DOI:10.32604/jnm.2022.031113

| Journal of New Media DOI:10.32604/jnm.2022.031113 |  |
| Article |
Semi-Supervised Medical Image Segmentation Based on Generative Adversarial Network
1Hunan Applied Technology University, Changde, 415000, China
2Central South University of Forestry and Technology, Changsha, 410004, China
3The Second Xiangya Hospital of Central South University, Changsha, 410011, China
*Corresponding Author: Jiaohua Qin. Email: qinjiaohua@163.com
Received: 10 April 2022; Accepted: 11 May 2022
Abstract: At present, segmentation for medical image is mainly based on fully supervised model training, which consumes a lot of time and labor for dataset labeling. To address this issue, we propose a semi-supervised medical image segmentation model based on a generative adversarial network framework for automated segmentation of arteries. The network is mainly composed of two parts: a segmentation network for medical image segmentation and a discriminant network for evaluating segmentation results. In the initial stage of network training, a fully supervised training method is adopted to make the segmentation network and the discrimination network have certain segmentation and discrimination capabilities. Then a semi-supervised method is adopted to train the model, in which the discriminant network will generate pseudo-labels on the results of the segmentation for semi-supervised training of the segmentation network. The proposed method can use a small part of annotated dataset to realize the segmentation of medical images and effectively solve the problem of insufficient medical image annotation data.
Keywords: Medical image; semi-supervised; U-net; generative adversarial network; image segmentation
In recent years, with the continuous optimization of the performance of convolutional neural networks, deep learning models have outperformed humans in some image processing tasks.
Computer Aided Diagnosis (CAD) uses computer technology to help medical staff deal with complex tasks. With the application of artificial intelligence technology in CAD, CAD can more effectively collect information from medical images of patients, provide doctors with diagnostic suggestions, and improve the efficiency of doctors’ diagnosis. Medical image segmentation and detection [1] is a basic task in CAD, which can effectively extract the focal organ region. At present, medical image segmentation methods based on deep learning can be composed of three categories: methods that take candidate regions, methods based on fully convolutional networks (FCNs), and weakly supervised segmentation methods [2]. Among the methods based on fully convolutional networks, the U-Net network proposed by Ronneberger et al. [3] in 2015 is often used for medical image semantic segmentation. It innovatively proposes U-shaped structure and Skip-Connect. During the upsampling process, high-level semantic features of different scales and low-level features are fused, so that the output of the network has more refined edge information. Hou et al. [4] improved medical image segmentation methods using attention mechanism and feature fusion. Zhou et al. [5] applied sequential feature pyramid and attention mechanism to multi-label segmentation and achieved state-of-the-art performance on their dataset.
Although fully supervised convolutional neural networks show remarkable results in segmentation tasks, they require many labor costs for data cleaning and labeling. Moreover, the complexity of medical image semantics makes its data annotation more time-consuming and labor-intensive than natural images. Semi-supervised algorithms can rely on a small amount of labeled data, and introduce easily obtained unlabeled data to complete the training of the model, which reduces the cost of data set production, and gradually shows its performance in image segmentation.
At present, the medical image segmentation based on weak supervision mainly adopts the structure of Mean Teacher [6] to design. Sun et al. [7] completed weakly supervised training on liver lesion segmentation using a teacher-student architecture. Goodfellow et al. [8] proposed generative Adversarial Network (GAN) in 2014, and this structure uses adversarial learning to train the model. Son et al. [9] used GAN to segment retinal blood vessels in 2017, and achieved good results. Liu et al. [10] proposed a semi-supervised segmentation model, CDR-GANs, to split the optic disc and optic cup segmentation task of fundus maps into two independent stages, each of which was trained separately using GAN. Its performance is close to that of fully supervised learning models.
This work draws on the structural framework of GAN and introduces U-Net based on literature [11] to construct a new semi-supervised semantic segmentation network. The network utilizes the excellent performance of U-Net in medical image segmentation to achieve semi-supervised segmentation of medical images, which can effectively solve the problem of insufficient labeling data for medical images. In this work, a CT image dataset of aortic angiography is constructed, which contains a number of 2598 arterial images. The research uses the dataset to train the model, and finally realizes the automatic segmentation of pulmonary artery and ascending and descending aorta multi-classification tasks.
The overall framework of the implemented semi-supervised semantic segmentation model is shown in Fig. 1.
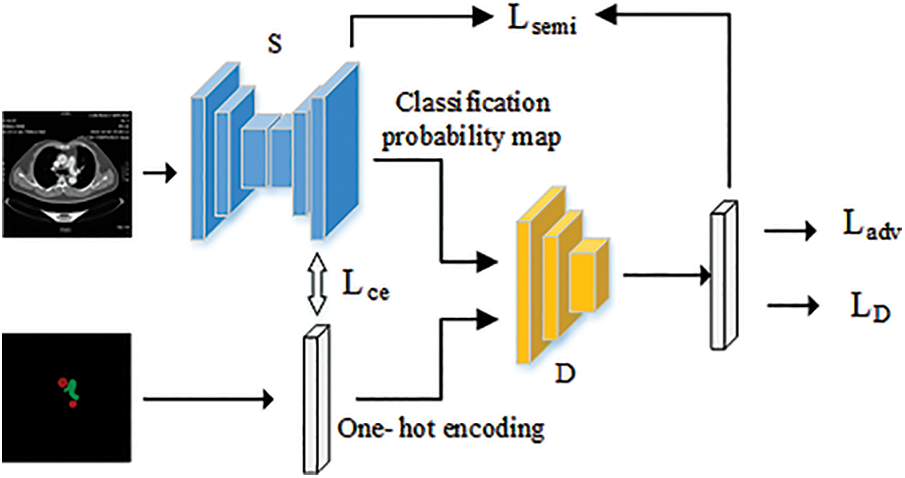
Figure 1: Framework diagram of semi-supervised segmentation model based on GAN framework
The framework consists of two parts, a segmentation network S based on the U-Net [12] network and a discriminative network D that discriminates the segmentation map. The segmentation network inputs a size of (512, 512, 3) medical image and outputs a classification probability map for a size of (512, 512, 3) medical image, where the output of 3 represents the number of categories that need to be output by the segmentation network. The (i, j, k)∈(512, 512, 3) in the probability map represent the probability that the location pixel point in the original image belongs to category k. The input of the discriminative network is the ground-truth label information or the classification probability map output by the U-Net segmentation network. The output of the discriminative network is a confidence map.
We use fully supervised training in the early stage to make it have a certain segmentation ability, and use semi-supervised training in the later stage. The cross-entropy Lce of the segmentation results with the ground truth and the Ladv under the adversarial framework are used to optimize the fully supervised training of the segmentation network. Since label data is not used during semi-supervised training, we train the model using the high confidence maps output by the discriminator as pseudo labels. With the addition of pseudo-labels, the loss entropy of the loss function of the segmentation network becomes composed of the segmentation result, the loss entropy of the pseudo-labels Lsemi and the result of the discriminant network Ladv component. The training of the discriminant network only needs to use labeled data for fully supervised training.
U-Net is a semantic segmentation model based on full convolutional neural network. The encoder-decoder constitutes a unique U-shaped structure, and an innovative jumper structure is proposed to save feature information more effectively. The basic structure is shown in Fig. 2.
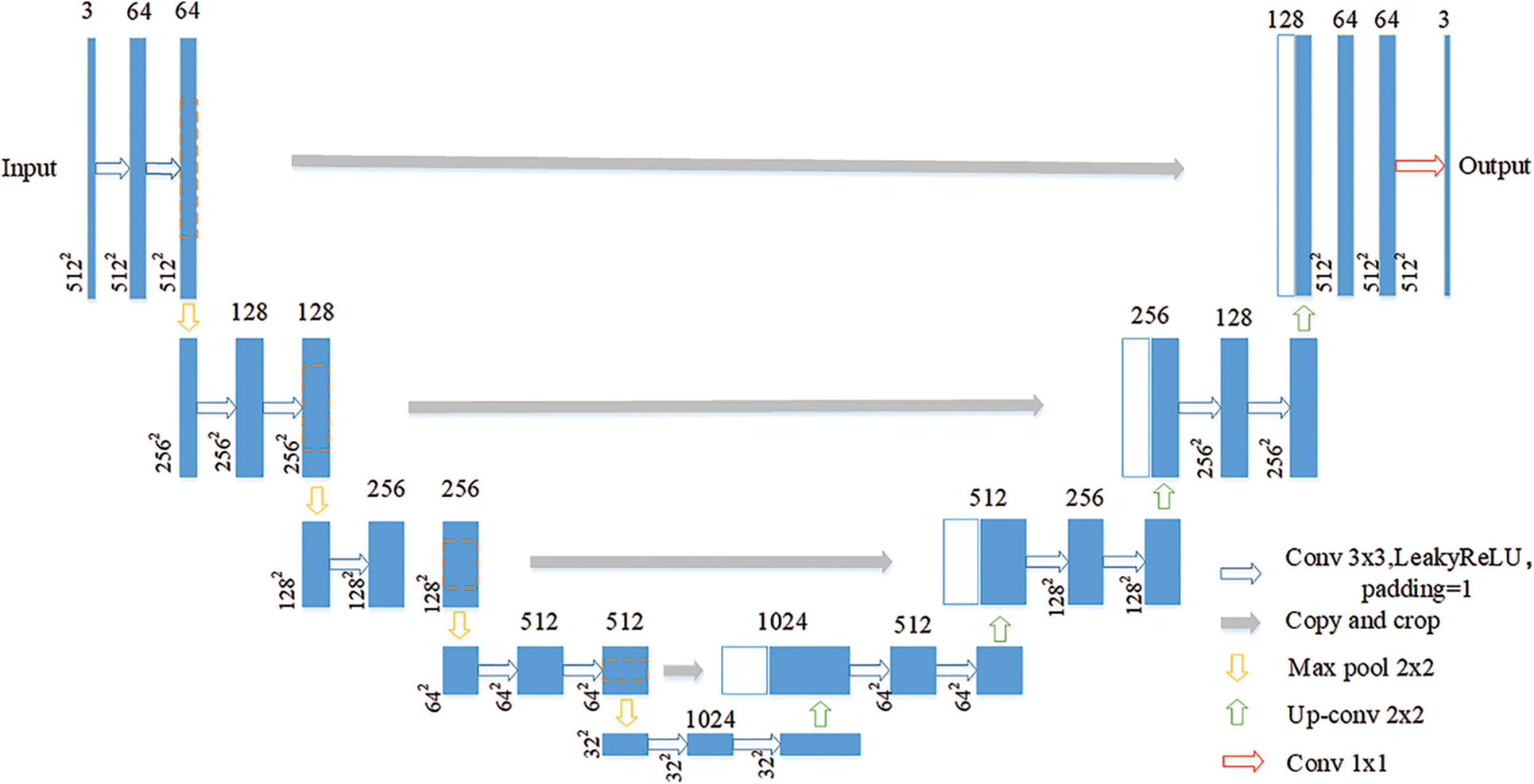
Figure 2: U-Net network structure
The segmentation network adopts the U-Net network as the backbone network due to its segmentation capability in the field of medical images, which is locally perceptive and can be trained with fewer samples. The U-Net network consists of two paths, the left path is the Contracting Path, which is mainly used to down sample the image to extract the high-dimensional features of the image, and the right Expansive Path, which aims to fuse the feature map back to the same size as the original image.
The setting of the discriminative network is based on the discriminative structure design in DCGAN [13]. The discriminator receives a 512 × 512 × 3 classification probability map or one-hot encoded label output by the segmentation network. The backbone of the network consists of 6 convolutional layers with the same settings as the DCGAN discriminator network, as see in Fig. 3. The pooling layer operation and batch normalization are cancelled, a 4 × 4 convolution kernel is used for convolution and pooling with a stride 2 convolution operation, and Leaky Relu [14] is used as the activation function, so that the discriminant network can be trained stably. Finally, the output of the discriminant network is upsampled by an interpolation operation in order to be able to calculate the loss value.
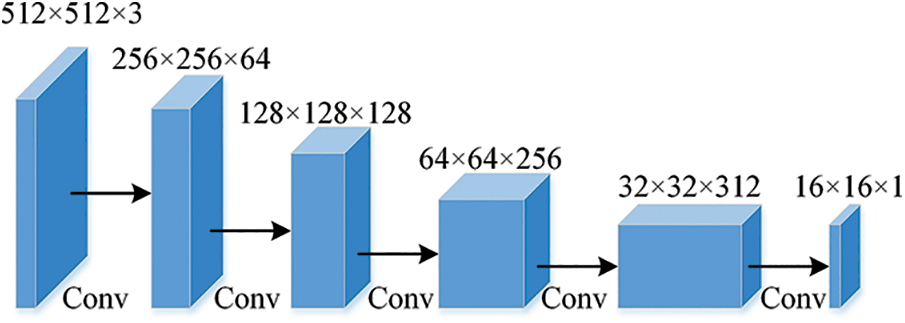
Figure 3: Network structure of discriminator
3 Experiment and Result Analysis
The dataset used in this work is the dataset of arterial angiography, which consists of 2608 images in total. The mask label is completed under the guidance of medical experts. The dataset images mainly include the aorta (AA) and pulmonary artery (PA), and the image size is 512 × 512. The image and annotation label visualization results of the constructed arterial dataset are shown in Fig. 4. The data image at the top and the corresponding labels at the bottom. The red area in the label map is AA and the green area is PA.
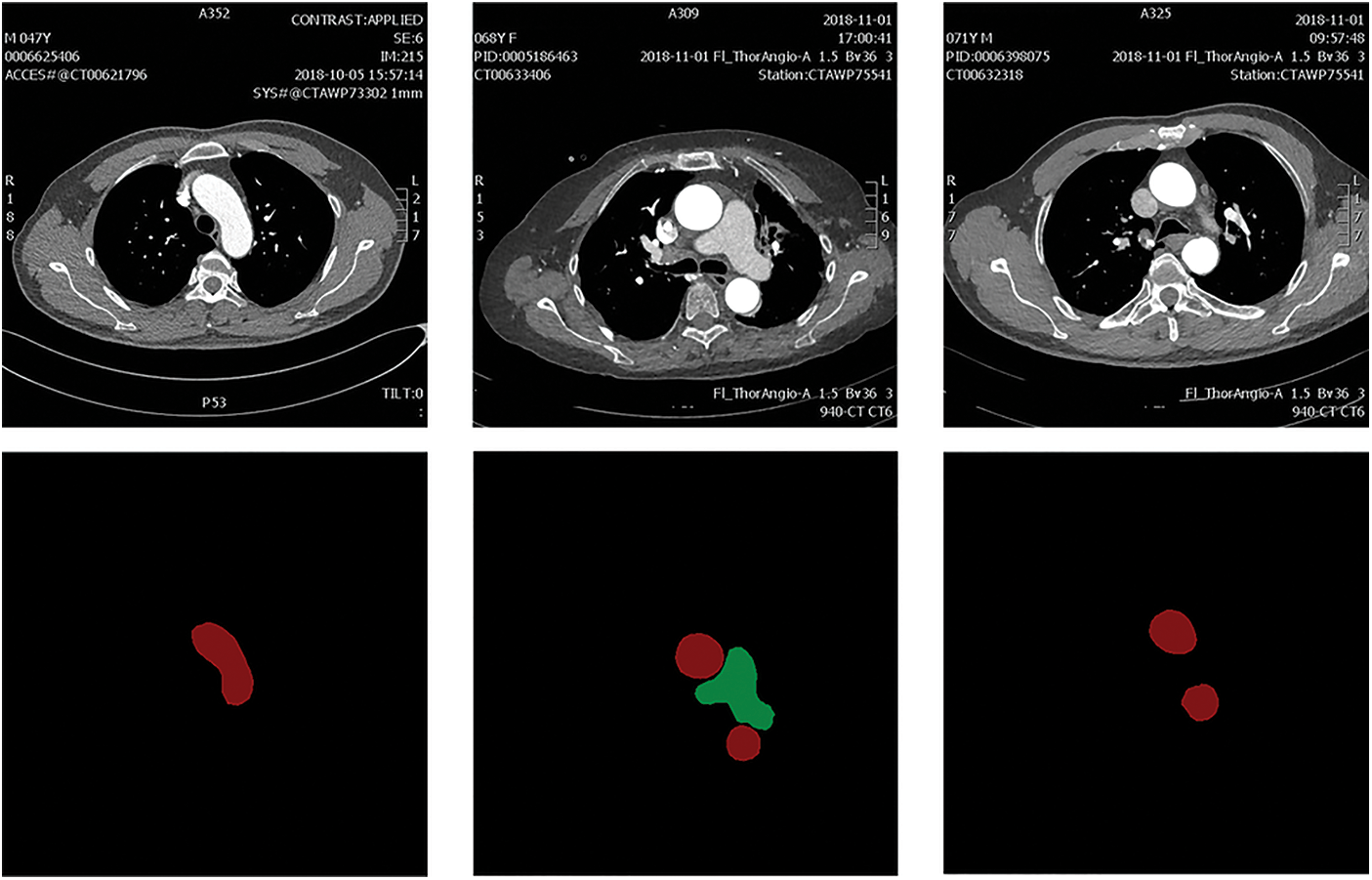
Figure 4: The constructed arterial dataset
3.2 Training of Discriminative Networks
The discriminator proposed in this work is mainly used to distinguish whether the input image Xn is from the output of the segmentation network S(Xn) or the ground truth label Yn. In each training epoch, we alternately train the segmentation network which denote by S(⋅) and the discriminant network D(⋅), and for the discriminant network, we train the discriminator using the segmentation map generated by the segmentation network and the real label map, respectively. When the output probability value of the discriminator network is closer to 1, it means that the image Xn input to the discriminator is closer to the real segmented image, otherwise the Xn is a fake image with the output probability value of the D(⋅) is closer to 0. We employ a binary cross-entropy loss denoted as Lbce to detect each pixel of the input image, which is used to constrain the training of the network. The loss function LD of the discriminant network is shown in Eq. (1), where D(⋅)(h, w) represents the confidence map output at position (h, w). We use gradient descent for network training in order to achieve rapid convergence.
3.3 Training of Segmentation Networks
3.3.1 Fully Supervised Training
During GAN network training, the purpose of the segmentation network is to generate as realistic a label map as possible to fool the discriminator. The segmentation network and the identification network are always in a process of confrontation. In order to make the two more reasonably confrontational training, a fully supervised training method is adopted for the segmentation network in the early stage. The loss function for fully supervised training of the segmentation network denoted as
The Lce in Eq. (2) represents the multi-class cross entropy loss function, and Ladv in Eq. (2) is denoted as the loss of the adversarial network, as shown in Eq. (4). We add the adversarial loss to the semantic segmentation network based on the GAN framework, and adjust the proportion of the adversarial loss in the loss function of the segmentation network through hyper parameters λadv to train the segmentation network.
For fully supervised training of the segmentation network, the segmentation result of the input image and the ground truth label Yn encoded by One-Hot are computed as losses Lce in Eq. (3), where C represents the pulmonary artery, aorta and background. The
3.3.2 Semi-Supervised Training
Since unlabeled data accounts for the majority of medical image datasets, we perform fully supervised training on the segmentation network in the first 1500 epochs to enable the segmentation network to have preliminary segmentation capabilities. At the same time, the discriminative network produces discriminative power in alternate training. Finally, the training is continued in a semi-supervised manner.
After fully supervised training, the discriminative network is able to generate a probability map with high confidence in the results generated by the segmentation network. Therefore, the unlabeled data can utilize the GAN network to generate a high-quality confidence map, combined with the corresponding binarization process to generate a pseudo-label for supervised model training. The semi-supervised segmentation network loss
where the Lce and Ladv is the same as the loss function for fully supervised training, and Lsemi denotes the multi-class cross-loss entropy during semi-supervised training. Because pseudo-labels are introduced to update the network parameters, a hyper parameter λsemi is needed to reduce the impact of incorrect labels on the network to be adjusted to prevent large errors from being generated.
The semi-supervised multi-class crossover loss function
where Tsemi is the threshold to binarize the confidence graph of the discriminative network and I(⋅) is used to filter the high confidence in the confidence graph for classification to perform pseudo-label construction.
3.4 Experimental Environment and Parameter Settings
The model code is written based on the Pytorch framework, and the model is trained on a 3060 Nvidia GPU with 12G memory. The specific parameter settings are shown in Tab. 1.

We randomly divided all data into 5 parts. For fully supervised training, one of them is selected as the test data in turn, and the other four are used as the training data. For semi-supervised training, one of the data is used as the test set in turn, two of the remaining data are used for fully supervised training, and the other two are used as unlabeled data for semi-supervised training.
In this work, the performance of the model is evaluated from multiple perspectives using multiple evaluation criteria: mean intersection ratio (MIOU), Dice coefficient, specificity, and accuracy rate. The semantic segmentation results can be expressed in Fig. 5, and the 0 and 1 represent the class of the sample, respectively.
Figure 5: Four cases of semantic segmentation results
Mean Intersection Over Union (MIOU), which calculates the ratio of the intersection to the union between the two sets of predicted and true values. As shown in Eq. (7).
The Dice coefficient (DICE) is an ensemble similarity measure function used to calculate the similarity between two samples. Its formula is given in Eq. (8). The value of Dice is taken between 0 and 1. Larger values indicate a larger proportion of overlapping regions and better prediction of segmentation.
The specificity (S) is the proportion of the predicted sample that is actually negative and is given in Eq. (9). Where the recall rate indicates how many of the predicted negative samples are truly negative samples.
Precision (P) is the proportion of true positives in a positive sample and is given in Eq. (10). Where the precision rate is mainly for how many of the samples predicted to be positive are positive samples.
We first evaluate the fully supervised training effect of the adversarial learning model on the arterial dataset with different neural networks, as shown in Tab. 2. MIOU, DICE, S, P represent mean intersection ratio, Dice coefficient, specificity, and precision, respectively. Compared with adding attention mechanism to U-net, the average results of three experiments of adversarial learning are improved in MIOU, DICE and P. This shows the advantages of adversarial learning.

Due to the small amount of data, we performed five-fold cross-validation for semi-supervised training. When the labeled data is reduced to 1/2, 1/4, and 1/8 of the original labeled data, the detection results are shown in Tab. 3. We can see that under the premise of the same amount of data, the proposed semi-supervised method achieves the performance of U-net’s full supervision when the training data is reduced to 1/2.

The experimental results in Tab. 3 verify the feasibility of the method and provide a new idea for medical image segmentation. Fig. 6 below shows the prediction results of this method under each model. We can see that 1/2 semi-supervised segmentation performance is comparable to full supervision, but 1/8 is inferior in semi-supervised performance, especially on the performance of pulmonary aorta segmentation. For slices containing only the aorta, the semi-supervised trained model at each scale can segment the aorta with good contours.

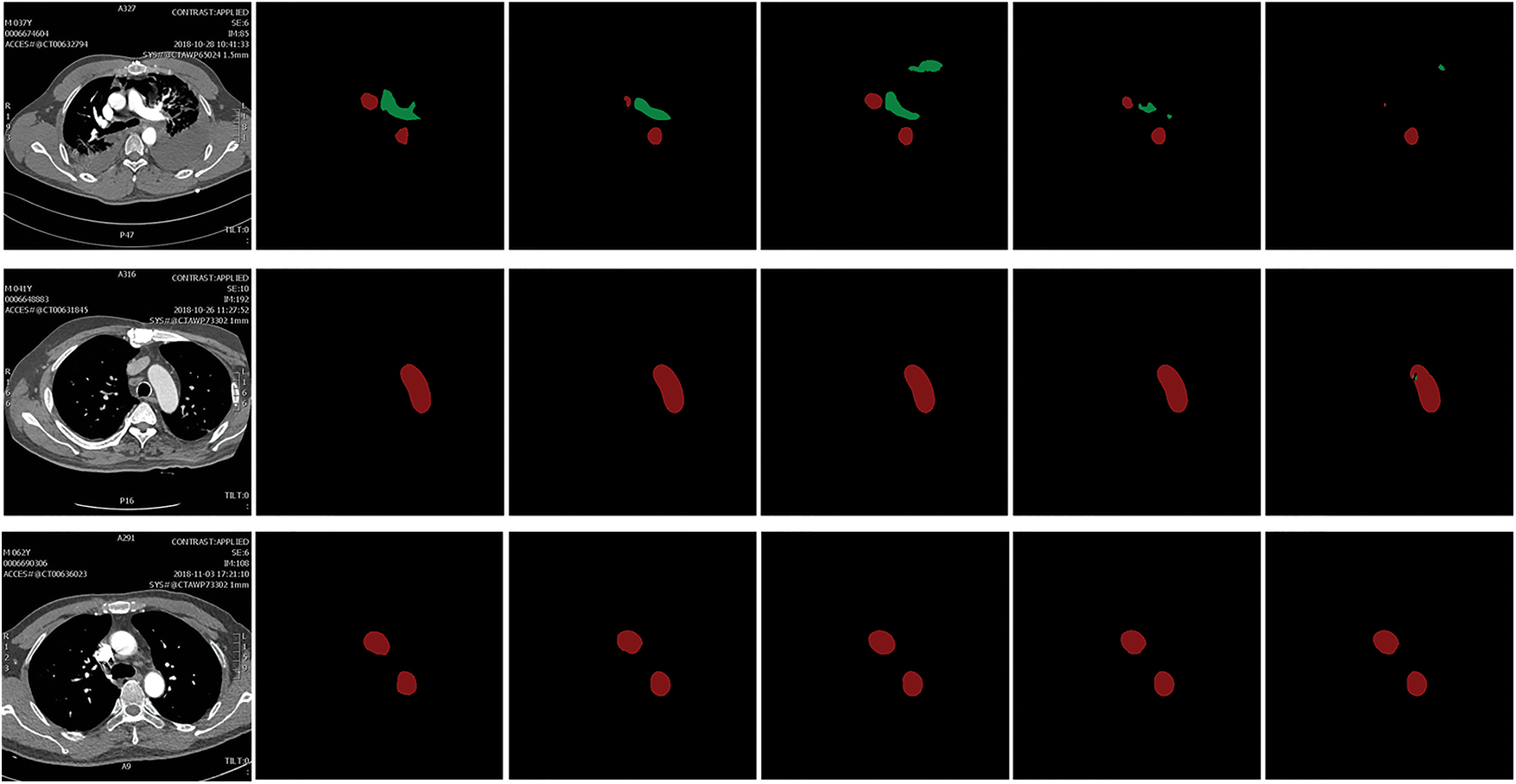
Figure 6: The prediction result of the proposed model. 1/2, 1/4, 1/8 represent the results of the semi-supervised training process reducing the labeled data to the proportion of the original fully supervised data, respectively
In this work, we adopt an adversarial learning framework for semi-supervised training of segmentation networks. The performance of the model trained with half the labeled data is close to the performance of the fully supervised training model, and certain results have been achieved. This also proves the advantages of semi-supervised learning in medical image segmentation, which can use a small amount of data to complete the training of the segmentation network. However, there is still a disadvantage that network training requires constant adjustment of thresholds to optimize pseudo-label generation.
Acknowledgement: The author would like to thank the support of Central South University of Forestry & Technology and the support of the Second Xiangya Hospital of Central South University.
Funding Statement: This work was supported in part by the National Natural Science Foundation of China (No. 62002392); in part by the Key Research and Development Plan of Hunan Province (No. 2019SK2022); in part by the Natural Science Foundation of Hunan Province (No. 2020JJ4140 and 2020JJ4141); in part by the Postgraduate Excellent teaching team Project of Hunan Province [Grant [2019] 370–133].
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Y. Tan, L. Tan, X. Y. Xiang, H. Tang, J. H. Qin et al., “Automatic detection of aortic dissection based on morphology and deep learning,” Computers, Materials & Continua, vol. 62, no. 3, pp. 1201–1215, 2020. [Google Scholar]
2. S. A. Taghanaki, K. Abhishek, J. P. Cohen and G. Hamarneh, “Deep semantic segmentation of natural and medical images: A review,” Artificial Intelligence Review, vol. 54, no. 1, pp. 137–178, 2021. [Google Scholar]
3. O. Ronneberger, P. Fischer and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in Proc. Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Springer, Cham, pp. 234–241, 2015. [Google Scholar]
4. G. Hou, J. Qin, X. Xiang, Y. Tan and N. N. Xiong, “AF-net: A medical image segmentation network based on attention mechanism and feature fusion,” Computers, Materials & Continua, vol. 69, no. 2, pp. 1877–1891, 2021. [Google Scholar]
5. Q. Y. Zhou, J. H. Qin, X. Y. Xiang, Y. Tan and Y. Ren, “MOLS-Net: Multi-organ and lesion segmentation network based on sequence feature pyramid and attention mechanism for aortic dissection diagnosis,” Knowledge-Based Systems, vol. 239, no. 2022, pp. 107853–107864, 2022. [Google Scholar]
6. A. Tarvainen and H. Valpola, “Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results,” in Proc. the 31st Int. Conf. on Neural Information Processing Systems, Long Beach, California, US, pp. 1195–1204, 2017. [Google Scholar]
7. L. Y. Sun, J. X. Wu, X. H. Ding, Y. Huang, G. S. Wang et al., “A teacher-student framework for semi-supervised medical image segmentation from mixed supervision,” arXiv preprint arXiv:2010.12219, 2020. [Google Scholar]
8. I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley et al., “Generative adversarial nets,” Advances in Neural Information Processing Systems, vol. 27, pp. 2672–2680, 2014. [Google Scholar]
9. J. Son, S. J. Park and K. H. Jung, “Towards accurate segmentation of retinal vessels and the optic disc in fundoscopic images with generative adversarial networks,” Journal of Digit Imaging, vol. 32, pp. 499–512, 2017. [Google Scholar]
10. S. P. Liu, J. M. Hong, J. P. Liang, X. P. Jia, J. Ouyang et al. “Medical image segmentation using semi-supervised conditional generative adversarial nets,” Ruan Jian Xue Bao/Journal of Software, vol. 31, no. 8, pp. 2588−2602, 2020. [Google Scholar]
11. G. French, T. Aila, S. Laine, M. Mackiewicz and G. Finlayson, “Semi-supervised semantic segmentation needs strong, varied perturbation,” arXiv preprint arXiv: 1906.01916, 2019. [Google Scholar]
12. Z. Zeng, W. Xie, Y. Zhang and Y. Lu, “RIC-Unet: An improved neural network based on unet for nuclei segmentation in histology images,” IEEE Access, vol. 7, pp. 21420–21428, 2019. [Google Scholar]
13. W. Fang, F. Zhang, V. S. Sheng and Y. W. Ding, “A method for improving CNN-based image recognition using DCGAN,” Computers, Materials & Continua, vol. 57, no. 1, pp. 167–178, 2018. [Google Scholar]
14. Y. Liu, X. Wang, L. Wang and D. L. Liu, “A modified leaky ReLU scheme (MLRS) for topology optimization with multiple materials,” Applied Mathematics and Computation, vol. 352, pp. 188–204, 2019. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |