DOI:10.32604/jcs.2022.028065

| Journal of Cyber Security DOI:10.32604/jcs.2022.028065 |  |
| Article |
CenterPicker: An Automated Cryo-EM Single-Particle Picking Method Based on Center Point Detection
1Key Laboratory of Intelligent Computing Information Processing, Ministry of Education, Computer Science College of Xiangtan University, Xiangtan, Hunan Province, 411100, China
2Department of Computer Science, the University of Georgia, Athens, Georgia, USA
*Corresponding Author: Jianquan Ouyang. Email: oyjq@xtu.edu.cn
Received: 01 May 2022; Accepted: 02 June 2022
Abstract: Cryo-electron microscopy (cryo-EM) has become one of the mainstream techniques for determining the structures of proteins and macromolecular complexes, with prospects for development and significance. Researchers must select hundreds of thousands of particles from micrographs to acquire the database for single-particle cryo-EM reconstruction. However, existing particle picking methods cannot ensure that the particles are in the center of the bounding box because the signal-to-noise ratio (SNR) of micrographs is extremely low, thereby directly affecting the efficiency and accuracy of 3D reconstruction. We propose an automated particle-picking method (CenterPicker) based on particle center point detection to automatically select a large number of high-quality particles from low signal-to-noise, low-contrast refrigerated microscopy images. The method uses a fully convolutional neural network to generate a keypoint heatmap. The heatmap value represents the probability that a micrograph pixel belongs to a particle center area. CenterPicker can process images of any size and can directly predict the center point and size of the particle. The network implements multiscale feature fusion and introduces an attention mechanism to improve the feature fusion part to obtain more accurate selection results. We have conducted a detailed evaluation of CenterPicker on a range of datasets, and results indicate that it excels in single-particle picking tasks.
Keywords: Cryo-electron microscope; deep learning; particle picking; object detection
The main experimental observation methods currently used to resolve the structures of biological protein macromolecules are X-ray crystallography, nuclear magnetic resonance spectroscopy, and cryo-electron microscopy. Cryo-electron microscopy (cryo-EM) is an experimental technique that captures images of biological samples at cryogenic temperatures using a transmission electron microscope [1]. The cryo-EM 3D reconstruction technique aims to determine the 3D structure of single particles using 2D electron microscopy images (or micrographs) [2]. To achieve 3D reconstruction, the researchers initially select a large number of single particles from micrographs. Then, 2D class averaging is performed on the selected single particles. The results of the 2D class average are used to complete the initial 3D model construction. Finally, a higher accurate 3D model is obtained. The pipeline for 3D reconstruction of cryo-EM is shown in Fig. 1. These high-resolution results that have been reconstructed rely on hundreds of thousands of high-quality particle images selected from the micrographs [3]. This paper presents a novel approach to one of the first computational problems in single-particle cryo-EM, namely, particle picking.
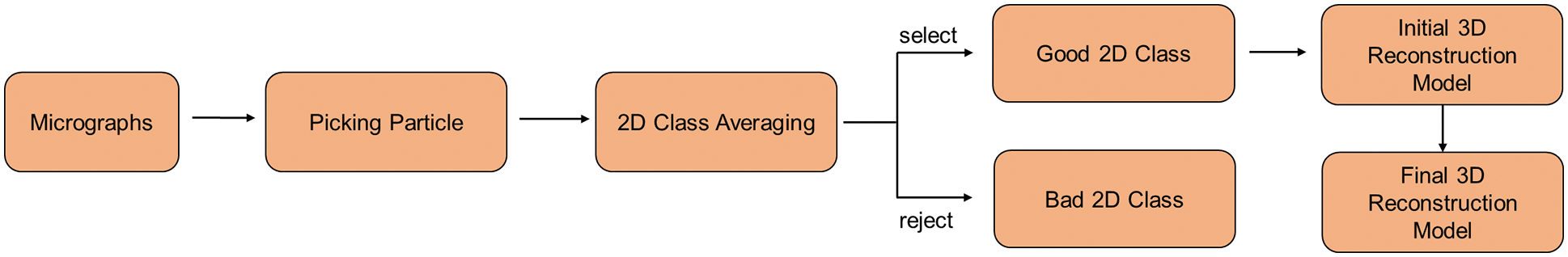
Figure 1: Pipeline for 3D reconstruction of cryo-EM
In particle picking, the goal is to locate individual particles in a micrograph while avoiding contaminants, malformed particles, and background regions [1]. However, the current particle selection has many challenges. One trouble is the low signal-to-noise ratio (SNR) of the electron microscopy images acquired by cryo-EM [4]. The high dose of electron beams in cryo-EM can considerably break the covalent bonds of particles during projection; thus, the dose size must be strictly limited, generating highly noisy images. In addition, the accuracy of the 3D reconstruction is directly influenced by the quality of the selected particles (accurate identification, particles in the center of the bounding box as much as possible). However, many of the present deep learning based single-particle extraction methods do not devote sufficient attention to the impact of noise. Most methods use fully connected layers, and the particle image should be resized when applying the model to new macromolecules of different sizes. Hence, it causes great limitations for particle picking and is vulnerable to noise that generates a larger border than the particle size. The efficiency and accuracy of these methods do not reach expectations. Thus, most researchers still use manual or semi-automated selection, which requires nearly a month or more to select the particles. Ensuring that the particles are in the center of the bounding box is challenging.
We build off a modern cryo-EM single-particle picking method based on object detection, in particular, the CenterNet [5], which enables anchor-free detection. However, different from CenterNet, we implement multiscale upsampling and introduce an attention module to improve the feature fusion process, referred to as the “CenterPicker.” The proposed method can fuse vital semantic information with more comprehensive spatial information and has certain noise immunity.
In summary, the contributions of this paper are as follows:
1. We propose a method for the direct detection of cryo-EM single-particle center, enabling automated end-to-end detection. The proposed method improves the accuracy and efficiency of cryo-EM single-particle picking. Meanwhile, it ensures that the single particles are in the center of the detection bounding box, providing better quality single-particle pictures for the subsequent cryo-EM 3D reconstruction step
2. We propose a high-resolution deconvolution module to compensate for the information loss during downsampling, and introduce attention mechanism to avoid the introduction of noise, thus improve the accuracy of the network.
3. Our results demonstrate the value of the new formulation, enabling performance improvements in efficiency and accuracy of single-particle picking.
We present the current state of research on single-particle picking algorithms in Section 2, and we describe our method in detail in Section 3. Finally, in Section 4, we have experimentally demonstrated that CenterPicker is an excellent single-particle detection algorithm for cryo-EM.
In the previous decades, many methods for automatic or semi-automatic particle picking have been proposed. They mainly include particle selection software, such as XMIPP [6], Picker [7], and RELION [8], most of which use computer vision techniques, namely, edge detection, feature extraction, and template matching. However, these methods are unsuitable for cryo-EM data with poor contrast and low SNR. The results of these approach degrade rapidly with the quality of micrographs, and the semi-automated methods that require the introduction of manual selection are time-consuming.
Subsequently, many cryo-EM single-particle detection methods based on deep learning have emerged to solve the problem of automated single-particle picking and improve the accuracy and efficiency of single-particle picking. The application of deep learning can show good robustness for images with low SNR. Others have also explored ways to improve fully automated particle picking tasks using traditional deep learning architectures. DeepPicker [9] proposed in 2016 uses VGG-Net [10] as the backbone, transforming particle picking into an image classification problem, which uses a sliding window to crop micrographs and classify these subimages as particles or backgrounds. In 2017, DeepEM was published. DeepEM [11] used a simple CNN architecture based on AlexNet [12] to train a model that can pick particles from unseen images of the same dataset. However, it is limited by the small number of training and test sets and performs poorly in terms of accuracy. The object detection model cleverly integrates the classification and localization tasks to achieve efficient detection.
Particle picking methods based on object detection have also been proposed successively given the advantages of object detection algorithms. FastParticlePicker [13] is presented; it used the Fast R-CNN [14] architecture with a simplified region proposal method to automate human-level particle picking tasks. However, it uses a sliding window to obtain the region of interest, and its performance mainly depends on the classification network. This search method greatly affects the detection efficiency and is susceptible to noise. After FastParticlePicker, crYOLO [15] improved on the target detection model YOLO [16] for particle detection, designed to automatically identify particles with high recall and high accuracy and to fully automate the data acquisition process. However, the experimental parts do not mention how the crYOLO framework detects single particles of other sizes and aspect ratios. Subsequently, many excellent object detection networks emerged, and they performed well in natural images (such as Faster R-CN [17], Mask R-CNN [18]). Researchers have used them for cryo-EM particle picking but did not obtain the desired results. The reason is that cryo-EM microscopy images are very different from natural images. These images are low-resolution, low-SNR images with a dense distribution of objects. Most current excellent target detection algorithms are anchor-based. When using different anchor sizes to generate regions of interest, the detectors and classifiers receive noise and neighboring particles to confuse the target objects. The network can only detect specific particles when the anchor scale is fixed. In addition, because they use the fully connected layer, they can only handle fixed-size images. Therefore, researchers have turned their attention to image segmentation networks.
In recent years, particle picking methods based on image segmentation networks have been proposed. PIXER [3] proposed a grid-based local maximum method to locate particles from probability density maps for cryo-EM data with low SNR, which can be as good as the semi-automatic method RELION in terms of accuracy. PARSED [19] proposed an automatic and fast deep learning framework for selecting cryo-EM single particles using fully convolutional networks (FCNs) as a baseline. However, the method requires a known 3D structure of the protein molecule and the production of a training set accordingly. This type of method initially generates probability density maps using a segmentation network and then computes the particle centers on the probability density maps by post-processing. The probability indicates the likelihood of one pixel belonging to a particle. Particle selection from probability density maps can produce more robust signals than direct selection from original noisy micrographs. However, they are not end-to-end methods. Post-processing affects the efficiency of detection. In addition, the probability value of the particle centroid position in the probability density map does not differ much from the probability value of its surrounding pixel positions. Therefore, the post-processing does not guarantee the accuracy of center point localization.
To address these problems, we propose the CenterPicker method. CenterPicker turns the particle picking problem into a center point estimation problem, and it is a fully convolutional network that can accept images of arbitrary size. This method will not be affected by noise and neighboring particles to generate excessively large boxes because our method does not use the anchor. At the same time, this method realizes multiscale feature fusion and introduces an attention mechanism, which can more accurately identify and locate particles. In addition, CenterPicker can directly predict the center point and size of the particle without post-processing, realizing real end-to-end detection. We have conducted a detailed evaluation of CenterPicker on a range of datasets, and the results show that our method has excellent performance.
CenterPicker is a network based on cryo-EM single-particle center point detection. First, compared with the cryo-EM particle picking methods based on object detection network, CenterPicker does not use the anchor to locate the particles but directly detects the key points (center points) of the particles and then returns to other particle attributes. The network become unaffected by noise to generate bounding box much larger than the particle. In addition, it is a fully convolutional network that can handle cryo-EM images of any size. Second, compared with the image segmentation network CenterPicker also generates a heatmap. Different from the single-particle picking method for cryo-EM based on image segmentation network, the value in the heatmap generated by CenterPicker represents the probability that the corresponding pixel area belongs to the center point of the particle. Therefore, it can directly predict particle position and size without post-processing, enabling end-to-end detection, and providing better quality single-particle for the subsequent cryo-EM 3D reconstruction step.
As shown in Fig. 2, our network is mainly divided into three parts, namely, Convolution module, Deconvolution module, and Detector. The cryo-EM micrograph is reduced to 1/32 times the original image through the Convolution module and then restored to 1/4 times through the Deconvolution module. Finally, the keypoint heatmap, particle bounding box size, and local offset are predicted through the three branches of the Detector.
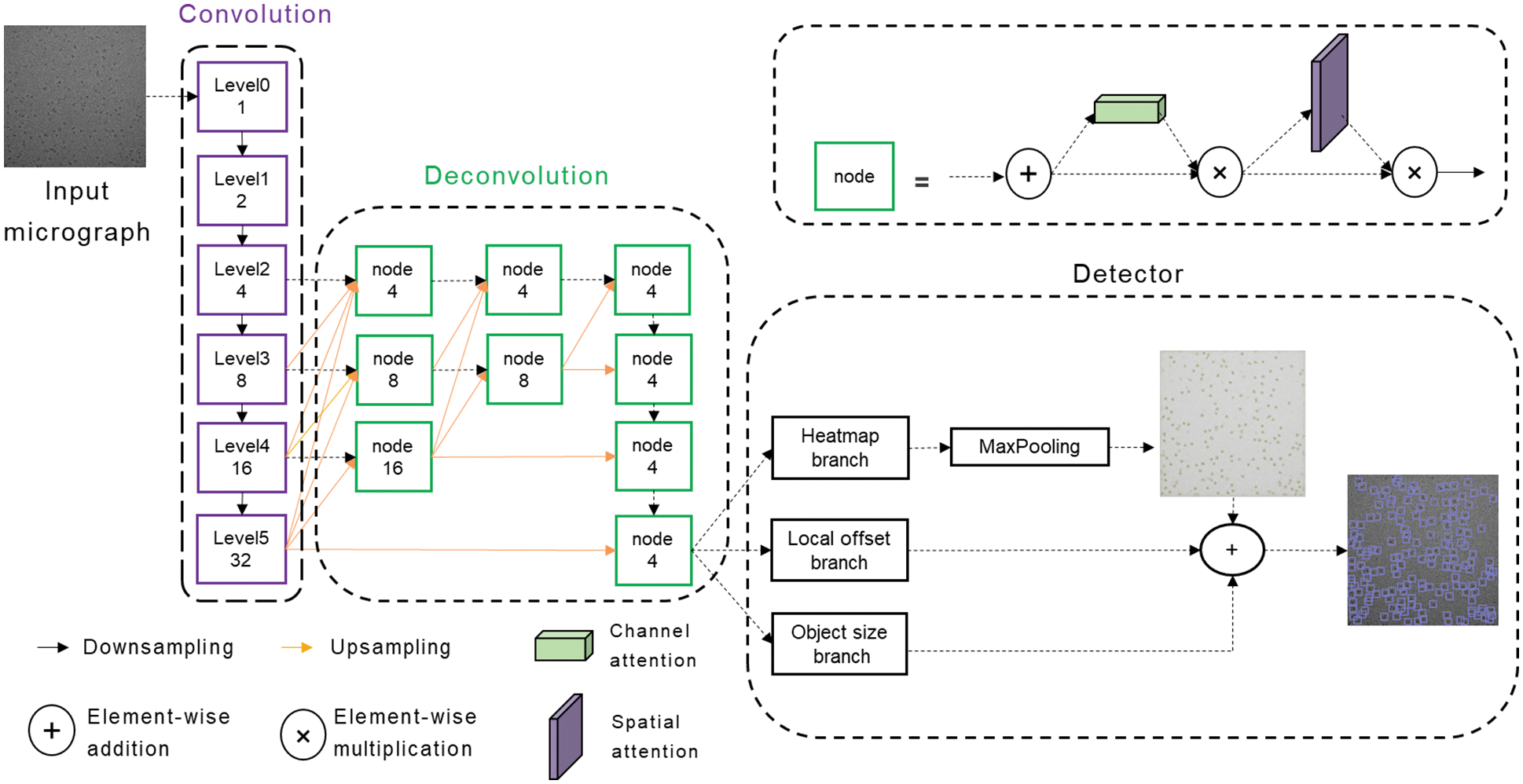
Figure 2: Schematic representation of the CenterPicker model
The Convolution module extracts features and reduces the feature map to 1/32 times of the input cryo-EM micrograph to obtain strong semantic information. In this module, we construct a variation of the DLA [20] network. The basic DLA network is designed for natural images, but cryo-EM micrographs are different from natural images. Cryo-EM micrographs have higher levels of noise and larger features, and the receptive field brought by the 3 × 3 filters is evidently insufficient for cryo-EM micrographs because the information must be aggregated over much larger spatial extents to perform effective detection. However, larger filters increase the computational cost and the number of parameters, easily leading to overfitting. Therefore, we replace the 3 × 3 convolution with a 2-dilated convolution, increasing the receptive field without introducing additional parameters. The variation of the DLA network can fuse semantic and spatial features effectively, whereas the receptive field is increased and more suitable for the single-particle picking task of the cryo-EM micrograph.
The downsampling rate of our network is 4, and the Deconvolution module outputs a feature map reduced by a factor of 4 through multiscale upsampling. The output feature map enters three parallel branches to predict the key point heat map, local offset, and particle size. If the input image size is 2048 × 2048, the output of the keypoint heatmap branch is 1 × 512 × 512, and the value of each position represents the probability that the corresponding original image area belongs to the particle center point area. The output of the Local offset branch is 2 × 512 × 512, and the two channels represent the offset of the center point x and y. Similarly, the output of the object size branch is 2 × 512 × 512, and the two channels are the w and h of the bounding box. Different from the pixel-by-pixel classification of the image segmentation network, our network is similar to a region-by-region classification. It searches for the center point area and can output the size and offset of the bounding-box corresponding to the center area through the two other branches. In the prediction phase, max pooling is used to detect the local maximum of the heatmap as the keypoint. Then the keypoint with the top K confidence score ranking is considered the particle center. Finally, the bounding box is generated by combining the results of the two other branches. At this point, CenterPicker has completed the particle picking task.
3.2 High-Resolution Deconvolution Module
As mentioned in Section 3.1, we used multiscale upsampling to restore the feature map from 1/32 to 1/4 times. For single-particle picking tasks, precise localization and accurate classification are required. Hence, the strong semantic information at the smaller scale and strong spatial information at the larger scale are extremely important. The commonly used method of recovering image resolution from low to high inevitably loses spatial information and is not the best option. Thus, we adopted a multiresolution parallel structure similar to a triangle (Fig. 2, orange arrow). High resolution is maintained throughout the process; thus, it is more precise in space. Moreover, the information is aggregated from low resolution to high resolution, thereby aggregating strong semantic information and shallow semantic information, which are beneficial to increase the classification accuracy. This structure preserves spatial information to a certain extent and can also allow the network to become suitable for the detection of small particles. The network performs detection directly on the high-resolution feature map, without a fully connected layer; it can accept cryo-EM images of any size within a reasonable range.
3.3 Attentional Feature Fusion
In 3.2, we achieved multiscale upsampling, but the problem is as follows: how do we merge the information transmitted from different scales? Cryo-EM micrographs are noisy and have low contrast. If each input is added directly, then the noise is inevitably incorporated, affecting the final detection and producing aliasing effects. Therefore, using the direct addition method for feature fusion is not feasible. The general approach is to use a convolution prior to fusion to alleviate the aliasing effect. In FPN, a 3 × 3 convolution is used. A 3 × 3 convolution is also used in our network to alleviate the aliasing effect. At the same time, considering the impact of noise on the network, we introduced a lightweight attention mechanism after the 3 × 3 convolution, allowing the network to focus on more important information and ignore useless information. In the node structure shown in Fig. 2, the attention module comprises a serial channel attention mechanism and a spatial attention mechanism. The feature map after 3 × 3 convolution fusion is adaptively adjusted in the two dimensions of channel and space. Such a feature fusion structure can alleviate the aliasing effect and reduce the introduction of noise while allowing the network to focus on important information rapidly, increasing the speed and stability of training and improving the network’s performance.
In this section, we conducted an ablation experiment to evaluate the effectiveness of our improvement. A comparison experiment with other methods was performed to verify the efficiency and accuracy of the network.
The dataset used for network training consists of the real cryo-EM micrograph dataset, as shown in Tab. 1. We used six different datasets to construct the training dataset, namely, beta-galactosidase (EMPIAR-10017 [21]), Plasmodium falciparum 80S ribosome (EMPIAR-10028 [22]), picornavirus (EMPIAR-10033 [23]), T20S proteasome (EMPIAR-10057 [24]), Nora virus (EMPIAR-10088 [25]), and TcdA1 (EMPIAR-10089 [26]), which contain particles of different sizes and various shapes, largely ensuring the diversity of the training dataset to enhance the generalization of the network. The dataset ratio of training to test is 8:2.
To evaluate whether our improvements are effective, we trained Faster R-CNN, CenterNet, CenterPicker-v1, CenterPicker-v2, and CenterPicker separately using the same dataset. Among them, CenterPicker-v1 represents the network with high-resolution deconvolution module. CenterPicker-v2 represents the network with attentional feature fusion. We propose evaluations based on a metric commonly used in the object detection literature, the precision-recall curve, and the area under it, also known as the Average Precision (AP). For completeness, we also use AP50, AP75, and AR as evaluation indicators. AP50 and AP70 represent the AP calculated when the IOU threshold is set to 0.5 and 0.75, respectively.
The results are shown in Tab. 2. Regardless of whether it only introduces high-resolution deconvolution module or only the attentional feature fusion, the network surpasses CenterNet on AP and AR, and their fusion achieves the best performance. CenterPicker’s AP and AR reached 0.520 and 0.628, respectively. CenterPicker has a larger receptive field than CenterNet and is more suitable for cryo-EM micrographs. The network can more effectively aggregate semantic information and spatial information while reducing the impact of noise to a certain extent by introducing the attention mechanism and multiscale feature fusion. Fig. 3 shows the changes in loss during the training of CenterNet and CenterPickerv2. Evidently, the network converges faster and more stable by introducing the attention mechanism.
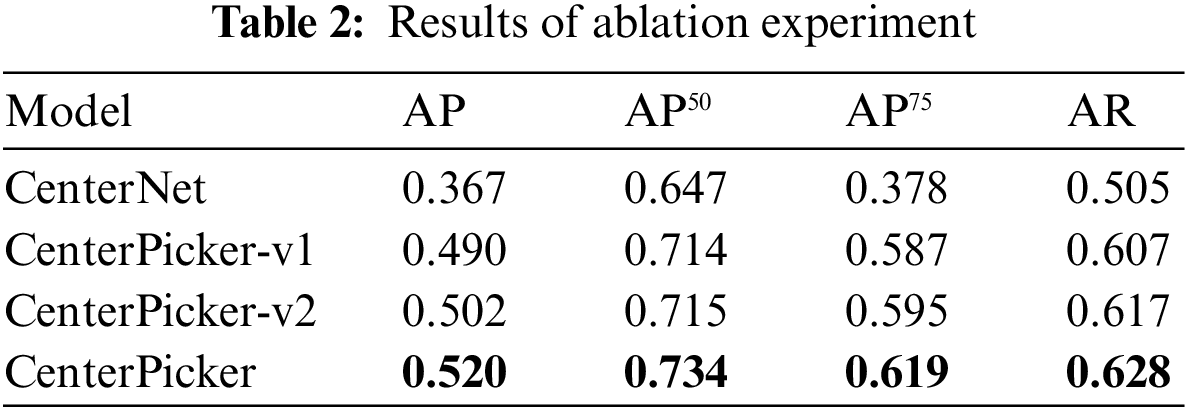
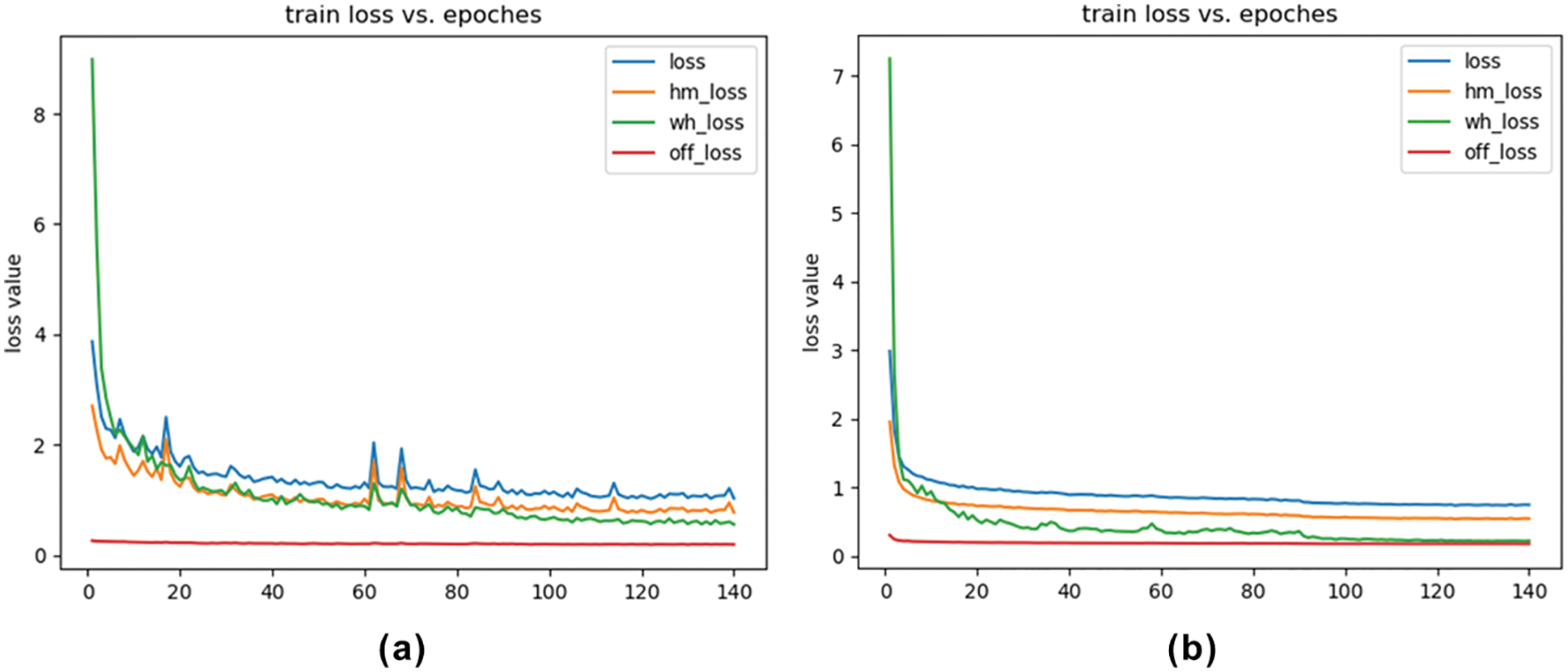
Figure 3: Loss curve. (a) Loss curve of CenterNet. (b) Loss curve of CenterPicker-v2
We also trained the Faster R-CNN with the same dataset. Faster R-CNN is an anchor-based detection method. It is susceptible to noise and surrounding particles to produce the bounding box larger than the particle size. In some situation, a box contains multiple particles. As shown in Tab. 3, the Faster R-CNN did not perform well in the cryo-EM single-particle picking task, CenterNet and CenterPicker performed better than it, and in some metrics, the CenterPicker even outperformed it by a factor of two. Fig. 4 shows the output comparison of Faster R-CNN (Fig. 4b) and CenterPicker (Fig. 4a) in EMPIAR-10081. For the same cryo-EM micrograph Faster R-CNN predicts a very small number of bounding boxes, and the size of the bounding box is significantly larger than the particles. However, CenterPicker has detected more particles, and the size and position of the bounding box are very accurate. The experimental results of CenterPicker and Faster R-CNN also prove that the anchor-free detection network is more suitable for cryo-EM micrograph picking tasks than the anchor-based detection network.
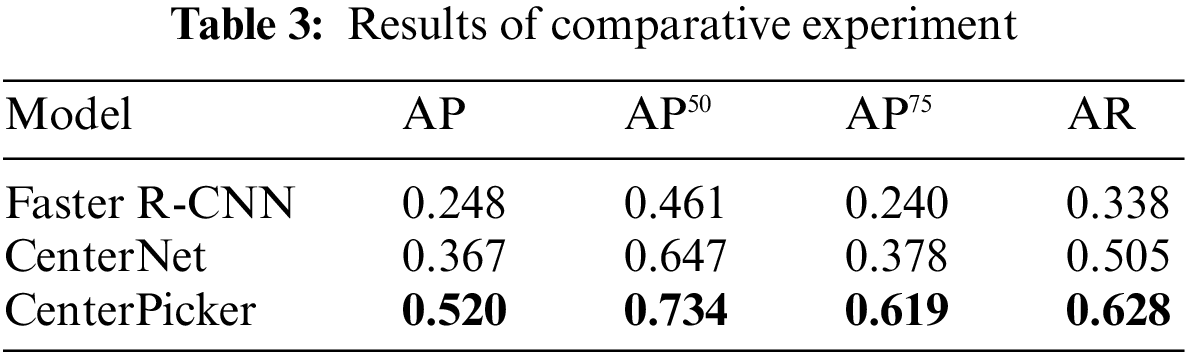
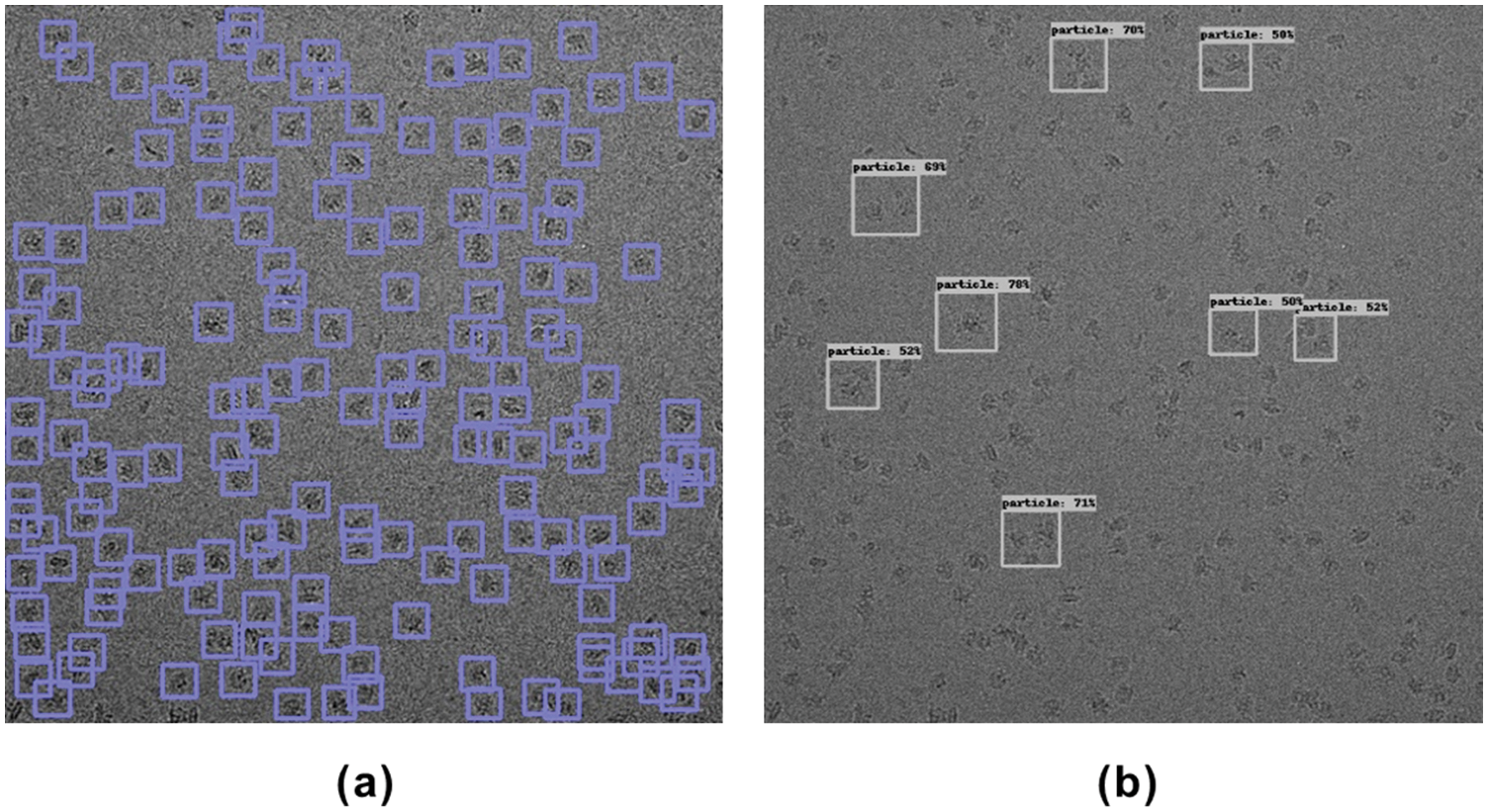
Figure 4: Particle picking results on EMPIAR-10081. (a) Particle picking results of CenterPicker on EMPIAR-10081. (b) Particle picking results of Faster R-CNN on EMPIAR-10081
4.3 Comparison with Other Picking Methods
Fig. 5 shows the intermediate results (keypoint heatmap) output by CenterPicker and the result of the final picking. The value in the keypoint heatmap represents the probability that the corresponding pixel position belongs to the center point of the particle, that is, the darkest position on the map. In the detector, we use 3 × 3 max pooling to process the keypoint heatmap, find the local maximum as the center point, and aggregate the results of the two other branches (object size and offset) to obtain the final bounding box, as shown in Fig. 5c. We used 80S Ribosome micrographs to test the Precision and Recall of CenterPicker, DeepPicker, and URDnet on the same computer to evaluate the particle picking performance of CenterPicker. In the experiment, for all these pickers, if the predicted bounding box and ground-truth IOU>0.5, then this particle is considered an accurately collected particle.
Figure 5: Particle picking in cryo-EM micrographs with CenterPicker. (a) Cryo-EM micrograph. (b) Keypoint heatmap. (c) Particle picking result
Precision measures the degree to which the prediction results are related to the ground-truth, and Recall measures the ability of the network to detect positive cases. As shown in Tab. 4, CenterPicker achieves a precision of 0.9087 and a recall of 0.9624, which are higher than the two other pickers, thereby confirming the high performance of our method in the particle picking task. URDnet is a U-shaped structure. It restores high-resolution representations from the low-resolution representations generated by the high-to-low resolution network. However, this structure inevitably causes the loss of spatial information, resulting in inaccurate spatial positioning. The upsampling process of our network maintains high resolution, which reduces the loss of spatial information to a certain extent. In addition, our network inputs cryo-EM micrograph and directly outputs particle borders without post-processing. The output of URDnet is a pixel prediction map, and post-processing (Connected Component Analysis) is required to determine the center point of the particle.
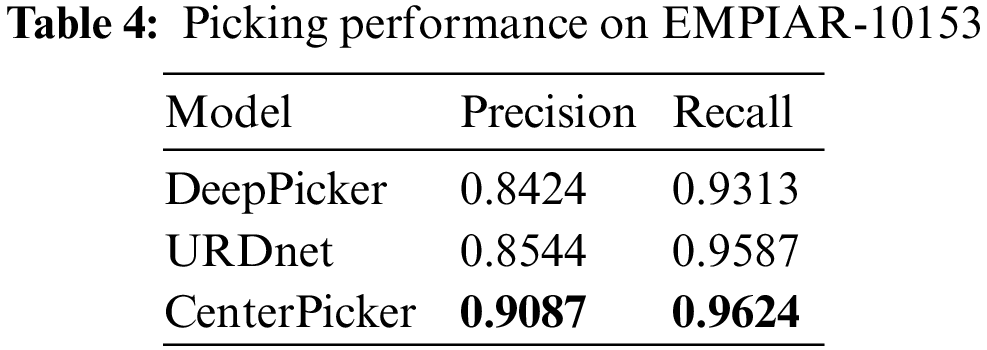
At the same time, to verify the ability of CenterPicker to generate heatmap, we calculated the center area of the training set in Tab. 1 as the particle area label and manually labeled the contaminated areas to generate the segmentation data set. Then, we used this dataset to train FCN, U-Net, and Attention U-Net. We randomly selected two cryo-EM micrographs to compare the prediction results of the three networks with the keypoint heatmap generated by the CenterPicker. The result is shown in Fig. 6. The left column is the original cryo-EM image, and the three middle columns are the heatmap predicted by the three image segmentation networks. In the heatmap, the dark green part is the contaminated area predicted by the segmentation network, and the light green part is the center point area of the particle. The rightmost column is the keypoint heatmap predicted by CenterPicker. Clearly, the FCN has the worst effect. It even predicts the entire picture as contaminated areas. U-Net has a more complex structure than FCN and has more upsampling and feature fusion processes; thus, his result is slightly better than FCN. The Attention U-Net has better results, but it is also affected by high-level noise and low contrast, resulting in many particles that cannot be accurately detected. The heatmap predicted by CenterPicker is the most accurate, and the detected particle is the most. This finding is also due to the introduction of multiscale upsampling and attention mechanisms. In an image with darker colors and low contrast, the remaining three segmentation networks mistakenly detect a large part of the image as contaminated areas because the three networks are insufficient to extract semantic information. At the same time, because of the addition of pollution categories, competition between classes exists, and the increase in the depth and width of the network may improve. For small particles, the structures of the three other networks are unsuitable. When the target is small, after a series of downsampling, its proportion in the feature map becomes very small or even disappear, and subsequent upsampling is also difficult to recover it; thus, detecting small particles is difficult. However, our network maintains a higher resolution throughout the process, so there is less loss of spatial information and have better performance in small particle detection.
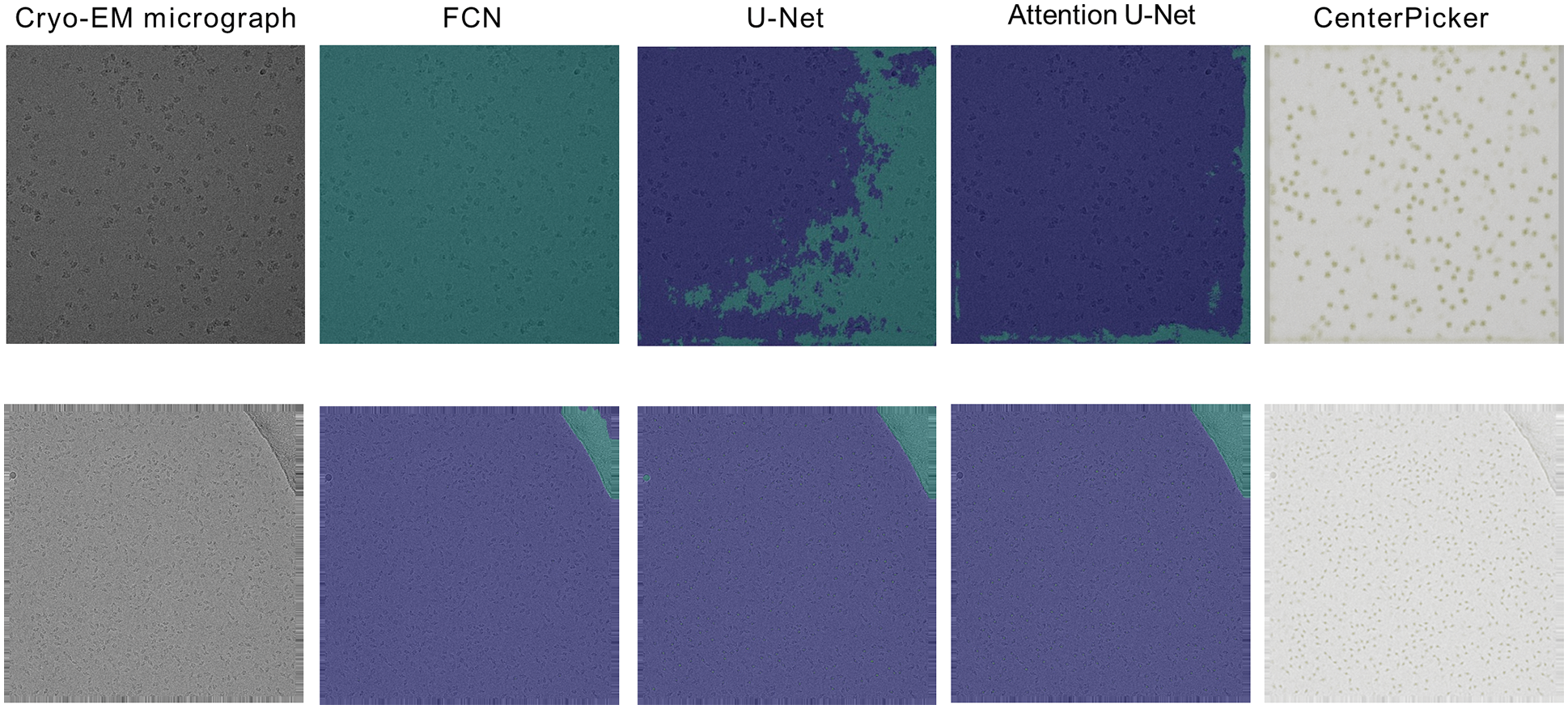
Figure 6: Experiments compared with segmented networks
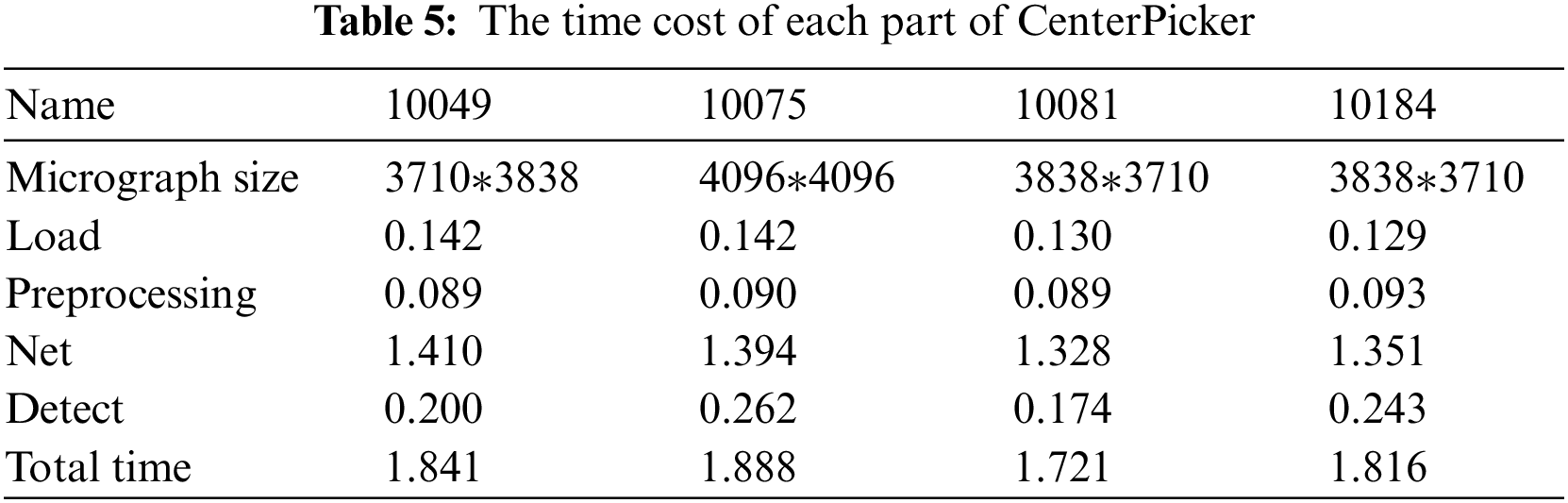
As for the picking speed, Tab. 5 records the average time cost of each substep (load, preprocessing, network, and detect). Generally, the processing time increases with the size of the micrograph. For any size of the images shown in Tab. 5, we can obtain the results within 2 s. CenterPicker uses a fully convolutional network without FC layers and no postprocessing. Thus, it is more efficient than other particle picking methods implemented based on target detection networks. Compared with the particle picking method based on the image segmentation network, CenterPicker realizes end-to-end detection. The network directly predicts the center of the particle and can predict a more accurate bounding box.
We have presented CenterPicker, a new method for particle picking in cryo-EM micrograph. Different from the object-detection particle picking method, the proposed method uses a fully convolutional network to input micrographs of any size and is more efficient. Compared with the particle picking method based on the segmentation network, our network can directly predict the center point and size of the particle to achieve end-to-end detection. To integrate strong semantic information and spatial information to improve the accuracy of the network, we have realized the fusion of multiscale features. At the same time, we introduced attention mechanism to enhance the feature fusion process. The results show that CenterPicker can break the bottleneck of particles picking and enable end-to-end, automated detection. However, using max pooling to detect the center point is not optimal. This aspect needs to be further explored. Furthermore, we have not designed an algorithm to eliminate particles with incomplete information which will be focus on our future work.
Acknowledgement: We thank the relevant teachers for their support in the experiments, the students in the lab for their contributions, and the National Key Research and Development Project Team for their support in the completion of this article.
Funding Statement: This research has been supported by Key Projects of the Ministry of Science and Technology of the People Republic of China (2018AAA0102301).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. A. Masoumzadeh and M. Brubaker, “HydraPicker: Fully automated particle picking in cryo-EM by utilizing dataset bias in single shot detection,” in Proc. BMVC, Britain, 2019. [Google Scholar]
2. J. L. Milne, M. J. Borgnia, A. Bartesaghi, E. E. Tran, L. A. Earl et al., “Cryo-electron microscopy–A primer for the non-microscopist,” The FEBS Journal, vol. 280, pp. 28–45, 2013. [Google Scholar]
3. J. Zhang, Z. Wang, Y. Chen, R. Han, Z. Liu et al., “PIXER: An automated particle-selection method based on segmentation using a deep neural network,” BMC Bioinformatics, vol. 20, pp. 1–14, 2019. [Google Scholar]
4. J. Ouyang, Y. He, H. Tang and Z. Fu, “Research on denoising of cryo-EM images based on deep learning,” Journal of Information Hiding and Privacy Protection, vol. 2, no. 1, pp. 1, 2020. [Google Scholar]
5. X. Zhou, D. Wang and P. Krähenbühl, “Objects as points,” arXiv preprint arXiv:1904.07850, 2019. [Google Scholar]
6. J. De la Rosa-Trevín, J. Otón, R. Marabini, A. Zaldivar, J. Vargas et al., “Xmipp 3.0: An improved software suite for image processing in electron microscopy,” Journal of Structural Biology, vol. 184, pp. 321–328, 2013. [Google Scholar]
7. F. Zhang, Y. Chen, F. Ren, X. Wang, Z. Liu et al., “A two-phase improved correlation method for automatic particle selection in cryo-EM,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 14, pp. 316–325, 2015. [Google Scholar]
8. S. H. Scheres, “RELION: Implementation of a Bayesian approach to cryo-EM structure determination,” Journal of Structural Biology, vol. 180, pp. 519–530, 2012. [Google Scholar]
9. F. Wang, H. Gong, G. Liu, M. Li, C. Yan et al., “DeepPicker: A deep learning approach for fully automated particle picking in cryo-EM,” Journal of Structural Biology, vol. 195, pp. 325–336, 2016. [Google Scholar]
10. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv preprint arXiv:1409.1556, 2014. [Google Scholar]
11. Y. Zhu, Q. Ouyang and Y. Mao, “A deep convolutional neural network approach to single-particle recognition in cryo-electron microscopy,” BMC Bioinformatics, vol. 18, pp. 1–10, 2017. [Google Scholar]
12. R. Langlois and J. Frank, “A clarification of the terms used in comparing semi-automated particle selection algorithms in cryo-EM,” Journal of Structural Biology, vol. 175, pp. 348–352, 2011. [Google Scholar]
13. Y. Xiao and G. Yang, “A fast method for particle picking in cryo-electron micrographs based on fast R-CNN.” in Proc. AIP Conf. Proc. AIP Publishing LLC, America, 2017. [Google Scholar]
14. R. Girshick, “Fast R-CNN.,” in Proc. Proc. of the IEEE Int. Conf. on Computer Vision, The CentroParque Convention Center in Santiago, Chile, 2015. [Google Scholar]
15. T. Wagner, F. Merino, M. Stabrin, T. Moriya, C. Antoni et al., “SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM,” Communications Biology, vol. 2, pp. 1–13, 2019. [Google Scholar]
16. J. Redmon, S. Divvala, R. Girshick and A. Farhadi, “You only look once: Unified, real-time object detection.” in Proc. Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Santiago, 2016. [Google Scholar]
17. S. Ren, K. He, R. Girshick and J. Sun, “Faster R-CNN: Towards real-time object detection with region proposal networks,” Advances in Neural Information Processing Systems, vol. 28, pp. 91–99, 2015. [Google Scholar]
18. K. He, G. Gkioxari, P. Dollár and R. Girshick, “Mask R-CNN.” in Proc. Proc. of the IEEE Int. Conf. on Computer Vision, Venice, 2017. [Google Scholar]
19. R. Yao, J. Qian and Q. Huang, “Deep-learning with synthetic data enables automated picking of cryo-EM particle images of biological macromolecules,” Bioinformatics, vol. 36, pp. 1252–1259, 2020. [Google Scholar]
20. F. Yu, D. Wang, E. Shelhamer and T. Darrell, “Deep layer aggregation,” in Proc. Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Venice. [Google Scholar]
21. S. H. Scheres, “Semi-automated selection of cryo-EM particles in RELION-1.3,” Journal of Structural Biology, vol. 189, pp. 114–122, 2015. [Google Scholar]
22. W. Wong, X. -C. Bai, A. Brown, I. S. Fernandez, E. Hanssen et al., “Cryo-EM structure of the plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine,” Elife, vol. 3, pp. e03080, 2014. [Google Scholar]
23. S. Shakeel, B. M. Westerhuis, A. Domanska, R. I. Koning, R. Matadeen et al., “Multiple capsid-stabilizing interactions revealed in a high-resolution structure of an emerging picornavirus causing neonatal sepsis,” Nature Communications, vol. 7, pp. 1–8, 2016. [Google Scholar]
24. R. Danev and W. Baumeister, “Cryo-EM single particle analysis with the volta phase plate,” Elife, vol. 5, pp. e13046, 2016. [Google Scholar]
25. P. Laurinmäki, S. Shakeel, J. -O. Ekström, P. Mohammadi, D. Hultmark et al., “Structure of nora virus at 2.7 Å resolution and implications for receptor binding, capsid stability and taxonomy,” Scientific Reports, vol. 10, pp. 1–11, 2020. [Google Scholar]
26. T. Moriya, M. Saur, M. Stabrin, F. Merino, H. Voicu et al., “High-resolution single particle analysis from electron cryo-microscopy images using SPHIRE,” JoVE (Journal of Visualized Experimentspp. e55448, 2017. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |