.1016/j.critrevonc.2003.11.002. [Google Scholar] [CrossRef]
Strudwick S, Borden KL. (2002). The emerging roles of translation factor eIF4E in the nucleus. Differentiation; Research in Biological Diversity 70: 10–22. DOI 10.1046/j.1432-0436.2002.700102.x. [Google Scholar] [CrossRef]
Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C, Rosner MR. (2013). HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proceedings of the National Academy of Sciences of the United States of America 110: 9920–9925. DOI 10.1073/pnas.1305172110. [Google Scholar] [CrossRef]
Tang SW, Chang WH, Su YC, Chen YC, Lai YH, Wu PT, Hsu CI, Lin WC, Lai MK, Lin JY. (2009). MYC pathway is activated in clear cell renal cell carcinoma and essential for proliferation of clear cell renal cell carcinoma cells. Cancer Letters 273: 35–43. DOI 10.1016/j.canlet.2008.07.038. [Google Scholar] [CrossRef]
Wang GG, Pasillas MP, Kamps MP. (2005). Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood 106: 254–264. DOI 10.1182/blood-2004-12-4664. [Google Scholar] [CrossRef]
Wang J, Jia Y, Zhao S, Zhang X, Wang X, Han X, Wang Y, Ma M, Shi J, Liu L. (2017). BIN1 reverses PD-L1-mediated immune escape by inactivating the c-MYC and EGFR/MAPK signaling pathways in non-small cell lung cancer. Oncogene 36: 6235–6243. DOI 10.1038/onc.2017.217. [Google Scholar] [CrossRef]
Xu X, Vatsyayan J, Gao C, Bakkenist CJ, Hu J. (2010). Sumoylation of eIF4E activates mRNA translation. EMBO Reports 11: 299–304. DOI 10.1038/embor.2010.18. [Google Scholar] [CrossRef]
Zhou H, Zhang J, Gu Y, Gan X, Gan Y, Zheng W, Kim BW, Xu X, Lu X, Dong Q, Zheng S, Huang W, Xu R. (2016). Identification of a novel RNA giant nuclear body in cancer cells. Oncotarget 7: 4724–4734. DOI 10.18632/oncotarget.6619. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |