2021 45(1): 17-26
DOI:10.32604/biocell.2021.013973

www.techscience.com/journal/biocell
| BIOCELL 2021 45(1): 17-26 DOI:10.32604/biocell.2021.013973 |  www.techscience.com/journal/biocell |
PathVisio Analysis: An Application Targeting the miRNA Network Associated with the p53 Signaling Pathway in Osteosarcoma
1Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, Canada
2Department of Sciences for Health Promotion and Mother & Child Care, Section of Anatomic Pathology, University of Palermo, Palermo, Italy
3Department of Orthopedics, TianYou Hospital, Wuhan University of Science and Technology, Wuhan, China
4Division of Pediatric Hematology/Oncology, Stollery Children’s Hospital, Edmonton, Canada
5Department of Oncology, University of Alberta, Edmonton, Canada
6Department of Pediatrics, University of Alberta, Edmonton, Canada
*Address correspondence to: Consolato Sergi, sergi@ualberta.ca
Received: 27 August 2020; Accepted: 19 October 2020
Abstract: MicroRNAs (miRNAs) are small single-stranded, non-coding RNA molecules involved in the pathogenesis and progression of cancer, including osteosarcoma. We aimed to clarify the pathways involving miRNAs using new bioinformatics tools. We applied WikiPathways and PathVisio, two open-source platforms, to analyze miRNAs in osteosarcoma using miRTar and ONCO.IO as integration tools. We found 1298 records of osteosarcoma papers associated with the word “miRNA”. In osteosarcoma patients with good response to chemotherapy, miR-92a, miR-99b, miR-193a-5p, and miR-422a expression is increased, while miR-132 is decreased. All identified miRNAs seem to be centered on the TP53 network. This is the first application of PathVisio to determine miRNA pathways in osteosarcoma. MiRNAs have the potential to become a useful diagnostic and prognostic tool in the management of osteosarcoma. PathVisio is a full pathway editor with the potentiality to illustrate the biological events, augment graphical elements, and elucidate all the physical structures and interactions with standard external database identifiers.
Keywords: MiRNA; Osteosarcoma; p53; Carcinogenesis; Oncology; Cancer; Bone tumor; Bioinformatics
Osteosarcoma (OS) is the most common primary malignant bone tumor, comprising about 20% of primary bone sarcomas. It is a high-grade malignant tumor characterized by the cells forming immature bone or osteoid. The tumor is considered primary when the underlying bone is normal and secondary when it is altered by a pre-existing condition such as prior irradiation or Paget disease (Osasan et al., 2016; Sergi and Zwerschke, 2008). OS is slightly more prevalent in males (male:female = 3:2) and has a bimodal age distribution with a preference for the adolescent and geriatric age groups, with most of the primary OS cases (60–70%) affecting adolescents and young adults (from 15 to 25 years of age). In the elderly, OS is usually associated with Paget disease of the bone, post-radiation sarcoma, and dedifferentiated chondrosarcomas (Sergi and Zwerschke, 2008). Primary OS may arise in any bone, generally in the long bones of the appendicular skeleton (80–90%), most commonly in the distal femur, proximal tibia, and proximal humerus. Within the long bones, the tumor is usually located in the metaphysis and arises as an enlarging and palpable mass, which results in progressive pain. OS originating in the mid-shaft of bones is uncommon. Conversely, tumors arise e often in the epiphysis where the growth plate is located. Less than 1% of OS is found in the bones of the hands and feet. There is an increase in osteosarcoma’s relative incidence in non-long bones, including the jaws, pelvis, spine, and skull within the senior age group. The standard first-line treatment regimens for OS include surgery and multi-agent chemotherapy. Almost all patients receive a neoadjuvant intravenous combination of doxorubicin and cisplatin with or without methotrexate as the initial chemotherapy regimen. In cases where surgical resection is not feasible or the margins are inadequate, the use of radiation therapy may improve local control, but this is not considered a standard of care in pediatric and young adult patients. There has been a significant increase in the 5-year survival rates of patients with OS due to the advances in patients’ clinical management. Most centers’ survival rates now exceed 50%, but patients presenting with metastatic and recurrent disease have a survival rate of below 20%. The lung is the leading site of metastatic deposits (Abarrategi et al., 2016; Chen et al., 2016b).
Some genetic syndromes are associated with an increased risk of OS. They include hereditary retinoblastoma (germline mutation of the Rb gene), Li-Fraumeni syndrome (germline mutation of the TP53 gene), Bloom syndrome (germline mutation of the RECQL2 gene), Werner syndrome (germline mutation of the RECQL3 gene), and Rothmund-Thomson syndrome (germline mutation of the RECQL4 gene) (Osasan et al., 2016). The two most prominent genes that harbor germline mutations in patients with OS are the retinoblastoma (Rb 1) and the TP53 tumor suppressor genes. Most OS demonstrate inactivation of both the retinoblastoma (Rb) and p53 pathways. OS has a disorganized genome characterized by complex, unbalanced karyotypes with varying patterns of abnormalities. The most consistent finding beyond the TP53 and RB genes’ dysregulation is significant aneuploidy with some evidence of chromothripsis. Chromothripsis is the phenomenon by which up to hundreds to thousands of clustered chromosomal rearrangements occur in a single event in localized and confined genomic regions in one or a few chromosomes (Ly and Cleveland, 2017; Poot, 2017; Smida et al., 2017). These findings suggest an early defect in DNA repair/surveillance as a mechanism for the pathogenesis of OS (Behjati et al., 2017). Tumor suppressor genes function to control cell growth by inhibiting cell proliferation and tumor development. Also, they play a role in cell repair and apoptosis. When tumor suppressors mutate, resulting in a loss or reduction in function, there is an increase in the likelihood of developing cancer. The retinoblastoma (RB) was the first tumor suppressor gene described and encodes a protein that functions as a negative regulator of the cell cycle (Ren and Gu, 2017). This protein stabilizes constitutive heterochromatin to maintain overall chromatin structure. RB1 is the checkpoint that binds the E2F family of transcription factors and inhibits cell cycle progression. Defects in this gene are associated with retinoblastoma, urinary bladder cancers, and OS. The RB gene is critical for the regulation of the G1 to S cell cycle transition. In the absence of mitogenic stimuli, Rb remains dephosphorylated and binds to E2F family transcription factors, preventing their activation of the cell cycle. Mutations that result in the loss of function of the RB protein occur in approximately 70% of OS, mostly due to a loss of heterozygosity. Structural rearrangements and point mutations in the RB gene can also occur (Ren and Gu, 2017). The TP53 gene functions as a tumor suppressor in essentially all tumors. It encodes a tumor suppressor protein, which contains transcriptional activation, DNA binding, and oligomerization domains. This protein plays a crucial role in maintaining genomic stability functioning as a transcription factor that regulates the expression of various genes involved in cell cycle arrest, DNA repair, changes in metabolism, and apoptosis. Mutations in this gene are associated with a wide variety of cancers, including OS. The function of p53 can be affected by mutations in the gene itself or by mutations to up- or downstream mediators of its activity. Mutations that result in the loss of function of the p53 gene occur in approximately 75% of OS cases. The mutations in the TP53 gene include allelic loss (75–80%), rearrangements (10–20%), and point mutations (20–30%) (Braithwaite et al., 2017; Duffy et al., 2017; Gold, 2017; Guha and Malkin, 2017; Kastenhuber and Lowe, 2017; Merkel et al., 2017). In the RB pathway setting, E2F3 and CDK4, both of which counteract RB control of cell cycle progression, are estimated to possess gain of function mutations. E2F3 is found in 60% of tumors, while CDK4 is found in 10% of tumors (Sampson et al., 2015). Within the p53 pathway, MDM2 is an E3 ubiquitin ligase that acts as a negative regulator of p53. The MDM2 gene is amplified in 3–25% of OS. COPS3 promotes the proteasomal degradation of p53, and COPS3 amplification is seen in 20–80% of OS cases. In the c-Myc pathway, the c-Myc gene is a key transcription factor that functions as a general amplifier of gene expression (Iaccarino, 2017). It enhances the transcription of essentially all genes with active promoters within the cell. This gene is amplified in 7–67% of OS cases and over-expressed in at least 30% of tumors (Morrow and Khanna, 2015; Sampson et al., 2015).
MicroRNAs (miRNAs) are a small single-stranded, non-coding RNA molecule (from 18 to 25 nucleotides in length), which are usually found in eukaryotic cells. They are involved in various biological processes that regulate differentiation, apoptosis, and proliferation of numerous non-neoplastic and neoplastic diseases (Dong et al., 2016; Hashimoto and Tanaka, 2017; Leichter et al., 2017; Nugent, 2014; Ram Kumar et al., 2016; Sampson et al., 2015; Sergi et al., 2017a; Sergi et al., 2017b; Zhao et al., 2013). This process is achieved by complementarily pairing with the 3’ untranslated region (3’ UTR) or 5’ untranslated region (5’ UTR) of target genes, thus inhibiting the mRNA translation of these genes. In 1993, miRNA was first discovered in the nematode species C. elegans, and the first molecule was named lin-4. Since this discovery, it has been estimated that as many as 1000 miRNAs exist in the human genome, with more than 30% of the human genome regulated by miRNAs that simultaneously target multiple genes. In the last decade, it became clear that miRNAs are implicated in the pathogenesis of cancer, including OS (Ram Kumar et al., 2016). This aspect was demonstrated by the differences in the miRNA expression profiles detected between normal and cancer cells. The expression of many different types of miRNA was found to be altered (either over-expressed or reduced) in malignancy. MiRNAs can function as tumor suppressors, oncogenes, or both. The dysregulation of miRNA expression may contribute to cancer development through the loss of controls of biological processes. These natural and physical properties can make miRNAs useful diagnostic and prognostic tools in the management of various cancers, including OS and non-oncological diseases (Agarwal et al., 2015; Chen et al., 2016a; Chen et al., 2013; Dong et al., 2016; Hsu et al., 2011; Jones et al., 2012; Kobayashi et al., 2012; Leichter et al., 2017; Lin et al., 2016; Nugent, 2014; Ram Kumar et al., 2016; Sampson et al., 2015; Zhao et al., 2013; Zhou et al., 2016). There is increasing evidence that multiple miRNAs may play a role in determining the response to chemotherapy in the treatment of OS (Ram Kumar et al., 2016; Sampson et al., 2015).
In the last two decades, numerous bioinformatics tools have been developed to manage the increasing abundance of data. The massive flow of miRNA data can be handled effectively and efficiently using specific bioinformatics tools. In targeting miRNAs, we can address the identification, expression, and analysis of explicit and multiple miRNAs, establish miRNA regulatory networks, miRNA metabolic and signaling pathways, and miRNA-transcription factor interplay, thereby linking miRNAs to particular diseases or status of the disease. WikiPathways is an open, collaborative platform for drawing, editing, and sharing biological pathways, built using the same software underlying Wikipedia. This platform can be used to integrate, visualize, and analyze system-wide transcriptomics, proteomics, and metabolomics data. Several studies have demonstrated miRNAs’ involvement in the pathogenesis, diagnostic potential, and therapeutics of OS. As indicated above, these miRNAs have been re-emphasized most recently because they intrinsically regulate the expression of different genes that play essential roles in tumorigenesis, cell invasion, migration, and metastasis. In this review, we aimed to discuss the current knowledge of miRNAs’ role and their target genes in OS and attempt to develop an OS pathway involving miRNA integrating WikiPathways and other bioinformatic tools.
PubMed, Scopus, and Google Scholar were used to systematically search for reviewed publications that investigated the functions of miRNA in the pathogenesis, treatment, and prognosis of osteosarcoma. Publications in the time frame “2008-2018” and targeting “miRNA” and “Osteosarcoma” were retrieved from the archives. The findings from these publications were used to compile a list of miRNAs that are associated with OS. This study relies on a systematic search, but it does not comply with PRISMA eligibility criteria (Preferred Reporting Items for Systematic Reviews and Meta-Analyses).
PathVisio, a no-cost open-source pathway editor, visualization, and analysis software, has significantly enhanced the capacity to explore large scale data. It provides an invaluable tool for investigating genes, proteins, and metabolites in both the healthy and diseased states of complex tissues and related diseases, including OS. We used PathVisio as a pathway editor, visualization, and analysis software. Since the first publication of PathVisio in 2008, the software has been cited more than 170 times and used in many different biological studies. As an online editor, PathVisio is also integrated into the community curated pathway database WikiPathways. Wikipathways is one of the most popular freely available databases for assessing biological pathways. It is an open, collaborative platform used to create and share paths and is known as a plugin for PathVisio. PathVisio 3 is a free open-source pathway editor, visualization, and analysis toolbox implemented in Java, a class-based, object-oriented programming language able to run on all major operating systems (Bhat et al., 2018; Kutmon et al., 2015). The miRTar bioanalysis tool was used to determine miRNAs’ interaction with genes in the TP53 pathway (Hsu et al., 2011). In particular, the miRTar tool adopts seven scenarios to identify putative miRNA target sites of the gene transcripts. It illustrates the biological functions of miRNAs concerning their targets in metabolic pathways. The prediction system helps biologists to quickly identify the regulatory relationships between crucial miRNAs and their targets.
The results were used in assembling the pathway for OS. Common miRNAs that have been previously identified in studies to have a role in the development and progression of OS were selected from the literature and imputed into this tool to identify the targeted genes. A pathway network was constructed using the ONCO.IO micro-analysis tool. A pathway for miRNAs linked to OS was then built using PathVisio and the Wikipathways plugin. The URLs of the website platforms we used are https://onco.io/, http://mirtar.mbc.nctu.edu.tw/human/, https://www.pathvisio.org/, and https://www.wikipathways.org/index.php/WikiPathways.
There is a significant number of miRNAs that we found to be associated with OS. We found 1298 records of osteosarcoma papers associated with the word “miRNA”. Three studies were substantially selected from which miRNAs associated with osteosarcoma were used for further detailed analysis (Chen et al., 2016a; Kobayashi et al., 2012; Nugent, 2014). In these studies,
Table 1: Experimental groups highlighting the MiRNAs associated with osteosarcoma (Kutmon et al., 2015)
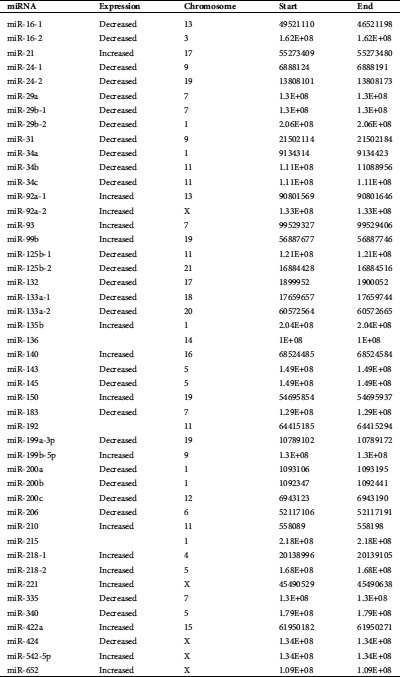
Table 2: MiRNAs with increased expression in osteosarcoma (Hsu et al., 2011; Nugent, 2014)
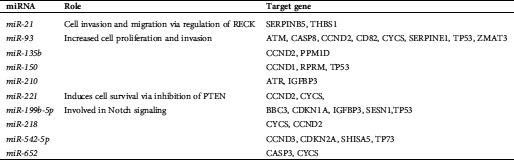
Table 3: MiRNAs with decreased expression in osteosarcoma

Table 4: MiRNAs with intrinsic regulation in osteosarcoma
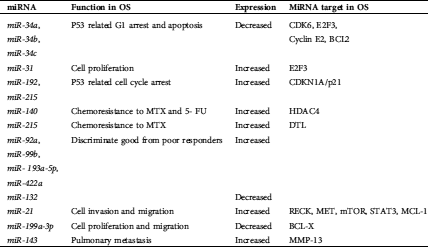
A total of 6 miRNAs were found on chromosome 1, making chromosome 1 the most popular miRNA location. Chromosomes X and 11 were the second most frequent miRNA locations, with each chromosome being responsible for five miRNAs. The third most common chromosomal location for miRNAs is chromosome 19, responsible for four miRNAs. In addition, miRNAs are also located on chromosomes 3, 4, 5, 6, 7, 9, 13, 14, 15, 16, 17, 18, 20, and 21. All types of cellular pathways from development to oncogenesis are affected by miRNAs. Tab. 1 highlights the miRNAs associated with OS. Tabs. 2 and 3 recapitulate the roles and target genes of miRNAs in OS, with Tab. 2 displaying those with increased expressions and Tab. 3 displaying those with decreased expressions. A careful perusal of the literature showed that OS has increased expression of miR-21, miR-93, miR-135b, miR-150, miR-210, miR-221, miR-199b-5p, miR-218, miR-542-5p, and miR-652. While target genes are known for each of these miRNAs, the role in which they play is only known for miR-21, miR-93, miR-221, and miR-199b-5p. Conversely, there was decreased expression of miR-16, miR-24, miR-29a, miR-29b, miR-31, miR-34a, miR-34b, miR-34c, miR-125b, miR-132, miR-133a, miR-143, miR145, miR-183, miR-199a-3p, miR-200, miR-206, miR-335, miR-340 and miR-424. Roles are defined for all the miRNAs except miR-29b, miR-34b, and miR143. Target genes have been identified for miR-16, miR-29a, miR-29b, miR-31, miR-34a, miR-34b, miR-133a and miR-206. The interconnection of these miRNAs with signaling pathways was the next step in our analysis and the miRNAs with OS intrinsic regulation are key and displayed in Tab. 4.
Fig. 1 shows the TP53 Network built by ONCO.IO, a bioinformatic tool on PathVisio software, and the miRNA-regulation of TP53 in OS, respectively. The weight of miR34 for transcriptional regulation is prominent.
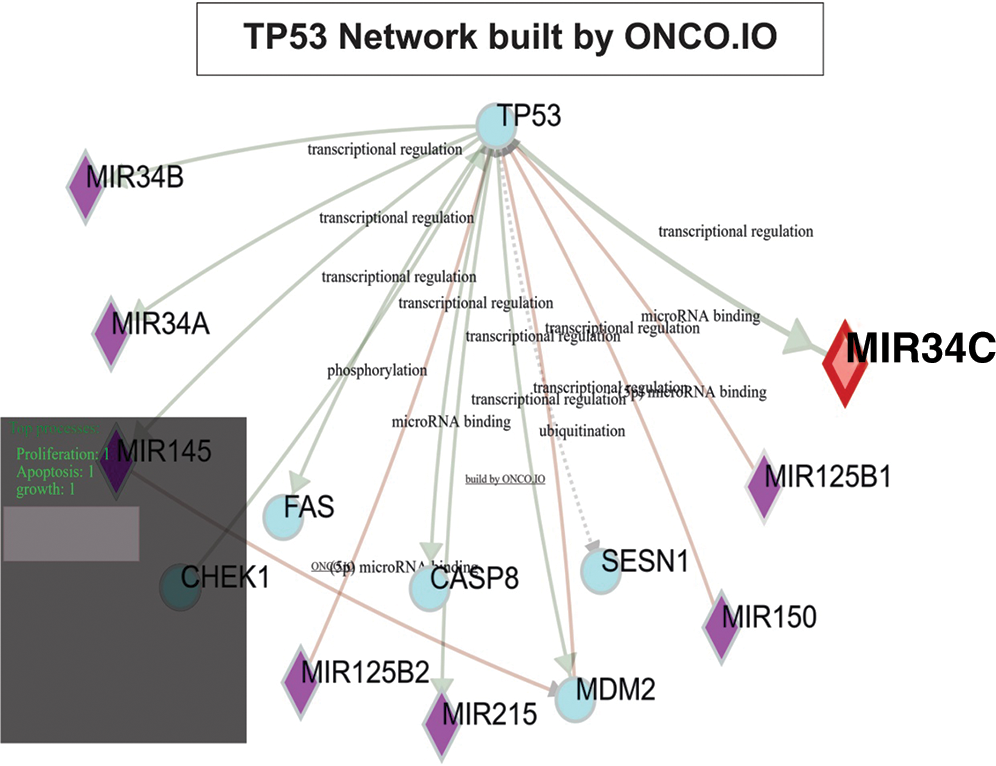
Figure 1: TP53 results are intimately linked to MiR34A, MiR34B, MiR125B1, MiR125B2, MiR145, MiR150, and MiR215.
The purpose of this study was to attempt to construct a pathway involving the miRNA regulation of the p53 signaling pathway for OS. No unique, perpetual, and solid pathway involving miRNAs for OS was found, but there are multiple pathways related to the TP53 gene which are associated with different conditions. Data regarding miRNA and target genes involved in the development and progression of OS corresponded to information that was published in previous studies. This data was imputed into onco.io to generate a signaling pathway for p53 that shows miRNA regulation. Fig. 1 is exclusively an example of multiple genetic interactions that can be revealed using PathVisio. It does not mean that it is comprehensive of all genes-miRNAs interaction as networks. The gray shadow of the left corner of Fig. 1 is supposed to polarize the attention on some molecules of interest, but it can be displayed in other locations according to different research questions.
The mechanism of action of miRNAs in OS remains not clearly understood. However, the TP53 gene is mutated in more than 20% of OS, with mutations demonstrated to be involved in tumorigenesis. MiRNAs are involved in the control of many cellular processes, and the dysregulation of miRNA expression can influence carcinogenesis once tumor suppressor genes or oncogenes encode the relevant target mRNAs. Even a small variation can have significant implications for the cell since each miRNA can have many targets. In humans, it has been established that many miRNA genes are located in cancer-associated regions or at the fragile sites of chromosomes, which are prone to deletion, amplification, and mutations in cancer cells. Since miRNAs can function as negative regulators of gene expression, an over-expression of oncogenic miRNAs can contribute to tumor development by promoting cellular proliferation and evasion of apoptosis. A similar effect will occur if there is a reduction in the expression of tumor-suppressive miRNAs. Research has demonstrated both increases and decreases in the expression of specific miRNAs in cancer. These appear to vary depending on the particular tissue and the cancer type (He et al., 2007; Kao et al., 2012; Kobayashi et al., 2012). Several miRNAs have been identified as direct targets of p53.
The miR-34 family (miR-34a, miR-34b, and miR-34c) has been an important component of the p53 tumor suppressor pathway. P53 induces the expression of these miRNAs in response to DNA damage and oncogenic stress in many cancers. He et al. (2007) reported that the miR-34 family induces G1 arrest and apoptosis via their targets CDK6, E2F3, Cyclin E2, and BCL2 in a p53-dependent manner in OS cells (Bhat et al., 2018). The expression of miR-34 is decreased in OS, and miR-34 enhances p53 mediated cell cycle arrest and apoptosis. Also, p53 induces the upregulation of miR-192, miR-194, and miR215 in U2OS cells, which carry the wild-type p53. The loss of miR-31 is associated with defects in the p53 pathway, while overexpression of miR-31 significantly inhibits OS cells’ proliferation. Moreover, miR-31 seems to have the potential to prevent disease progression or the development of pulmonary metastasis in OS (Kao et al., 2012; Kobayashi et al., 2012).
Biological pathways are descriptive. Sometimes complex diagrams are used to summarize and describe physical processes. These pathways show the potential interaction among genes, proteins, and metabolites. Path diagrams are a common way to graph the wealth of information available on these biological processes. To the best of our knowledge, no established pathways involving miRNAs for OS has been confirmed so far. However, there are multiple pathways related to TP53, which are associated with different disease conditions. The purpose of this study was to construct a path involving the miRNA regulation of the p53 signaling pathway for OS using PathVisio. Data regarding miRNA and target genes involved in the development and progression of OS corresponds to information that is available in the biomedical research literature. There is significant involvement of miRNAs in the development, progression, and metastasis of OS. The involvement spans from gene expression to epigenetics.
MiRNAs and their identified target genes are associated with multiple biological pathways and functions related to bone biology and cancer development and progression. Dysregulation of miRNAs is thereby associated with tumorigenesis in OS. A study by Andersen et al. (2018) investigated the miRNA expression in 101 OS samples (Andersen et al., 2018). A total of 752 miRNAs were profiled, with 33 of these being identified as deregulated in OS. Andersen et al. (2018) found a significant role of miRNAs in the tumorigenesis of OS and that 29 deregulated miRNAs were strongly correlated with cancer development and progression. MiR-221 and miR-222 are associated significantly with time to metastasis. Significant downregulated miRNAs were identified as miR-100-5p, miR-125b-5p, miR-127-3p, miR-370-3p, miR-335-5p and miR-411-5p. Scott et al. (2007) and Sempere et al. (2004) showed that miR-125b is an important regulator of both proliferation and differentiation of different cell types. At the same time, Mizuno et al. (2008) indicated that miR-125b inhibits normal OB proliferation in mouse cells and plays a role in bone development and OS tumorigenesis. Andersen et al. (2018) also identified miR-181a-5p, miR-181c-5p, miR-223-3p and miR-342-3p as being significantly upregulated in OS.
Our study was done to summarise and further increase our understanding of the roles played by various miRNAs at various stages of the signaling pathway regulated by TP53 in OS. Improved knowledge would allow for the development of specific miRNAs as biomarkers for diagnosis, disease monitoring, and OS progression. The possibility exists that miRNAs may have a therapeutic role in managing OS in the nearest future, particularly with the adoption of protocols of personalized medicine, renewed gene technologies, and digital pathology (Burnett et al., 2020; Jin et al., 2020; Sergi, 2019). MiRNA-directed gene regulation will pave the way for improving traditional gene therapy approaches to cancer, including OS. Presently, validation of miRNA pathways and targets in metastatic osteosarcoma has not been determined. Still, miRNA plays a role in the progression of OS by regulating proliferation, invasion, adhesion, metastasis, apoptosis, and angiogenesis. Identifying dysregulated miRNAs in patients with OS may contribute to the development of biomarkers for diagnosis and prognosis. There are challenges faced in identifying all the targets of miRNAs and establishing their contribution towards malignancy. Circulating miRNAs are considered predictive biomarkers for various types of cancers. They can be used as non-invasive disease biomarkers in cancer since they exist in human serum and plasma in remarkably stable forms. Comprehensive screening of miRNA profiles would allow for earlier detection of OS, as well as nullify the need for the collection of tissue samples through invasive procedures such as biopsies. Despite the clinical potential for the use of miRNAs as diagnostic biomarkers, several limitations are present. In most studies, the cohort of patients used has been relatively small, and therefore evaluations of large, long-term sample sizes with long-term follow-up are required. There is a lack of standardized approaches in the methodology of the normalization of circulating miRNAs. A refined approach is needed in future studies to establish miRNAs as circulating biomarkers for clinical use. The role of miRNAs in OS has been studied in detail, but it is not clear whether it can be utilized to treat patients with OS. The involvement of miRNA function in the progression of OS has raised the possibility of the utilization of miRNA as a novel therapy. Extensive toxicity studies and preclinical safety trials would need to be conducted before considering a miRNA-based therapeutic approach. A greater understanding of the roles that different miRNAs play in the development and progression of OS could ultimately improve this tumor (Abarrategi et al., 2016; Bhat et al., 2018; He et al., 2007; Jones et al., 2012; Kao et al., 2012; Kobayashi et al., 2012; Kutmon et al., 2015; Leichter et al., 2017; Nugent, 2014; Ram Kumar et al., 2016).
Moreover, the EIMMO, MicroInspector, miRU, MMIA, RNA22, StarMir, and MMIA are additional tools with variable data from biology scientists. They are web-based and specific for identifying miRNA binding sites (Hsu et al., 2011).
There are a few additional limitations to our study. First, the most common weakness of bioinformatics tools is the generation of large amounts of false-positive data. We considered the other open-source tools, such as DIANA, TARGETSCAN, and MIRANDA, but we chose to use miRTar because of the familiarity with this tool. Although based on available scientific data, many of the proposed gene interactions in these databases may be speculative. Second, the current method of pathway analysis depends on existing databases. Not all the miRNAs and genes linked to OS were found in the ONCO.IO miRNA analysis tool database, which was used to construct the pathway network. Third, the interpretation of results based on pathway analysis tools needs to be interpreted with caution because the miRNA field is an evolving platform spanning from genomics to proteomics.
In conclusion, although the field of miRNA research is still relatively new, its rapid expansion has the potential to use these small molecules in the management of cancer. The PathVisio analysis of Wikipathways may be a useful bioinformatic tool for cancer research. Several miRNAs have been involved in OS, with some demonstrated to be overexpressed while others are downregulated. Our analysis indicates that there is indeed potential for miRNAs to play a critical role in the management of OS both as promising diagnostic biomarkers and either predictive or prognostic indicators. Bioinformatics speed has increased daily, and we expect that the PathVisio analysis on Wikipathways may be a useful tool for cancer research readily available for cancer research investigators worldwide. The miRTar bioanalysis tool can be used to determine the interaction of miRNAs with genes in the TP53 pathway, and the ONCO.IO miRNA analysis tool database was used to identify miRNAs and OS. In OS patients considered good responders to chemotherapy, miR-92a, miR-99b, miR-193a-5p, and miR-422a expression increased, while miR-132 decreased. This is the first application of PathVisio to determine miRNA pathways in osteosarcoma to the best of our knowledge. PathVisio is a full pathway editor with the potentiality to illustrate the biological events, augment graphical elements, and elucidate all the biological structures and interactions with standard external database identifiers. MiRNAs have the potential to become a useful diagnostic and prognostic tool in the management of OS.
Acknowledgement: The authors would like to recognize the physicians, nurses, and other allied healthcare workers who are responsible for the care of our patients at the Pediatric Oncological Departments of the Stollery Children’s Hospital, University of Alberta Hospital (Canada), University of Palermo (Italy), and TianYou Hospital, Wuhan University of Science and Technology (China).
Authors’ Contributions: MB contributed to the research and summarised relevant data, developed the osteosarcoma pathway using PathVisio. First author of the manuscript with support from the other contributors; VR, FS, RL, MZ, DE reviewed the manuscript and made necessary corrections and suggestions. FS is involved in R application and data science in our research group. CS contributed to the project's design and implementation, collecting funds, and revising the manuscript thoroughly. All authors approved the final version of the manuscript.
Availability of Data and Materials: Project Name: PathVisio Analysis: An Application Targeting the miRNA Network in Osteosarcoma and Review on its Tumorigenesis; Project Home Page: https://www.pathvisio.org/ Operating System: Platform Independent; Programming Language: Java; Other Requirements: Java 7 (update 51); License: Apache License, Version 2.0. The data that support the findings of this study are available from the open-sources platforms described in this paper. We would like to foster and extend our availability to any research cooperation useful to strengthen bone cancer research and improve therapy protocols against osteosarcoma.
Financial Support and Sponsorship: This research has been funded by the generosity of the Stollery Children’s Hospital Foundation and supporters of the Lois Hole Hospital for Women through the Women and Children’s Health Research Institute (WCHRI Grant Application ID #: 2096), Austrian Tyrolean Cancer Research Institute (Tiroler Krebsforschungsinstitut, Innsbruck, Austria), Austrian Research Fund (Fonds zur Förderung der wissenschaftlichen Forschung, FWF), and the Saudi Cultural Bureau, Ottawa, Canada. The role of the funding body in the experiment design, collection, analysis and interpretation of data, and writing of the manuscript should be declared:
Ethical Approval and Consent to Participate: This research does not involve human data but belongs to an Osteosarcoma Research Project approved by the University of Alberta, Alberta, Canada (Pro72708).
Consent for Publication: Not applicable.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abarrategi A, Tornin J, Martinez-Cruzado L, Hamilton A, Martinez-Campos E, Rodrigo JP, González M, Baldini N, Garcia-Castro J, Rodriguez R. (2016). Osteosarcoma: Cells-of-origin, cancer stem cells, and targeted therapies. Stem Cells International 2016: 3631764. DOI 10.1155/2016/3631764. [Google Scholar] [CrossRef]
Agarwal V, Bell G, Nam J, Bartel D. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4: e05005. DOI 10.7554/eLife.05005. [Google Scholar] [CrossRef]
Andersen GB, Knudsen A, Hager H, Hansen LL, Tost J. (2018). mi RNA profiling identifies deregulated mi RNA s associated with osteosarcoma development and time to metastasis in two large cohorts. Molecular Oncology 12: 114–131. DOI 10.1002/1878-0261.12154. [Google Scholar] [CrossRef]
Behjati S, Tarpey PS, Haase K, Ye H, Young MD, Alexandrov LB, Farndon SJ, Collord G, Wedge DC, Martincorena I. (2017). Recurrent mutation of IGF signalling genes and distinct patterns of genomic rearrangement in osteosarcoma. Nature Communications 8: 15936. DOI 10.1038/ncomms15936. [Google Scholar] [CrossRef]
Bhat MY, Solanki HS, Advani J, Khan AA, Prasad TK, Gowda H, Thiyagarajan S, Chatterjee A. (2018). Comprehensive network map of interferon gamma signaling. Journal of Cell Communication and Signaling 12: 745–751. DOI 10.1007/s12079-018-0486-y. [Google Scholar] [CrossRef]
Braithwaite A, Ballinger M, Baran-Marszak F, Bond GL, Concin N, Donehower L, El-Deiry W. (2017). Recommended guidelines for validation, quality control, and reporting of TP53 variants in clinical practice. Cancer Research 77: 1250–1260. [Google Scholar]
Burnett M, Abuetabh Y, Wronski A, Shen F, Persad S, Leng R, Eisenstat D, Sergi C. (2020). Graphene oxide nanoparticles induce apoptosis in wild-type and CRISPR/Cas9-IGF/IGFBP3 knocked-out osteosarcoma cells. Journal of Cancer 11: 5007–5023. DOI 10.7150/jca.46464. [Google Scholar] [CrossRef]
Chen G, Fang T, Huang Z, Qi Y, Du S, Di T, Lei Z, Zhang X, Yan W (2016a). MicroRNA-133a inhibits osteosarcoma cells proliferation and invasion via targeting IGF-1R. Cellular Physiology and Biochemistry 38: 598–608. DOI 10.1159/000438653. [Google Scholar] [CrossRef]
Chen L, Wang Q, Wang GD, Wang H-S, Huang Y, Liu XM, Cai XH. (2013). miR-16 inhibits cell proliferation by targeting IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS Letters 587: 1366–1372. DOI 10.1016/j.febslet.2013.03.007. [Google Scholar] [CrossRef]
Chen Y, Yu XC, Xu SF, Xu M, Song RX (2016b). Impacts of tumor location, nature and bone destruction of extremity osteosarcoma on selection of limb salvage operative procedure. Orthopaedic Surgery 8: 139–149. DOI 10.1111/os.12237. [Google Scholar] [CrossRef]
Dong J, Liu Y, Liao W, Liu R, Shi P, Wang L. (2016). miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. Journal of Bone Oncology 5: 74–79. DOI 10.1016/j.jbo.2016.05.001. [Google Scholar] [CrossRef]
Duffy MJ, Synnott NC, Crown J. (2017). Mutant p53 as a target for cancer treatment. European Journal of Cancer 83: 258–265. DOI 10.1016/j.ejca.2017.06.023. [Google Scholar] [CrossRef]
Gold B. (2017). Somatic mutations in cancer: Stochastic versus predictable. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 814: 37–46. DOI 10.1016/j.mrgentox.2016.12.006. [Google Scholar] [CrossRef]
Guha T, Malkin D. (2017). Inherited TP53 mutations and the Li-Fraumeni syndrome. Cold Spring Harbor Perspectives in Medicine 7: a026187. DOI 10.1101/cshperspect.a026187. [Google Scholar] [CrossRef]
Hashimoto N, Tanaka T. (2017). Role of miRNAs in the pathogenesis and susceptibility of diabetes mellitus. Journal of Human Genetics 62: 141–150. DOI 10.1038/jhg.2016.150. [Google Scholar] [CrossRef]
He L, He X, Lowe SW, Hannon GJ. (2007). microRNAs join the p53 network — another piece in the tumour-suppression puzzle. Nature Reviews Cancer 7: 819–822. DOI 10.1038/nrc2232. [Google Scholar] [CrossRef]
Hsu JBK, Chiu CM, Hsu SD, Huang WY, Chien CH, Lee TY, Huang HD. (2011). miRTar: An integrated system for identifying miRNA-target interactions in human. BMC Bioinformatics 12: 300. DOI 10.1186/1471-2105-12-300. [Google Scholar] [CrossRef]
Iaccarino I. (2017). lncRNAs and MYC: An intricate relationship. International Journal of Molecular Sciences 18: 1497. DOI 10.3390/ijms18071497. [Google Scholar] [CrossRef]
Jin L, Shen F, Weinfeld M, Sergi C. (2020). Insulin Growth Factor Binding Protein 7 (IGFBP7)-related cancer and IGFBP3 and IGFBP7 crosstalk. Frontiers in Oncology 10: 727. DOI 10.3389/fonc.2020.00727. [Google Scholar] [CrossRef]
Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, Lovat F, Leblanc K, Palatini J, Randall RL. (2012). miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Research 72: 1865–1877. DOI 10.1158/0008-5472.CAN-11-2663. [Google Scholar] [CrossRef]
Kao S, Shiau CK, Gu DL, Ho CM, Su WH, Chen CF, Lin CH, Jou YS. (2012). IGDB. NSCLC: Integrated genomic database of non-small cell lung cancer. Nucleic Acids Research 40: D972–D977. DOI 10.1093/nar/gkr1183. [Google Scholar] [CrossRef]
Kastenhuber ER, Lowe SW. (2017). Putting p53 in context. Cell 170: 1062–1078. DOI 10.1016/j.cell.2017.08.028. [Google Scholar] [CrossRef]
Kobayashi E, Hornicek FJ, Duan Z. (2012). MicroRNA involvement in osteosarcoma. Sarcoma 2012: 359739. DOI 10.1155/2012/359739. [Google Scholar] [CrossRef]
Kutmon M, Van Iersel MP, Bohler A, Kelder T, Nunes N, Pico AR, Evelo CT. (2015). PathVisio 3: An extendable pathway analysis toolbox. PLoS Computational Biology 11: e1004085. DOI 10.1371/journal.pcbi.1004085. [Google Scholar] [CrossRef]
Leichter AL, Sullivan MJ, Eccles MR, Chatterjee A. (2017). MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Molecular Cancer 16: 15. DOI 10.1186/s12943-017-0584-0. [Google Scholar] [CrossRef]
Lin Z, Song D, Wei H, Yang X, Liu T, Yan W, Xiao J. (2016). TGF-β1-induced miR-202 mediates drug resistance by inhibiting apoptosis in human osteosarcoma. Journal of Cancer Research and Clinical Oncology 142: 239–246. DOI 10.1007/s00432-015-2028-9. [Google Scholar] [CrossRef]
Ly P, Cleveland DW. (2017). Rebuilding chromosomes after catastrophe: Emerging mechanisms of chromothripsis. Trends in Cell Biology 27: 917–930. DOI 10.1016/j.tcb.2017.08.005. [Google Scholar] [CrossRef]
Merkel O, Taylor N, Prutsch N, Staber PB, Moriggl R, Turner SD, Kenner L. (2017). When the guardian sleeps: Reactivation of the p53 pathway in cancer. Mutation Research/Reviews in Mutation Research 773: 1–13. DOI 10.1016/j.mrrev.2017.02.003. [Google Scholar] [CrossRef]
Mizuno Y, Yagi K, Tokuzawa Y, Kanesaki-Yatsuka Y, Suda T, Katagiri T, Fukuda T, Maruyama M, Okuda A, Amemiya T. (2008). miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochemical and Biophysical Research Communications 368: 267–272. DOI 10.1016/j.bbrc.2008.01.073. [Google Scholar] [CrossRef]
Morrow JJ, Khanna C. (2015). Osteosarcoma genetics and epigenetics: Emerging biology and candidate therapies. Critical Reviews in Oncogenesis 20: 173–197. DOI 10.1615/CritRevOncog.2015013713. [Google Scholar] [CrossRef]
Nugent M. (2014). MicroRNA function and dysregulation in bone tumors: The evidence to date. Cancer Management and Research 6: 15. DOI 10.2147/CMAR.S53928. [Google Scholar] [CrossRef]
Osasan S, Zhang M, Shen F, Paul PJ, Persad S, Sergi C. (2016). Osteogenic sarcoma: A 21st century review. Anticancer Research 36: 4391–4398. DOI 10.21873/anticanres.10982. [Google Scholar] [CrossRef]
Poot M. (2017). Of simple and complex genome rearrangements, chromothripsis, chromoanasynthesis, and chromosome chaos. Molecular Syndromology 8: 115–117. DOI 10.1159/000454964. [Google Scholar] [CrossRef]
Ram Kumar RM, Boro A, Fuchs B. (2016). Involvement and clinical aspects of microRNA in osteosarcoma. International Journal of Molecular Sciences 17: 877. DOI 10.3390/ijms17060877. [Google Scholar] [CrossRef]
Ren W, Gu G. (2017). Prognostic implications of RB1 tumour suppressor gene alterations in the clinical outcome of human osteosarcoma: A meta-analysis. European Journal of Cancer Care 26: e12401. DOI 10.1111/ecc.12401. [Google Scholar] [CrossRef]
Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. (2015). MicroRNAs and potential targets in osteosarcoma: Review. Frontiers in Pediatrics 3: 69. DOI 10.3389/fped.2015.00069. [Google Scholar] [CrossRef]
Scott G, Goga A, Bhaumik D, Berger C, Sullivan C, Benz C. (2007). Coordinate suppression of erBB2 and erBB3 by enforced expression of micro-rna mir-125a or mir-125b. Journal of Biological Chemistry 282: 1479–1486. DOI 10.1074/jbc.M609383200. [Google Scholar] [CrossRef]
Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. (2004). Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biology 5: R13. DOI 10.1186/gb-2004-5-3-r13. [Google Scholar] [CrossRef]
Sergi C, Shen F, Lim DW, Liu W, Zhang M, Chiu B, Anand V, Sun Z (2017a). Cardiovascular dysfunction in sepsis at the dawn of emerging mediators. Biomedicine & Pharmacotherapy 95: 153–160. [Google Scholar]
Sergi C, Zwerschke W. (2008). Osteogenic sarcoma (osteosarcoma) in the elderly: Tumor delineation and predisposing conditions. Experimental Gerontology 43: 1039–1043. DOI 10.1016/j.exger.2008.09.009. [Google Scholar] [CrossRef]
Sergi CM. (2019). Digital pathology: The time is now to bridge the gap between medicine and technological singularity. In: Cvetković D, eds. Interactive Multimedia-Multimedia Production and Digital Storytelling, IntechOpen. DOI 10.5772/intechopen.84329. [Google Scholar] [CrossRef]
Sergi CM, Caluseriu O, Mccoll H, Eisenstat DD (2017b). Hirschsprung’s disease: Clinical dysmorphology, genes, micro-RNAs, and future perspectives. Pediatric Research 81: 177–191. DOI 10.1038/pr.2016.202. [Google Scholar] [CrossRef]
Smida J, Xu H, Zhang Y, Baumhoer D, Ribi S, Kovac M, Von Luettichau I, Bielack S, O’leary VB, Leib-Mösch C. (2017). Genome-wide analysis of somatic copy number alterations and chromosomal breakages in osteosarcoma. International Journal of Cancer 141: 816–828. DOI 10.1002/ijc.30778. [Google Scholar] [CrossRef]
Zhao H, Li M, Li L, Yang X, Lan G, Zhang Y. (2013). MiR-133b is down-regulated in human osteosarcoma and inhibits osteosarcoma cells proliferation, migration and invasion, and promotes apoptosis. PLoS One 8: e83571. DOI 10.1371/journal.pone.0083571. [Google Scholar] [CrossRef]
Zhou C, Tan W, Lv H, Gao F, Sun J. (2016). Hypoxia-inducible microRNA-488 regulates apoptosis by targeting Bim in osteosarcoma. Cellular Oncology 39: 463–471. DOI 10.1007/s13402-016-0288-2. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |