2021 45(1): 41-48
DOI:10.32604/biocell.2021.010454

www.techscience.com/journal/biocell
| BIOCELL 2021 45(1): 41-48 DOI:10.32604/biocell.2021.010454 |  www.techscience.com/journal/biocell |
LncRNA NKILA suppresses airway hyper reactivity via interfering the facilitation of MUC5AC and MUC5B mediated by GALNT2
1Intensive Medicine Departments, Central Hospital of Baoji City, Baoji, 721000, China
2Emergency Department, Tangdu Hospital of Fourth Military Medical University, Xi’an, 710000, China
3Emergency Department, Central Hospital of Baoji City, Baoji, 721000, China
*Address correspondence to: Pengchong Liang, lpc68599677@163.com
Received: 05 March 2020; Accepted: 18 May 2020
#Yongli Zhang and Yizhan Cao have contributed equally to this work.
Abstract: Glycosylation of mucins mediated by N-acetylgalactosaminyltransferases (GALNTs) is closely related to respiratory diseases such as asthma and chronic obstructive pulmonary disease (COPD). In addition, long non-coding RNAs (LncRNAs) participate in physiological and pathological processes through various epigenetic mechanisms. In this study, we found that a novel LncRNA named NKILA combined with multiple mucins and GALNTs potentially by several bioinformatics methods, and we used quantitative real-time PCR (RT-qPCR) to detect the expressions of NKILA, MUC5AC, MUC5B, and GALNT2 mRNA in 50 cases of asthma samples and 19 cases of normal samples, whose results showed that the expression of NKILA was significantly decreased in asthmatic samples, negatively correlated with the severity of asthma and the expressions of MUC5AC and MUC5B, while GALNT2 was significantly increased in asthmatic tissues, and positively correlated with the severity of asthma and the expressions of MUC5AC and MUC5B. In vitro, we used transient transfection technology to overexpress or interfere with NKILA and GALNT2 and then detected the expressions of MUC5AC and MUC5B via RT-qPCR and Western blot, which demonstrated GALNT2 can promote the expressions of MUC5AC and MUC5B protein, while NKILA could inhibit this effect. Furthermore, co-immunoprecipitation results showed that GALNT2 could bind to MUC5AC and MUC5B protein. RNA immunoprecipitation and RNA pull-down experiments showed that NKILA could bind to GALNT2. These evidences suggested that there are correlations among the expression of NKILA, GALNT2, MUC5AC, and MUC5B proteins in asthmatic patients. Mechanically, we concluded that NKILA can suppress the O-linked glycosylation of MUC5AC and MUC5B proteins by binding to GALNT2 and inhibit the expression of MUC5AC and MUC5B proteins. Our researches provided a potential therapeutic target for AHR.
Keywords: Airway hyper reactivity; LncRNA NKILA; MUC5AC; MUC5B; GALNT2
According to the statistics in 2015, there are about 174.5 million chronic obstructive pulmonary disease (COPD) patients and 358.2 million bronchial asthma patients worldwide, which cause 3.2 million and 0.4 million related deaths each year respectively and are major components of the global burden of disease (Han et al., 2017). Airway hyperreactivity (AHR) is a critical factor leading to chronic bronchitis and bronchial asthma, which are induced by mucus hypersecretion from hyperplasia and metaplasia of goblet cell, and the hypersecretion from numerous goblet cells plays a critical role in the mucus plugging and airway obstruction (Aron and Akbari, 2017; Sung et al., 2017). Excessive respiratory mucus develops in coordination with persistent infections, leading to increased morbidity and mortality of respiratory diseases (Dunican et al., 2018).
The central macromolecular components of mucus are the mucin glycoproteins, which are essential for the formation of AHR. There are three classes of mucins in the airways: secreted non-polymerized mucin (Muc7), secreted polymerized mucins (MUC5AC and MUC5B) and transmembrane mucins (Muc1, Muc4, Muc16, and Muc20) (Zhou-Suckow et al., 2017). Among these, MUC5AC and MUC5B is the common and major respiratory mucin in goblet cell, which were reported to participate in occurrence and development of AHR closely (Bonser et al., 2016; Yokota et al., 2016).
Mucins are regulated at transcriptional, post-transcriptional and epigenetic levels. Glycosylation, as a kind of post-translational modification, is of great significance for maintaining the structure and function of mucins, involving in solubility and protein charge (Magalhães et al., 2016; Welsh et al., 2017). N-acetylgalactosaminyltransferases (GALNTs) initiate mucin-type O-linked glycosylation in the Golgi apparatus by catalyzing the transfer of N-acetylgalactosamine to serine and threonine residues on target proteins, which are key enzymes for the synthesis of mucins O-linked glycosylation (Roth et al., 1994; Ikehara et al., 2006). However, the relationship between GALNTs and AHR-related diseases is still a lack of systematic research.
Long non-coding RNAs (LncRNAs) has become a hotspot in the field of epigenetics in recent years, which were involved in a wide range of biological processes, such as promoter transcription, chromatin remodeling, and gene splicing. Abnormal expression and regulation of LncRNAs were also found to partake of occurrence and progress of AHR (Bi et al., 2015), including LncRNA COPDA1, MIR155HG, and MEG3 (Zheng et al., 2019; Song et al., 2020; Qiu et al., 2019). NF-κB interacting LncRNA (NKILA) was involved in a series of biological processes, such as oxygen-glucose deprivation/re-oxygenation, inflammation and EMT of various cancer (Wang et al., 2018; Zhu et al., 2019a; Wu et al., 2018; Huang et al., 2016), while re-oxygenation and inflammation are central factors in AHR (Skuljec et al., 2017; Plihalova et al., 2016), which indicated the potential research and application value of NKILA in the diagnosis and treatment of AHR-related diseases.
In this study, we revealed the expression characteristics of LncRNA NKILA in AHR tissues and its interaction with GALNT2 in pulmonary epithelial cells by bioinformatics and molecular biology technology. Our results suggested that NKILA is low expressed in asthmatic tissues and binds to GALNT2. At the same time, GALNT2 promotes the expression of mucins MUC5AC and MUC5B, which was significantly inhibited by NKILA. Mechanically, we found that NKILA can inhibit the expression of MUC5AC and MUC5B via blocking the O-glycosylation induced by GALNT2.
A total of 50 cases with asthma were recruited from The Central Hospital of Baoji City from January to October 2018. According to criteria for asthma (Lommatzsch and Virchow, 2014), all the tissues from asthmatics were divided into median (n = 23) and severe (n = 27) groups, at the same time, tissues from patients without respiratory or allergic history were selected as the control group (n = 19). All 69 patients underwent research fibreoptic bronchoscopy with BALF collection and endobronchial biopsy. Each case was divided into two parts: one for pathological examination, formalin immobilization immediately, paraffin embedding standard procedure, and the other for RNA and protein extraction. All patients were in a stable phase of the disease, with no evidence of exacerbation in the past 4 weeks. Exclusion criteria: significant nonasthma pulmonary disease or other medical problems. All patients provided written informed consent for the use of the tissues for clinical research, and this study was approved by The Central Hospital of Baoji City Medical Ethics Committee.
The Human bronchial epithelial cells (HBEs) are ATCC (American Type Culture Collection) sources and were purchased from the Chinese Academy of Sciences (Shanghai, China), which were cultured in RPMI-1640 medium (Thermo Scientific, USA) with 10% Fetal Bovine Serum (FBS, HyClone, Australia) and 1% penicillin-streptomycin (Gibco Company, USA). Supportive culture environment: 37°C, 5% CO2 with 100% relative humidity. Transient transfection method for over-expressing or interfering NKILA and GALNT2: HBEs were cultured in 6-well plates or 10 cm culture dishes with 80% cell density. 1.5 μg or 7.5 μg Vector, NKILA full-length, GALNT2 full-length, shControl or shGALNT2#1-4 plasmid were blended in 2 mL RPMI-1640 medium with 7.5 μL or 37.5 μL (10 cm dishes) Lipofectamine 2000 (Invitrogen, USA) for 20 min. HBEs were cultured by the mixture for 6 h after PBS treated three times, and then by 10%FBS + RPMI-1640 medium for 48 h to extract of total RNA and protein from cells.
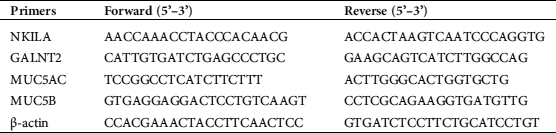
Quantitative real-time PCR (RT-qPCR)
Total RNA was extracted from cells or tissues by RNAiso Plus (Takara, Japan). Nanodrop 2000 (Thermo Scientific, USA) was used to detect the concentration of RNA. 1 μg RNA was reverse transcribed to cDNA by SuperScript IV (Invitrogen, USA). Specific primers were synthesized by Sangon Biotech (Shanghai, China). 0.5 μM Primers and 5 μL cDNA were added into 10 μL SYBR Green I (Takara, Japan) according to Takara kit instruction. Quantitative fluorescence analysis was carried out in the fluorescence quantitative PCR instrument (7500, Applied Biosystems). The RT-qPCR primers are listed in Tab. 1. Results were normalized to the expression levels of β-actin. The ΔCt was used to show the gene expression levels, and relative gene expression was calculated by using the 2 − ΔΔCt method (ΔCt = Ct target – Ct β-actin).
RIPA buffer (Beyotime, shanghai) with a cocktail (Beyotime, shanghai) lysed cells (5 × 106) and tissues (1 mg, ground in liquid nitrogen) at 0°C for 20 min to extract total protein. Pierce™ Rapid Gold BCA (Thermo Scientific, USA) was used to detect the concentration of total protein. 50 μg protein of were separated by 10% (for GALNT2, Muc5ac, and β-Actin) or 6% (for Muc5b) SDS-PAGE electrophoresis and transferred to PVDF (0.45 μm, Thermo Scientific). 5% of skim milk was used to block membranes for 2 h. The relevant first antibodies incubated purpose strip at 4°C overnight, including GALNT2 (1:1000 diluted, 65 KDa, Invitrogen, PA580653); MUC5AC (1:1000 diluted, 130 kDa, Abcam, ab24071); MUC5B (1:1000 diluted, 590 kDa, Abcam, ab77995), β-Actin (1:2000, 42 KDa, Abcam, ab8226). The relevant second rabbit anti-mouse or anti-rabbit antibody (1:2000 diluted, Abcam, ab6728 or ab6721) incubated for 1 h at 37°C, and then ECL luminescence reagent (Solarbio, Beijing) incubated for 10 sec. GelDoc XR (Bio-Rad, USA) was used to detect the intensity of the strip signal.
Co-Immunoprecipitation (Co-IP) and RNA immunoprecipitation (RNA-IP) assay
Co-IP: HBEs transfected GALNT2 plasmid were cultured in three 10 cm-culture dishes were lysed by IP lysis buffer (Beyotime, shanghai) for 2 h at 0°C. The lysate was divided into three groups, containing Input, IgG, and GALNT2 named respectively, at 3 of centrifugal tube. 2 μg Normal IgG (Abcam, ab188776 or CST, #2729S) or GALNT2 antibody incubated the lysate overnight at 4°C. 40 μL Protein A/G Magnetic Beads (Thermo Scientific Pierce, USA) were cultured with lysate overnight at 4°C. Centrifuge magnetic beads and remove the supernatant, and wash the beads by IP lysis buffer three times. 40 μL mixture of loading buffer and IP lysis buffer was added into the beads and boiling for 5 min. After removing the magnetic beads by the magnetic frame, the supernatant was used for 10% PAGE.
RNA-IP: RIP buffer (Promega, USA) was used to treat the lysate that was divided into three groups, containing Input, IgG and GALNT2, MUC5AC and MUC5B as Co-IP assay. After incubation of antibodies and pull-down of magnetic beads, washing the beads by RIP buffer. Extraction of total RNA by TRIzol for RT-qPCR.
The RNA fragment of NKILA and its anti-sense (as negative control) were transcribed in vitro (Takara, Japan) and biotin-labeled via the Biotin RNA Labeling Mix (Sigma-Aldrich, USA) and T7 RNA polymerase (Thermo Scientific, USA). DNA was removed by Ambion DNase I (Thermo Scientific, USA). The fragments were purified by the RNeasy Plus Mini Kit (Qiagen, Germany). Preparation of magnetic beads: 500 μL RIP wash buffer washed DynaBeads M-270 Streptavidin three times. DNA fragments and magnetic beads were added to 50 μL RIP wash buffer overnight at 4°C. After 3000 rpm centrifugation for 1 min, RIP Wash Buffer was rinsed three times, then incubated at room temperature with cell lysate added to the magnetic beads-RNA mixture for 1 h. The supernatant was retrieved, and 10% PAGE and WB analysis were performed.
All statistical analyses were performed using IBM SPSS Statistics v19 software package (Armonk, New York, USA). The quantitative data were compared and analyzed with Student’s t-test and analysis of variance (ANOVA). Pearson correlation was used for correlation analysis. A two-tailed value of P < 0.05 was considered statistically significant. All experiments were done at least three times independently.
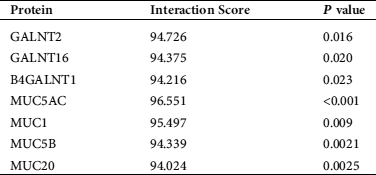
Bioinformatics prediction of NKILA and GALNT2 interacting proteins
Firstly, we found potential binding effects of NKILA with GALNTs and MUCs (Tab. 2) by retrieving bioinformatics website http://annolnc.cbi.pku.edu.cn/, while observed interaction between GALNT2 and MUCs (Fig. 1) via https://string-db.org./, which suggested that NKILA may participate in the regulation of AHR by influencing the interaction between GALNT2 and MUCs.
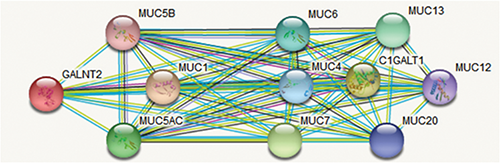
Figure 1: The predictive interaction between GALNT2 and MUCs.
Table 2: Prediction of NKILA interacting proteins
The expressions and relationships of MUC5AC, MUC5B, NKILA, and GALNT2 in asthma patients
To investigate the functions of NKILA and GALNT2 in asthma, we firstly observed the expressions of NKILA and GALNT2. Compared with patients without asthma, there were significant reductions of NKILA (Fig. 2c) and increases of MUC5AC, MUC5B, and GALNT2 in asthma patients (Figs. 2a, 2b, and 2d). Then, we were supposed to determine the expression correlation of MUC5AC, MUC5B, NKILA, and GALNT2 to providing clues for those interactions. The results of Pearson correlation showed the negative correlations between NKILA and MUC5AC and between NKILA and MUC5B (Figs. 2e and 2f), and the positive correlations between GALNT2 and MUC5AC and between GALNT2 and MUC5B (Figs. 2g and 2h), which further suggested the potential value of NKILA and GALNT2 in patients with AHR.
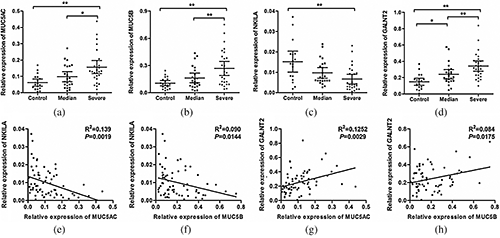
Figure 2: The expressions of NKILA and GALNT2 were related to MUC5AC and MUC5B.
The effect of overexpressing or interfering GALNT2 on MUC5AC and MUC5B
In order to elucidate the exact regulatory effects of GALNT2 on MUC5AC and MUC5B, HBE cells were used as in vitro models. RT-qPCR and Western Blot experiments showed that exogenous up-regulation or interference of GALNT2 could increase or decrease the expression of MUC5AC and MUC5B protein (Figs. 3a and 3b), but had no significant effect on the expression of both mRNAs (Figs. 3c and 3d), suggesting that GALNT2 regulates the expression of MUC5AC and MUC5B at the post-transcriptional level. CO-IP experiments confirmed that GALNT2 binds significantly to MUC5AC and MUC5B proteins (Fig. 3e), indicating that the regulation of GALNT2 on MUC5AC and MUC5B may be based on the combination.
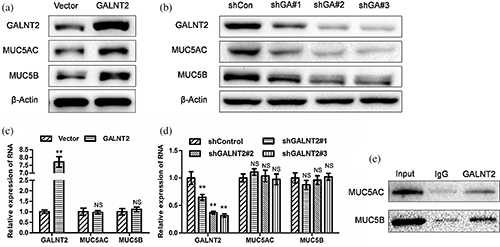
Figure 3: GALNT2 can up-regulate the expression of MUC5AC and MUC5B via binding to MUC5AC and MUC5B.
The effect of overexpressing NKILA on MUC5AC and MUC5B
The effects of NKILA on MUC5AC and MUC5B are also noteworthy. RT-qPCR and Western Blot experiments elucidated that NKILA can significantly reduce the expressions of MUC5AC and MUC5B protein (Fig. 4a), but has no significant effect on mRNA level (Fig. 4b), which indicated that NKILA also plays a regulatory role at the post-transcriptional level, just like GALNT2. In order to clarify whether NKILA is effective through GALNT2, we co-transfected NKILA and GALNT2 vector into HBE cells. We found that NKILA can significantly inhibit the up-regulation of MUC5AC and MUC5B induced by GALNT2 (Fig. 4c), and confirmed the exact combinations of NKILA and GALNT2 (Figs. 4d and 4e), NKILA and Muc5ac (Fig. 4f) and NKILA and Muc5b (Fig. 4g).
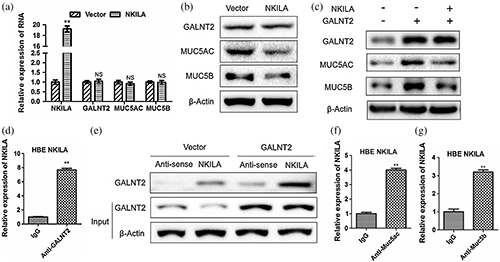
Figure 4: NKILA inhibits the protein expressions of MUC5AC and MUC5B via binding to GALNT2.
Mucins are essential for the occurrence of AHR-related diseases. MUC5AC and MUC5B, as pivotal mucins in the airway, are found to overproduce in respiratory disease, including COPD and asthma. Mucins can reduce pulmonary function by increasing the frequency and duration of infection (Boucherat et al., 2013; Livraghi-Butrico et al., 2017). Glycosylation is a critical factor in maintaining the structure and function of mucins (Symmes et al., 2018). GALNTs are the key enzymes of mucin O-glycosylation. As a member of GALNTs, GALNT2 coding gene is located on human chromosome 1q42.1. It was found that GALNT2 is involved in influencing triglyceride levels and in the development of type 2 diabetes mellitus and various cancers (Holleboom et al., 2011; Liu et al., 2016; Wu et al., 2011). However, the role of GALNT2 in AHR-related diseases remains unclear. Based on the close relationship between GALNTs and Mucins (Bennett et al., 2012), this study explored the potential relationship between GALNT2 and AHR. We detected the expression of GALNT2 in lung tissues from normal controls and asthmatic patients. The high expression of GALNT2 in severe AHR tissues and its positive correlation with the expressions of MUC5AC and MUC5B suggested the potential value of GALNT2 in the occurrence of AHR. Moreover, in vitro HBE cell model, we found that overexpression or interference of GALNT2 can significantly promote or inhibit the expression of MUC5AC and MUC5B proteins, and bind to both, which indicated that GALNT2 maintains the expression of MUC5AC and MUC5B proteins at the post-transcriptional level by direct combination.
As a momentous epigenetic molecule, LncRNAs take part in an extensive and complex biological process. Various LncRNAs were found to be related to airway inflammation and AHR. LncRNA LNC_000127 was highly expressed in eosinophilic samples before treatment and was increased in Jurkat cells and human CD4+ T cells following stimulation with PMA/CD28. Subsequent analyses revealed that LNC_000127 functions in the Th2 inflammation pathway, which indicated targeting LNC_000127 may be effective for reducing Th2 inflammation in eosinophilic asthma (Zhu et al., 2019b). A new study reported that lncRNA AK085865 served as a critical regulator of macrophages polarization and reduced the pathological progress of asthmatic airway inflammation (Pei et al., 2020). Moreover, LncRNA MIR155HG were identified to be related to the macrophage polarization in COPD, and functional experiments showed that MIR155HG deletion could reverse cigarette smoke extract exposure-induced apoptosis and inflammation in HPMECs (Song et al., 2020). NKILA, a LncRNA that interacts with NF-κB, can down-regulate the expressions of NF-κB and its downstream signaling pathway, be involved in inhibiting inflammation, anti-cancer and anti-cell hypoxia (Huang et al., 2018; Liu et al., 2015; Zhou et al., 2018). In addition, inflammation and hypoxia are the core factors causing AHR (Lee et al., 2017; Huerta-Yepez et al., 2011), and these clues suggested that NKILA may play a pivotal role in AHR.
Through a bioinformatics search, we found that NKILA potentially interacts with glycosylase GALNTs and MUCs, including GALNT2, MUC5AC, and MUC5B, which aroused our strong interest in the effects of NKILA on AHR. Therefore, we detected the expression of NKILA in asthmatic and normal lung tissues and found that the expression of NKILA was down-regulated in AHR tissues and negatively correlated with the expression of MUC5AC and MUC5B, which provided clues to reveal the relationship between NKILA and GALNT2/MUCs. Furthermore, exogenous overexpression of NKILA in HBE cells resulted in the suppression of MUC5AC and MUC5B protein expression. In order to determine whether this effect is produced by blocking the GALNT2/MUCs pathway. We supplemented NKILA in HBE cells on the basis of overexpression of GALNT2, which revealed that NKILA could decrease the expressions of MUC5AC and MUC5B in HBE cells with high GALNT2 level and restore the expressions to the wild type level. Meanwhile, the bindings of NKILA with GALNT2, MUC5AC, and MUC5B were confirmed, which suggested that NKILA could bind GALNT2/MUCs directly and form complex with GALNT2/MUCs, which interferes with the GALNT2-mediated facilitation on MUCs at the post-translational level. It is worth mentioning that the expressions of MUC5AC and MUC5B were increased during the development of AHR in clinical tissue samples, which may not only be due to the transcriptional activation of MUC5AC/MUC5B, but the glycosylation of their protein level is still one of the significant factors. Although we observed the correlation between NKILA/GALNT2 and MUC5AC/MUC5B expressions at mRNA level, following studies showed that NKILA/GALNT2 regulated MUC5AC/MUC5B expressions at protein level, while the ascending of MUC5AC/MUC5B mRNA level in clinical samples was not mediated by NKILA/GALNT2 but due to other factors, although the correlations of their expression occurred in the same pathological condition of AHR.
This study clarified the expression situations of NKILA and GALNT2 in asthmatic patients and revealed the mechanism that NKILA inhibits the expression of AHR-related molecules MUC5AC and MUC5B in bronchial epithelial cells in vitro by interfering with the interaction between GALNT2 and MUCs, thus building a bridge between inflammation-related molecules NKILA and mucus related pathway to mediate AHR, which provides further insights on the LncRNA the pathogenesis of AHR and a therapeutic strategy on AHR treatments.
Acknowledgement: Authors would like to express their gratitude to Fourth Military Medical University (Xi’an, China) for supporting this study.
Availability of Data and Materials: The datasets used or analyzed during the current study are available from the corresponding authors on reasonable request.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Aron JL, Akbari O. (2017). Regulatory T cells and type 2 innate lymphoid cell-dependent asthma. Allergy 72: 1148–1155. DOI 10.1111/all.13139. [Google Scholar] [CrossRef]
Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. (2012). Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22: 736–756. DOI 10.1093/glycob/cwr182. [Google Scholar] [CrossRef]
Bi H, Zhou J, Wu D, Gao W, Li L, Yu L, Liu F, Huang M, Adcock IM, Barnes PJ, Yao X. (2015). Microarray analysis of long non-coding RNAs in COPD lung tissue. Inflammation Research 64: 119–126. DOI 10.1007/s00011-014-0790-9. [Google Scholar] [CrossRef]
Bonser LR, Zlock L, Finkbeiner W, Erle DJ. (2016). Epithelial tethering of MUC5AC-rich mucus impairs mucociliary transport in asthma. Journal of Clinical Investigation 126: 2367–2371. DOI 10.1172/JCI84910. [Google Scholar] [CrossRef]
Boucherat O, Boczkowski J, Jeannotte L, Delacourt C. (2013). Cellular and molecular mechanisms of goblet cell metaplasia in the respiratory airways. Experimental Lung Research 39: 207–216. DOI 10.3109/01902148.2013.791733. [Google Scholar] [CrossRef]
Dunican EM, Elicker BM, Gierada DS, Nagle SK, Schiebler ML, Newell JD, Raymond WW, Lachowicz-Scroggins ME, Di Maio S, Hoffman EA, Castro M, Fain SB, Jarjour NN, Israel E, Levy BD, Erzurum SC, Wenzel SE, Meyers DA, Bleecker ER, Phillips BR, Mauger DT, Gordon ED, Woodruff PG, Peters MC, Fahy JV. (2018). Mucus plugs in patients with asthma linked to eosinophilia and airflow obstruction. Journal of Clinical Investigation 128: 997–1009. DOI 10.1172/JCI95693. [Google Scholar] [CrossRef]
Han MK, Quibrera PM, Carretta EE, Barr RG, Bleecker ER, Bowler RP, Cooper CB, Comellas A, Couper DJ, Curtis JL, Criner G, Dransfield MT, Hansel NN, Hoffman EA, Kanner RE, Krishnan JA, Martinez CH, Pirozzi CB, O’Neal WK, Rennard S, Tashkin DP, Wedzicha JA, Woodruff P, Paine R 3rd, Martinez FJ, SPIROMICS investigators. (2017). Frequency of exacerbations in patients with chronic obstructive pulmonary disease: an analysis of the SPIROMICS cohort. Lancet Respiratory Medicine 5: 619–626. [Google Scholar]
Holleboom AG, Karlsson H, Lin RS, Beres TM, Sierts JA, Herman DS, Stroes ES, Aerts JM, Kastelein JJ, Motazacker MM, Dallinga-Thie GM, Levels JH, Zwinderman AH, Seidman JG, Seidman CE, Ljunggren S, Lefeber DJ, Morava E, Wevers RA, Fritz TA, Tabak LA, Lindahl M, Hovingh GK, Kuivenhoven JA. (2011). Heterozygosity for a loss-of-function mutation in GALNT2 improves plasma triglyceride clearance in man. Cell Metabolism 14: 811–818. DOI 10.1016/j.cmet.2011.11.005. [Google Scholar] [CrossRef]
Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, Zhao J, Liu J, Lu Y, Xing Y, Chen F, Su F, Yao H, Liu Q, Su S, Song E. (2018). NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nature Immunology 19: 1112–1125. DOI 10.1038/s41590-018-0207-y. [Google Scholar] [CrossRef]
Huang W, Cui X, Chen J, Feng Y, Song E, Li J, Liu Y. (2016). Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget 7: 62520–62532. DOI 10.18632/oncotarget.11528. [Google Scholar] [CrossRef]
Huerta-Yepez S, Baay-Guzman GJ, Bebenek IG, Hernandez-Pando R, Vega MI, Chi L, Riedl M, Diaz-Sanchez D, Kleerup E, Tashkin DP, Gonzalez FJ, Bonavida B, Zeidler M, Hankinson O. (2011). Hypoxia inducible factor promotes murine allergic airway inflammation and is increased in asthma and rhinitis. Allergy 66: 909–918. DOI 10.1111/j.1398-9995.2011.02594.x. [Google Scholar] [CrossRef]
Ikehara Y, Sato T, Niwa T, Nakamura S, Gotoh M, Ikehara SK, Kiyohara K, Aoki C, Iwai T, Nakanishi H, Hirabayashi J, Tatematsu M, Narimatsu H. (2006). Apical Golgi localization of N'-diacetyllactosediamine synthase, β4GalNAc-T3, is responsible for LacdiNAc expression on gastric mucosa. Glycobiology 16: 777–785. DOI 10.1093/glycob/cwl005. [Google Scholar] [CrossRef]
Lee HY, Hur J, Kim IK, Kang JY, Yoon HK, Lee SY, Kwon SS, Kim YK, Rhee CK. (2017). Effect of nintedanib on airway inflammation and remodeling in a murine chronic asthma model. Experimental Lung Research 43: 187–196. DOI 10.1080/01902148.2017.1339141. [Google Scholar] [CrossRef]
Liu SY, Shun CT, Hung KY, Juan HF, Hsu CL, Huang MC, Lai IR. (2016). Mucin glycosylating enzyme GALNT2 suppresses malignancy in gastric adenocarcinoma by reducing MET phosphorylation. Oncotarget 7: 11251–11262. DOI 10.18632/oncotarget.7081. [Google Scholar] [CrossRef]
Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. (2015). A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27: 370–381. DOI 10.1016/j.ccell.2015.02.004. [Google Scholar] [CrossRef]
Livraghi-Butrico A, Grubb BR, Wilkinson KJ, Volmer AS, Burns KA, Evans CM, O’Neal WK, Boucher RC. (2017). Contribution of mucus concentration and secreted mucins Muc5ac and Muc5b to the pathogenesis of muco-obstructive lung disease. Mucosal Immunology 10: 395–407. DOI 10.1038/mi.2016.63. [Google Scholar] [CrossRef]
Lommatzsch M, Virchow JC. (2014). Severe asthma: definition, diagnosis and treatment. Deutsches Ärzteblatt International 111: 847–855. [Google Scholar]
Magalhães A, Rossez Y, Robbe-Masselot C, Maes E, Gomes J, Shevtsova A, Bugaytsova J, Borén T, Reis CA. (2016). Muc5ac gastric mucin glycosylation is shaped by FUT2 activity and functionally impacts Helicobacter pylori binding. Scientific Reports 6: 25575. DOI 10.1038/srep25575. [Google Scholar] [CrossRef]
Pei W, Zhang Y, Li X, Luo M, Chen T, Zhang M, Zhong M, Lv K. (2020). LncRNA AK085865 depletion ameliorates asthmatic airway inflammation by modulating macrophage polarization. International Immunopharmacology 83: 106450. DOI 10.1016/j.intimp.2020.106450. [Google Scholar] [CrossRef]
Plihalova A, Bartakova H, Vasakova M, Gulati S, deGlisezinski I, Stich V, Polak J. (2016). The effect of hypoxia and re-oxygenation on adipose tissue lipolysis in COPD patients. European Respiratory Journal 48: 1218–1220. DOI 10.1183/13993003.00602-2016. [Google Scholar] [CrossRef]
Qiu YY, Wu Y, Lin MJ, Bian T, Xiao YL, Qin C. (2019). LncRNA-MEG3 functions as a competing endogenous RNA to regulate Treg/Th17 balance in patients with asthma by targeting microRNA-17/ RORγt. Biomedicine & Pharmacotherapy 111: 386–394. DOI 10.1016/j.biopha.2018.12.080. [Google Scholar] [CrossRef]
Roth J, Wang Y, Eckhardt AE, Hill RL. (1994). Subcellular localization of the UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-mediated O-glycosylation reaction in the submaxillary gland. Proceedings of the National Academy of Sciences of the United States of America 91: 8935–8939. DOI 10.1073/pnas.91.19.8935. [Google Scholar] [CrossRef]
Skuljec J, Chmielewski M, Happle C, Habener A, Busse M, Abken H, Hansen G. (2017). Chimeric antigen receptor-redirected regulatory T cells suppress experimental allergic airway inflammation, a model of asthma. Frontiers in Immunology 8: 1125. DOI 10.3389/fimmu.2017.01125. [Google Scholar] [CrossRef]
Song J, Wang Q, Zong L. (2020). LncRNA MIR155HG contributes to smoke-related chronic obstructive pulmonary disease by targeting miR-128-5p/BRD4 axis. Bioscience Reports 40: BSR20192567. DOI 10.1042/BSR20192567. [Google Scholar] [CrossRef]
Sung JE, Lee HA, Kim JE, Yun WB, An BS, Yang SY, Kim DS, Lee CY, Lee HS, Bae CJ, Hwang DY. (2017). Saponin-enriched extract of Asparagus cochinchinensis alleviates airway inflammation and remodeling in ovalbumin-induced asthma model. International Journal of Molecular Medicine 40: 1365–1376. DOI 10.3892/ijmm.2017.3147. [Google Scholar] [CrossRef]
Symmes BA, Stefanski AL, Magin CM, Evans CM. (2018). Role of mucins in lung homeostasis: regulated expression and biosynthesis in health and disease. Biochemical Society Transactions 46: 707–719. DOI 10.1042/BST20170455. [Google Scholar] [CrossRef]
Wang M, Jiang YM, Xia LY, Wang Y, Li WY, Jin T. (2018). LncRNA NKILA upregulation mediates oxygen glucose deprivation/re-oxygenation-induced neuronal cell death by inhibiting NF-κB signaling. Biochemical and Biophysical Research Communications 503: 2524–2530. DOI 10.1016/j.bbrc.2018.07.010. [Google Scholar] [CrossRef]
Welsh KG, Rousseau K, Fisher G, Bonser LR, Bradding P, Brightling CE, Thornton DJ, Gaillard EA. (2017). MUC5AC and a glycosylated variant of MUC5B alter mucin composition in children with acute asthma. Chest 152: 771–779. DOI 10.1016/j.chest.2017.07.001. [Google Scholar] [CrossRef]
Wu W, Chen F, Cui X, Yang L, Chen J, Zhao J, Huang D, Liu J, Yang L, Zeng J, Zeng Z, Pan Y, Su F, Cai J, Ying Z, Zhao Q, Song E, Su S. (2018). LncRNA NKILA suppresses TGF-β-induced epithelial-mesenchymal transition by blocking NF-κB signaling in breast cancer. International Journal of Cancer 143: 2213–2224. DOI 10.1002/ijc.31605. [Google Scholar] [CrossRef]
Wu YM, Liu CH, Hu RH, Huang MJ, Lee JJ, Chen CH, Huang J, Lai HS, Lee PH, Hsu WM, Huang HC, Huang MC. (2011). Mucin glycosylating enzyme GALNT2 regulates the malignant character of hepatocellular carcinoma by modifying the EGF receptor. Cancer Research 71: 7270–7279. DOI 10.1158/0008-5472.CAN-11-1161. [Google Scholar] [CrossRef]
Yokota M, Tamachi T, Yokoyama Y, Maezawa Y, Takatori H, Suto A, Suzuki K, Hirose K, Takeda K, Nakajima H. (2016). IκBNS induces Muc5ac expression in epithelial cells and causes airway hyper-responsiveness in murine asthma models. Allergy 72: 1043–1053. DOI 10.1111/all.13079. [Google Scholar] [CrossRef]
Zheng M, Hong W, Gao M, Yi E, Zhang J, Hao B, Liang C, Li X, Li C, Ye X, Liao B, He F, Zhou Y, Li B, Ran P. (2019). LncRNA COPDA1 promotes airway smooth muscle cell proliferation in chronic obstructive pulmonary disease. American Journal of Respiratory Cell and Molecular Biology 61: 584–596. DOI 10.1165/rcmb.2018-0269OC. [Google Scholar] [CrossRef]
Zhou Q, Zhou L, Qian J, Yuan ZL, Chen ZJ. (2018). NKILA inhibition protects retinal pigment epithelium cells from hypoxia by facilitating NFκB activation. Biochemical and Biophysical Research Communications 503: 3134–3141. DOI 10.1016/j.bbrc.2018.08.105. [Google Scholar] [CrossRef]
Zhou-Suckow Z, Duerr J, Hagner M, Agrawal R, Mall MA. (2017). Airway mucus, inflammation and remodeling: emerging links in the pathogenesis of chronic lung diseases. Cell and Tissue Research 367: 537–550. DOI 10.1007/s00441-016-2562-z. [Google Scholar] [CrossRef]
Zhu X, Du J, Yu J, Guo R, Feng Y, Qiao L, Xu Z, Yang F, Zhong G, Liu F, Cheng F, Chu M, Lin J (2019a). LncRNA NKILA regulates endothelium inflammation by controlling a NF-κB/KLF4 positive feedback loop. Journal of Molecular and Cellular Cardiology 126: 60–69. DOI 10.1016/j.yjmcc.2018.11.001. [Google Scholar] [CrossRef]
Zhu Y, Mao D, Gao W, Han G, Hu H (2019b). Analysis of lncRNA expression in patients with eosinophilic and neutrophilic asthma focusing on LNC_000127. Frontiers in Genetics 10: 141. DOI 10.3389/fgene.2019.00141. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |