 Open Access
Open Access
ARTICLE
Statistical Inference for Kumaraswamy Distribution under Generalized Progressive Hybrid Censoring Scheme with Application
Department of Statistics and Operations Research, College of Science, King Saud University, Riyadh, 11451, Saudi Arabia
* Corresponding Author: Magdy Nagy. Email:
Computer Modeling in Engineering & Sciences 2025, 143(1), 185-223. https://doi.org/10.32604/cmes.2025.061865
Received 05 December 2024; Accepted 14 February 2025; Issue published 11 April 2025
Abstract
In this present work, we propose the expected Bayesian and hierarchical Bayesian approaches to estimate the shape parameter and hazard rate under a generalized progressive hybrid censoring scheme for the Kumaraswamy distribution. These estimates have been obtained using gamma priors based on various loss functions such as squared error, entropy, weighted balance, and minimum expected loss functions. An investigation is carried out using Monte Carlo simulation to evaluate the effectiveness of the suggested estimators. The simulation provides a quantitative assessment of the estimates accuracy and efficiency under various conditions by comparing them in terms of mean squared error. Additionally, the monthly water capacity of the Shasta reservoir is examined to offer real-world examples of how the suggested estimations may be used and performed.Keywords
Kumaraswamy [1] created a novel two-parameter distribution with hydrological concerns in mind because the conventional probability distributions functions such as beta, normal, log-normal, Student-t, and other empirical distributions do not suit hydrological data well. For further details on how the Kumaraswamy distribution fits well with several natural occurrences, including engineering modeling, daily rainfall, water flows, and other pertinent fields, see [2–4], and [5]. This is particularly true for results that have upper and lower boundaries, such as economic statistics, test scores, human height, and air temperatures. Although the Kumaraswamy distribution has a closed form cumulative distribution function (CDF) in the closed interval [0, 1], it is comparable to the beta distribution. The probability density function (PDF) of the Kumaraswamy distribution, CDF, reliability function (RF), and hazard rate function (HRF) can be written in exponential form, respectively, as follows:
where
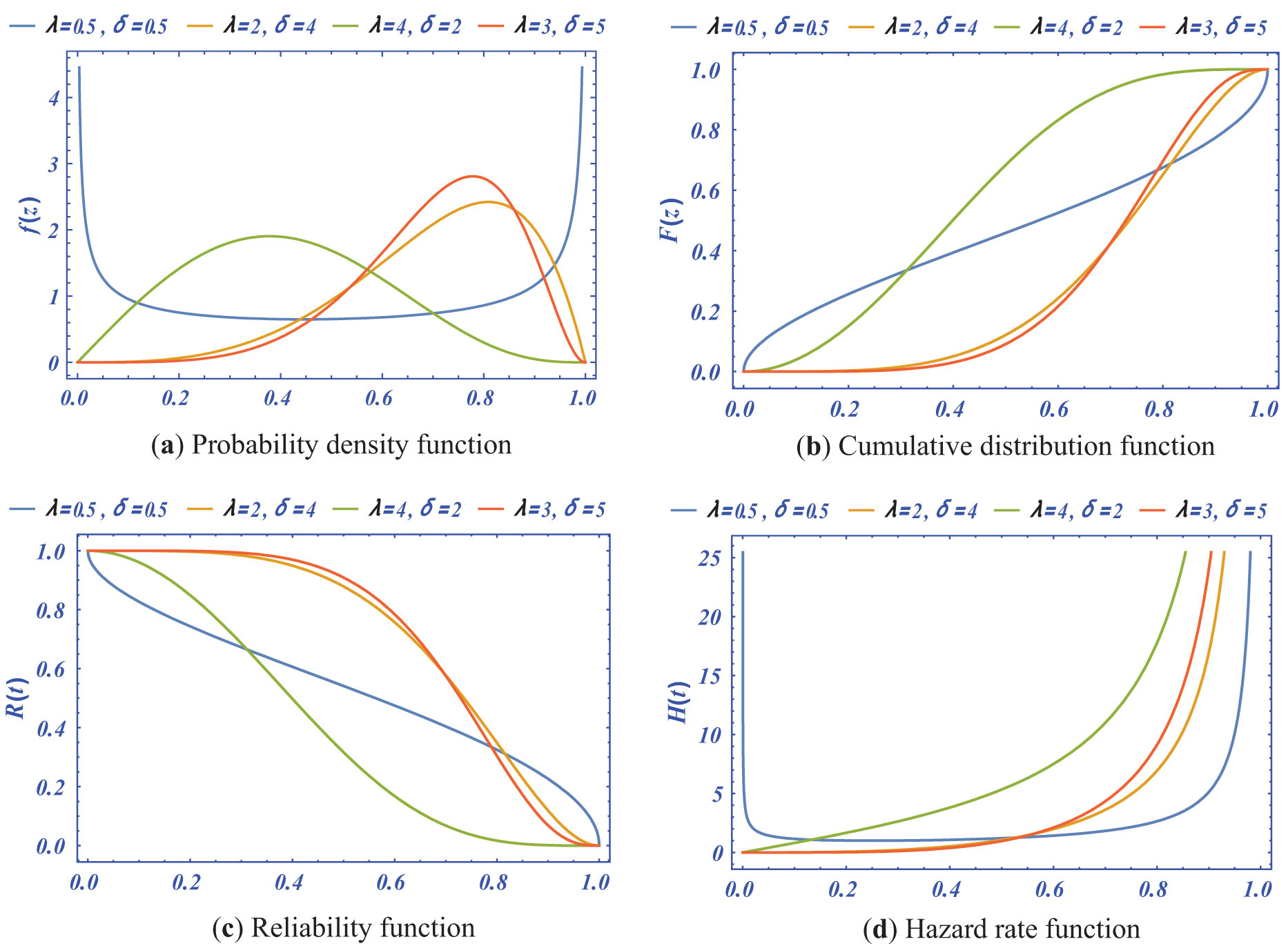
Figure 1: The PDF, CDF, RF and HRF for the Kumaraswamy distribution with different values of shape and scale parameters
Due to time, cost, and resource limitations, quality assessments that use lifetime data may encounter issues in practice. To address these problems, censoring schemes (CSs) have been employed. The progressive censoring schemes (PCSs) technique has become the norm due to its versatility and the range of data it provides. PCS states that in an experiment on
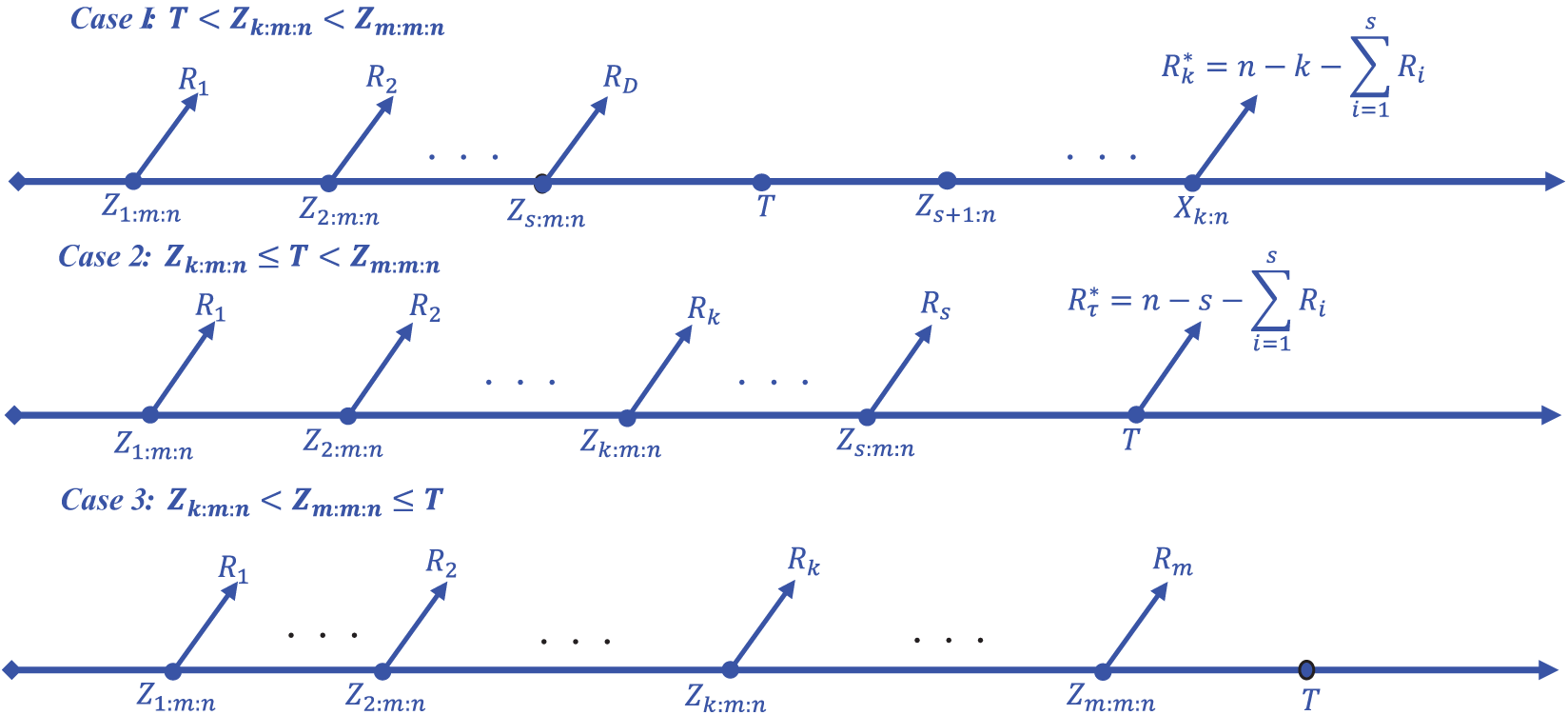
Figure 2: The Layout representation of generalized progressive hybrid censoring scheme
In the recent years, GPHCS has gained a lot of attention in reliability and survival analysis. Based on GPHCS from various distributions, Bayesian estimation has been studied by Nagy et al. in [12,13], and Nagy et al. in [14].
Bayesian estimation involves updating prior beliefs with observed data to obtain a posterior distribution. In expected Bayesian (E-Bayesian), we impose certain restrictions on our hyperparameters; it provides more stable and reliable estimates, especially in situations with uncertainty about the appropriate prior specification. The hierarchical Bayesian (H-Bayesian) method is more robust because it entails a two-stage process for constructing the prior distribution. However, a drawback of this approach lies in the intricate integrals within the estimator expression. These integrals often necessitate resolution through numerous numerical approximation techniques, making calculations tedious and time-consuming.
In the recent past, E- and H-Bayesian estimations have been considered for the parameters and the reliability characteristics under different censored data. Han [15] discussed the E-Bayesian and H-Bayesian estimations of the parameter derived from the Pareto distribution under different loss functions. Okasha et al. [16] discussed the E-Bayesian estimation for geometric distribution. Yousefzadeh [17] considered the E- and H-Bayesian estimates for Pascal distribution. Rabie et al. [18] discussed the Bayesian and E-Bayesian estimation methods of the parameter and the reliability function of the Burr-X distribution based on a generalized Type-I hybrid censoring scheme. Yaghoobzadeh [19] conducted research on E-Bayesian and H-Bayesian estimation of a scalar parameter in the Gompertz distribution under type-II censoring schemes based on fuzzy data. Their study focused on developing estimation techniques that account for uncertainty and imprecision in the data. Nassar et al. [20] conducted research on E-Bayesian estimation and associated properties of the simple step-stress model for the exponential distribution based on type-II censoring. Their study focused on developing estimation methods that account for the censoring scheme employed and investigating the properties of the estimated parameters. Nagy et al. [21] provided an E-Bayesian estimation for an exponential model based on simple step stress with Type I hybrid censored data. They developed estimation methods to obtain reliable parameter estimates considering the specific censoring scheme employed. Balakrishnan et al. [22] discussed the best linear unbiased estimation (BLUE) and maximum likelihood estimation (MLE) techniques for exponential distributions under general progressive type-II censored samples. They provided statistical methodologies for parameter estimation under this type of censoring scenario. They developed methodologies to analyze this specific censoring scheme and its impact on the estimation of the distribution’s parameters. Mohie El-Din et al. [23] proposed an E-Bayesian estimation approach for the parameters and HRF of the Gompertz distribution using type-II PCS. Their research aimed to provide reliable estimation techniques for this specific censoring scheme and demonstrated the application of these methods in practical scenarios.
Recent research has demonstrated that E- and H-Bayesian estimates are more effective than both classical and Bayesian approaches. To the best of our knowledge, no article can be found in the literature having both E- and H-Bayesian estimation for the Kumaraswamy distribution under GPHCS. Motivated by the effectiveness of E- and H-Bayesian estimates, the usefulness of the GPHCS, and the importance of the Kumaraswamy distribution in reliability applications. Our main objective in this paper is to estimate the shape parameter and the HRF of the Kumaraswamy distribution based on GPHCS using E-Bayesian and H-Bayesian approaches, which we believe would be of profound interest to applied statisticians and quality control engineers.
The structure of the article is as follows: Section 1 provides an introduction, outlining the research problem and objectives. Section 2 focuses on the MLE and Bayesian estimation techniques for the shape parameter and HRF, considering different loss functions. Section 3 presents the derivation of E-Bayesian estimators for the shape parameter under different loss functions. In Section 4, H-Bayesian estimators are obtained, again considering different loss functions. To assess the performance of the estimators, a simulation study is conducted in Section 5, where different loss functions are considered and the estimators are compared. Section 6 presents the analysis of a real-life data to demonstrate the practical application of the proposed estimators. In Section 7, the article concludes by summarizing the key findings and implications of the study. Finally, the properties of the E- and H-Bayesian estimators are discussed in the Appendix A of this paper.
2 Maximum Likelihood and Bayesian Estimation
In this section, we estimate the shape parameter
where
and
Let
where
We differentiate the above equation with respect to
The ML estimate of
In calculating statistical inference and estimation of scale parameters using the Bayesian method, this is done through integrations of non-closed formulas. Then, calculating the inference and estimation of these parameters using the E- and H-Bayesian approaches is a more complex integral formula. Therefore, methods are used to approximate these quantities, which often lead to large errors. Therefore, in this manuscript, we calculate the estimate of the shape parameter for the Kumaraswamy distribution because the Bayesian estimate of this parameter is a closed form. Therefore, calculating the estimates using the E- and H-Bayesian approaches is more accurate and efficient. Here, we have developed Bayesian estimators for the shape parameter
We are using gamma distribution as the prior distribution of
Here the hyper-parameters
where
2.2 Bayesian Estimate with SELF
The Bayes estimator of
provided if
The Bayes estimate of
2.3 Bayesian Estimate with ELF
Dey et al. [25] discussed the ELF and the Bayes estimator of
provided if
The Bayes estimate of
2.4 Bayesian Estimate with WBLF
The WBLF can be expressed as (see Nasir et al. [26]) where the Bayes estimator of
provided if E[
Hence, the Bayes estimator of
The Bayes estimate of
2.5 Bayesian Estimate with MELF
Tummala et al. [27] defined the MELF where the Bayes estimator of
provided that E[
Hence, by using MELF, the Bayes estimate is obtained as
The Bayes estimate of
According to Han [28], it is recommended to select the prior parameters
Specifically, it is suggested that for
The E-Bayesian estimate of
The specific distributions chosen for the hyper parameters
and
3.1 E-Bayesian Estimation under SELF
For a sample
For a time
where
3.2 E-Bayesian Estimation under ELF
For a sample
For a time
3.3 E-Bayesian Estimation under WBLF
For a sample
For a time
3.4 E-Bayesian Estimation under MELF
For a sample
For a time
In this part, the H-Bayesian estimates of the shape parameter of the Kumaraswamy distribution are obtained using different loss functions, namely SELF, ELF, WBLF, and MELF. Following the methodology proposed by Lindley et al. [29], we introduce hyper parameters denoted as a and b in the prior distribution Prior
and
Using Bayes theorem, likelihood function and Eqs. (36)–(38), the hierarchical posterior distributions of
and
4.1 H-Bayesian Estimation Based on SELF
Using SELF and the H-posterior distributions which are defined respectively in Eqs. (13), (36)–(38), the H-Bayesian estimates
Here,
For a time
4.2 H-Bayesian Estimation Based on ELF
Using ELF and the H-posterior distributions which are defined respectively in Eqs. (16), (36)–(38), the H-Bayesian estimates
Here,
For a time
4.3 H-Bayesian Estimation Based on WBLF
Assuming WBLF as defined in Eq. (19) and using the priors defined in Eqs. (36)–(38), the H-Bayes estimates
Here,
For a time
4.4 H-Bayesian Estimation Based on MELF
Assuming MELF as defined in Eq. (22), and using the priors defined in Eqs. (36)–(38), the H-Bayesian estimates
Then, we get
Also, the H-Bayesian estimates of
In the Appendix A of this paper, we review the properties of E-Bayesian and H-Bayesian estimators of parameter
In this section, Monte Carlo simulation study is performed to compare the performance of the proposed estimates based on GPHCS for the Kumaraswamy distribution. The simulation study has been conducted using the R software. To generate the data, the initial true values of
• Scheme 1:
• Scheme 2:
• Scheme 3:
The GPHCS has been generated by using the algorism in Nagy et al. [13]. Using gamma prior, the Bayes, E-Bayes and H-Bayes estimates are obtained under four different loss functions, as SELF, ELF, WBLF, and MELF. For these gamma priors, the hyperparameters
• As the values of
• The Bayesian, E-Bayesian and H-Bayesian estimates outperform MLE in terms of MSE.
• For any fixed loss function, the E-Bayesian estimates have smaller MSE than the Bayesian and H-Bayesian estimates.
• For fixed value of
• In most of the cases, the estimates under ELF performs better than the estimates using other loss functions.
• In most cases, Scheme 3 have minimum mean square error compared to Schemes 1 and 2 at the same time T.
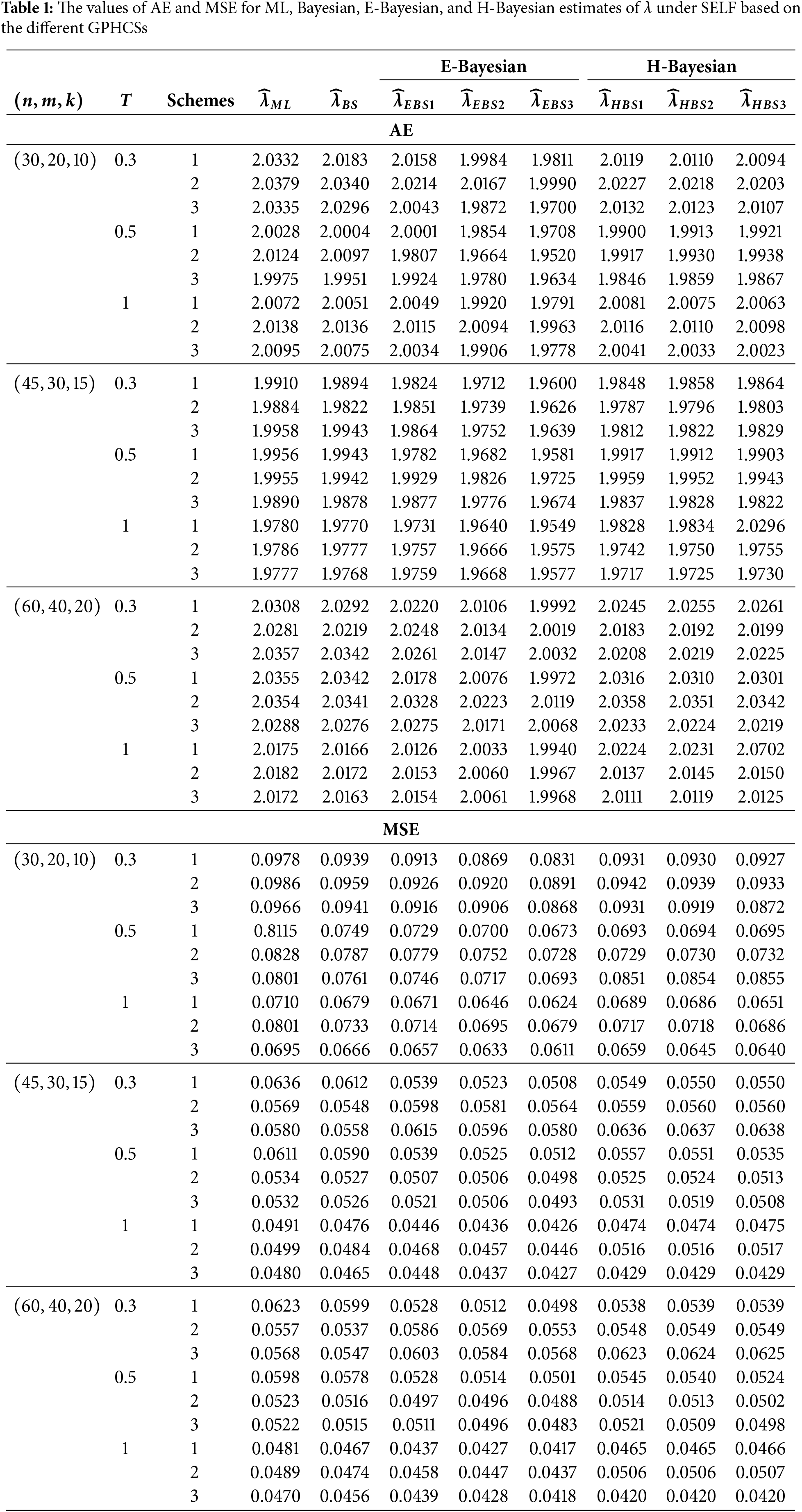
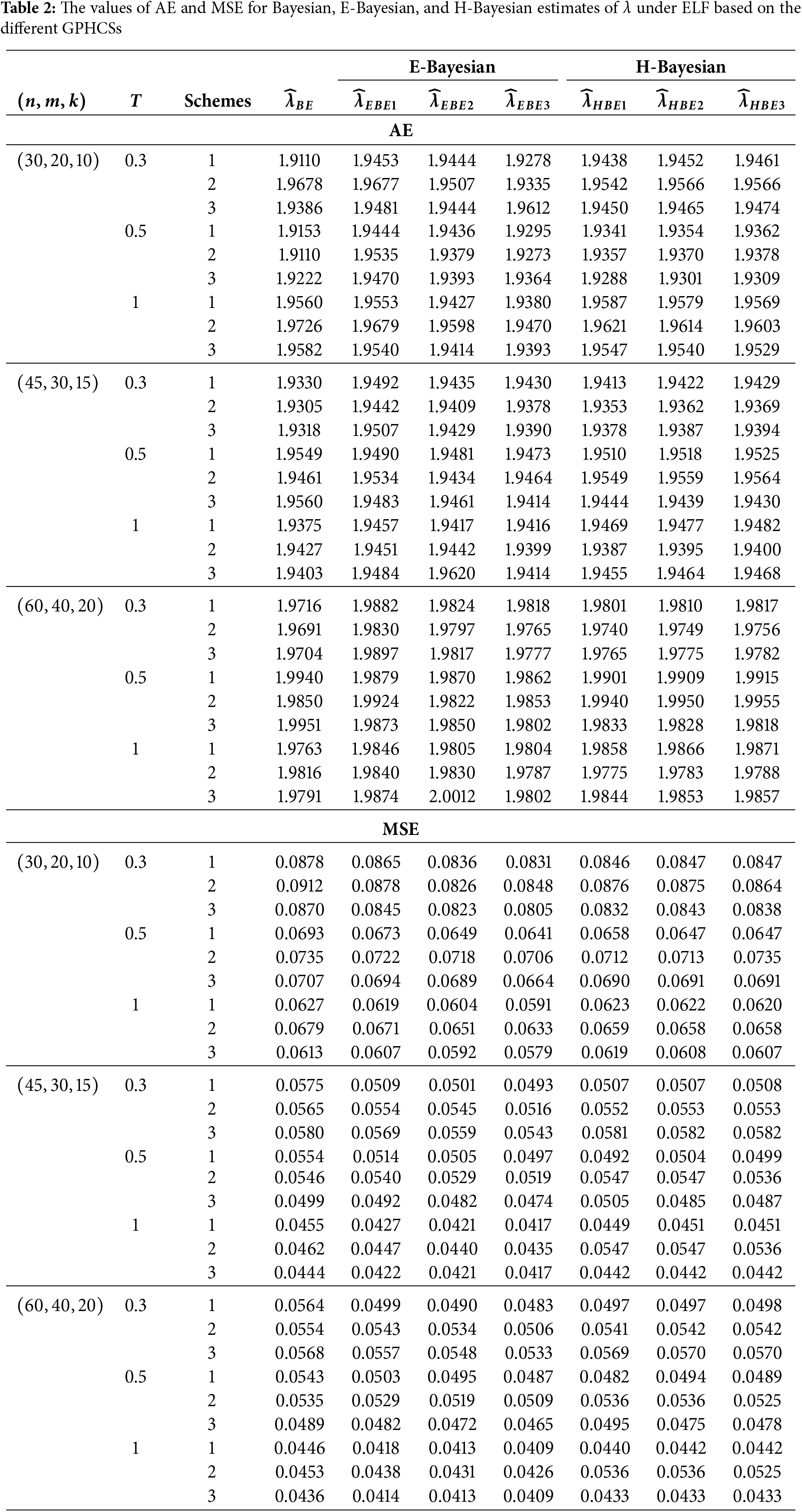
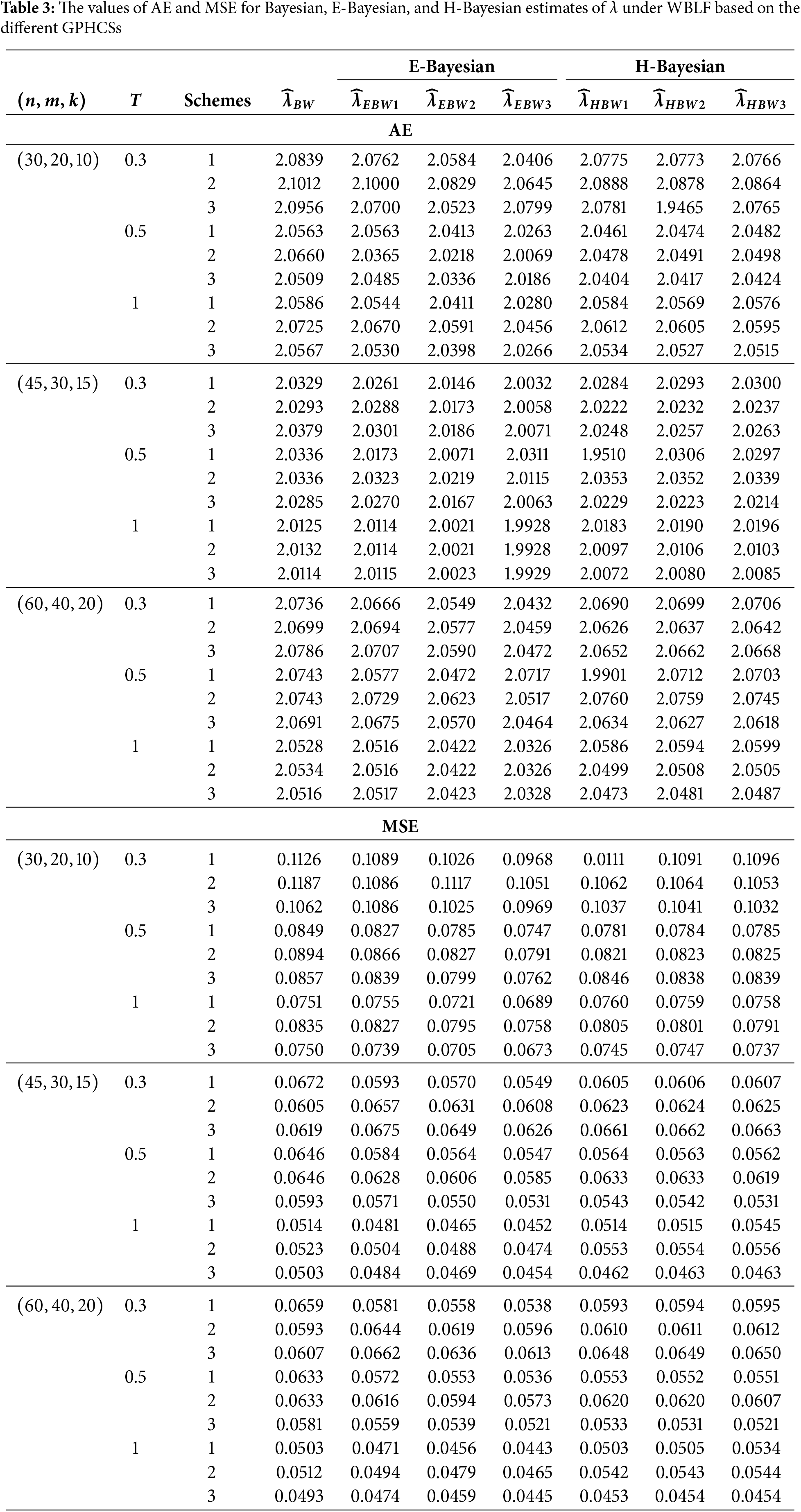
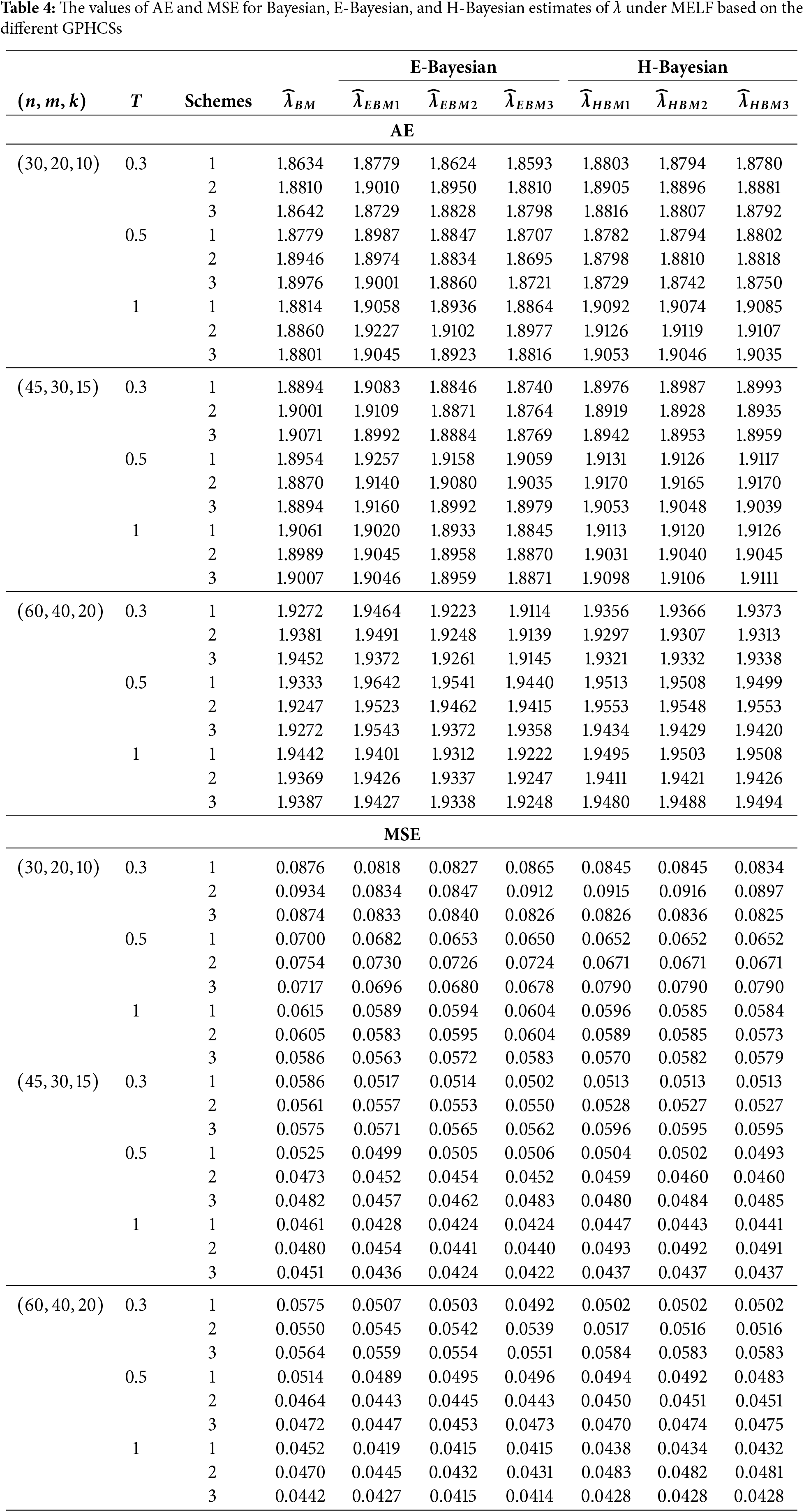

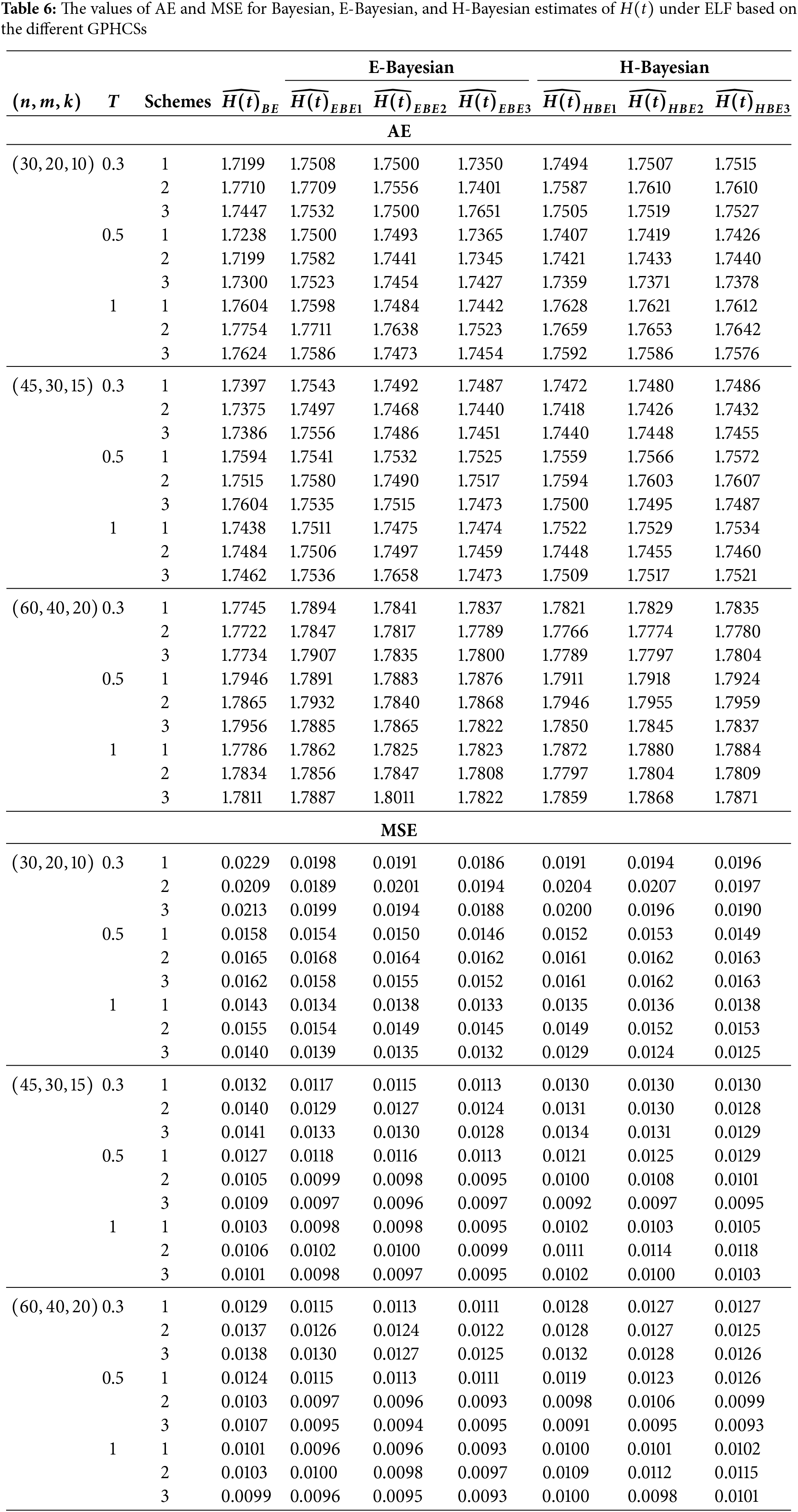

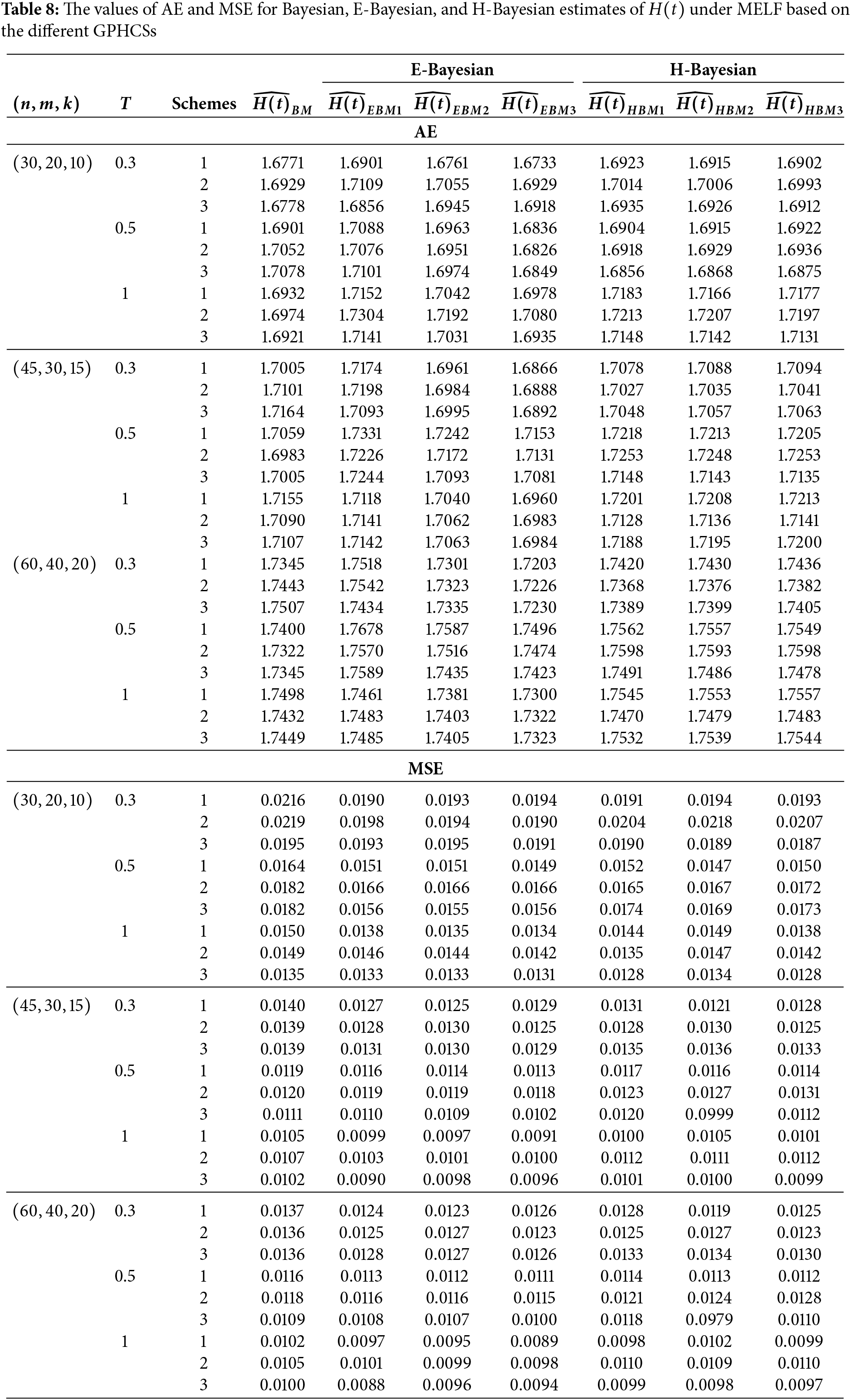
Combining all the above results, it is recommended to use the E-Bayesian technique to estimate the parameters and the HRF for GPHCS based on ELF, due to the better performance than other estimates in terms of MSE.
This section uses a real-world dataset as an example to analyze the applicability of the suggested estimation techniques. The following dataset, which spans the years 1975 to 2016, displays the monthly water capacity of the Shasta reservoir between the months of August and December. Some statisticians, including Kohansal [30] and Tu et al. [31], have previously exploited these data. We shall use these data to consider the following PCS: Suppose
• Scheme 1:
• Scheme 2:
• Scheme 3:
Table 9 shows the ML, Bayesian, E-Bayesian, and H-Bayesian estimates for the unknown parameter
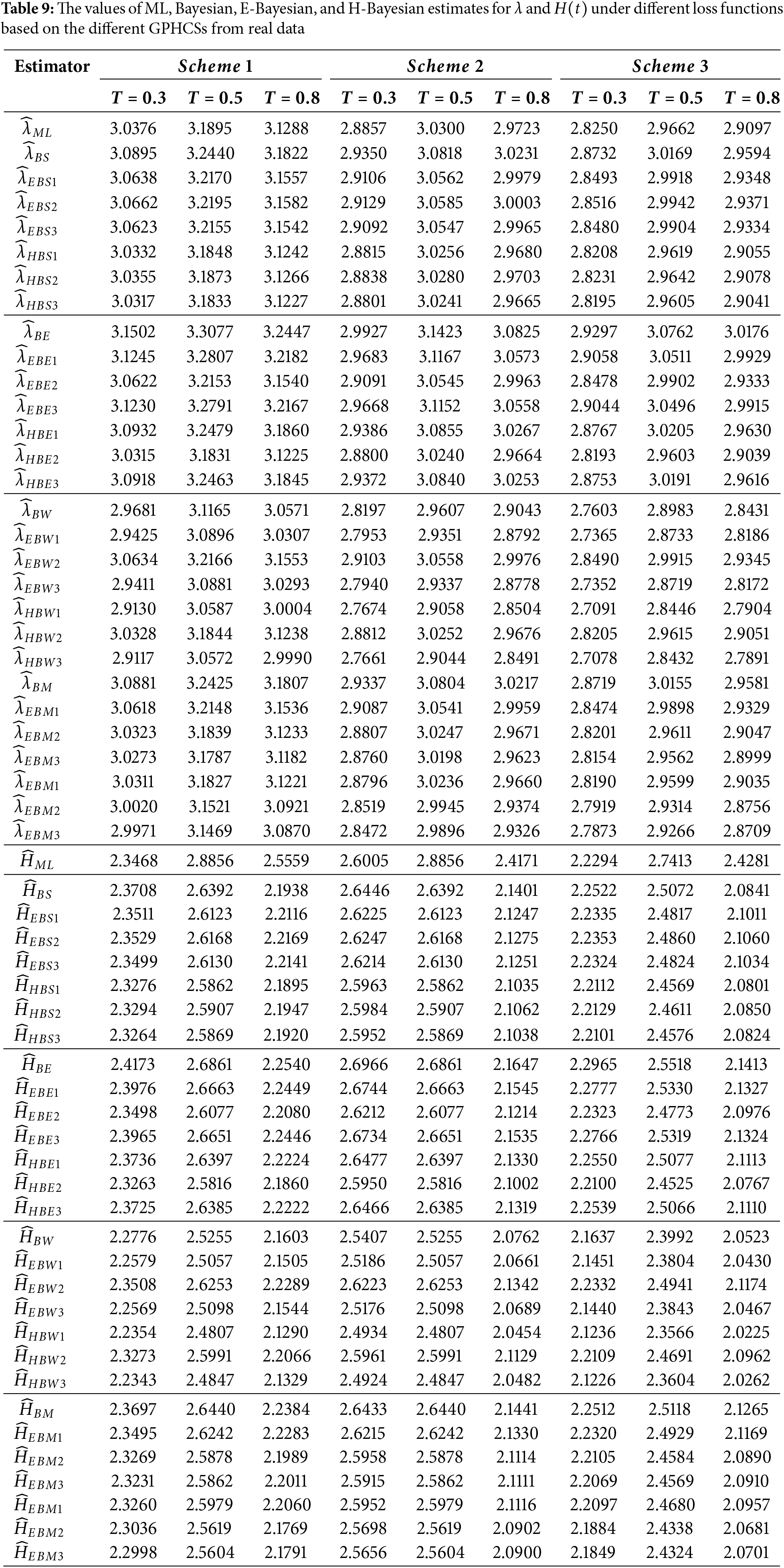
This paper proposes E-Bayesian and H-Bayesian estimations using GPHCS for the unknown shape parameter and HRF of the Kumaraswamy distribution. To obtain the Bayesian, E- and H-Bayesian estimates, the squared error, entropy, weighted balance, and minimum expected loss functions are introduced. A Monte Carlo simulation study has been performed to compare the performance of the estimates of parameters such as the shape parameter and HRF. In terms of AE and MSE, the simulation study yields that E-Bayesian estimates outperform all other estimates. Finally, a real data set has been analyzed to illustrate the applicability of the proposed estimates. After analyzing this data, it is concluded that E-Bayesian estimates for the parameters and the HRF perform better than other estimates; the sample size
Acknowledgement: The author is grateful to the editor and anonymous referees for their insightful comments and suggestions, which helped to improve the paper’s presentation. The author extends his appreciation to King Saud University for funding this work through Researchers Supporting Project, King Saud University, Riyadh, Saudi Arabia.
Funding Statement: This research was funded by Researchers Supporting Project number (RSPD2025R969), King Saud University, Riyadh, Saudi Arabia.
Availability of Data and Materials: The data was mentioned along the paper.
Ethics Approval: The author declares that this study don’t include human or animal subjects.
Conflicts of Interest: The author declares no conflicts of interest to report regarding the present study.
Appendix A Properties of E-Bayesian and H-Bayesian Estimation of λ: In this section, the properties of E-Bayesian estimates and the relations among the E-Bayesian and H-Bayesian estimates are discussed.
Appendix A.1 The Relations between the E-Bayesian Estimates under Different Loss Functions
Theorem A1: It follows from Eq. (28) that
(1)
(2)
(3)
Proof: (1) From Eq. (28), we have
and
Therefore,
For
Let
where
which yields that,
(2) From (1), we get
So it can be easily obtained that
(3) From Eq. (28) and from the proof of (1), we have
Using (2), we get
Theorem A2: It follows from Eq. (30) that
(1)
(2)
(3)
Proof: (1) From Eq. (30), we have
and
Therefore,
For
where
which yields that,
(2) From (1), we get
So it can be easily obtained that
(3) From Eq. (30) and from the proof of (1), we have
Using (2), we get
Theorem A3: It follows from Eq. (32) that
(1)
(2)
(3)
Proof: (1) From Eq. (32), we have
and
Therefore,
For
Then,
This shows that
(2) From (1), we get
This yields that,
Theorem A4: It follows from Eq. (34) that
(1)
(2)
(3)
Proof: (1) From Eq. (34), we have
and
For
Let
where
Thus,
That is
That is,
Similar relationships holds for the E-Bayesian estimates of
Appendix A.2 The Relations between the H-Bayesian Estimates under Different Loss Functions
Theorem A5: It follows from Eq. (39) that
Proof: Based on SELF, the H-Bayesian estimate of
Using the result
As
Therefore,
Taking limit as
Theorem A6: It follows from Eq. (41) that
Proof: Based on ELF, the H-Bayesian estimate of
Using the result
As
Therefore,
Taking limit as
Theorem A7: It follows from Eq. (43) that
Proof: Under WBLF, the H-Bayesian estimate of
Using the result,
As
Therefore,
Taking limit as
Theorem A8: It follows from Eq. (45) that
Proof: Under MELF, the H-Bayesian estimate of
Using the result
As
Taking limit as
Similar relationships holds for the H-Bayesian estimates of HRF under different loss functions.
References
1. Kumaraswamy P. A generalized probability density function for double-bounded random processes. J Hydrol. 1980;46(1–2):79–88. doi:10.1016/0022-1694(80)90036-0. [Google Scholar] [CrossRef]
2. Sundar V, Subbiah K. Application of double bounded probability density function for analysis of ocean waves. Ocean Eng. 1989;16(2):193–200. doi:10.1016/0029-8018(89)90005-X. [Google Scholar] [CrossRef]
3. Fletcher SG, Ponnambalam K. Estimation of reservoir yield and storage distribution using moments analysis. J Hydrol. 1996;182(1–4):259–75. doi:10.1016/0022-1694(95)02946-X. [Google Scholar] [CrossRef]
4. Mitnik PA. New properties of the Kumaraswamy distribution. Commun Statistcs-Theory Methods. 2013;42(5):741–55. doi:10.1080/03610926.2011.581782. [Google Scholar] [CrossRef]
5. Ponnambalam K, Seifi A, Vlach J. Probabilistic design of systems with general distributions of parameters. Int J Circuit Theory Appl. 2001;29(6):527–36. doi:10.1002/cta.173. [Google Scholar] [CrossRef]
6. Dey S, Mazucheli J, Nadarajah S. Kumaraswamy distribution: different methods of estimation. Comput Appl Math. 2018;37(2):2094–111. doi:10.1007/s40314-017-0441-1. [Google Scholar] [CrossRef]
7. Jamal F, Arslan Nasir M, Ozel G, Elgarhy M, Mamode Khan N. Generalized inverted Kumaraswamy generated family of distributions: theory and applications. J Appl Stat. 2019;46(16):2927–44. doi:10.1080/02664763.2019.1623867. [Google Scholar] [CrossRef]
8. Alshkaki R. A generalized modification of the Kumaraswamy distribution for modeling and analyzing real-life data. Statist, Optim Inform Comput. 2020;8(2):521–48. doi:10.19139/soic-2310-5070-869. [Google Scholar] [CrossRef]
9. Mahto AK, Lodhi C, Tripathi YM, Wang L. Inference for partially observed competing risks model for Kumaraswamy distribution under generalized progressive hybrid censoring. J Appl Stat. 2022;49(8):2064–92. doi:10.1080/02664763.2021.1889999. [Google Scholar] [PubMed] [CrossRef]
10. Alduais FS, Yassen MF, Almazah MM, Khan Z. Estimation of the Kumaraswamy distribution parameters using the E-Bayesian method. Alex Eng J. 2022;61(12):11099–110. doi:10.1016/j.aej.2022.04.040. [Google Scholar] [CrossRef]
11. Cho Y, Sun H, Lee K. Exact likelihood inference for an exponential parameter under generalized progressive hybrid censoring scheme. Stat Methodol. 2015;23:18–34. doi:10.1016/j.stamet.2014.09.002. [Google Scholar] [CrossRef]
12. Nagy M, Sultan KS, Abu-Moussa MH. Analysis of the generalized progressive hybrid censoring from Burr Type-XII lifetime model. AIMS Math. 2021;6(9):9675–704. doi:10.3934/math.2021564. [Google Scholar] [CrossRef]
13. Nagy M, Bakr ME, Alrasheedi AF. Analysis with applications of the generalized type-II progressive hybrid censoring sample from burr type-XII model. Math Probl Eng. 2022;2022(1):1241303. doi:10.1155/2022/1241303. [Google Scholar] [CrossRef]
14. Nagy M, Alrasheedi AF. The lifetime analysis of the Weibull model based on Generalized Type-I progressive hybrid censoring schemes. Math Biosci Eng. 2022;19(3):2330–54. doi:10.3934/mbe.2022108. [Google Scholar] [PubMed] [CrossRef]
15. Han M. The E-Bayesian and hierarchical Bayesian estimations of Pareto distribution parameter under different loss functions. J Stat Comput Simul. 2017;87(3):577–93. doi:10.1080/00949655.2016.1221408. [Google Scholar] [CrossRef]
16. Okasha HM, Wang J. E-Bayesian estimation for the geometric model based on record statistics. Appl Math Model. 2016;40(1):658–70. doi:10.1016/j.apm.2015.05.004. [Google Scholar] [CrossRef]
17. Yousefzadeh F. E-Bayesian and hierarchical Bayesian estimations for the system reliability parameter based on asymmetric loss function. Commun Statist-Theory Methods. 2017;46(1):1–8. doi:10.1080/03610926.2014.968736. [Google Scholar] [CrossRef]
18. Rabie A, Li J. E-Bayesian estimation for Burr-X distribution based on generalized type-I hybrid censoring scheme. Am J Math Manag Sci. 2020;39(1):41–55. doi:10.1080/01966324.2019.1579123. [Google Scholar] [CrossRef]
19. Yaghoobzadeh Shahrastani S. Estimating E-bayesian and hierarchical bayesian of scalar parameter of gompertz distribution under type II censoring schemes based on fuzzy data. Commun Stat–Theory Methods. 2019;48(4):831–40. doi:10.1080/03610926.2017.1417438. [Google Scholar] [CrossRef]
20. Nassar M, Okasha H, Albassam M. E-Bayesian estimation and associated properties of simple step-stress model for exponential distribution based on type-II censoring. Qual Reliab Eng Int. 2021;37(3):997–1016. doi:10.1002/qre.2778. [Google Scholar] [CrossRef]
21. Nagy M, Abu-Moussa M, Alrasheedi AF, Rabie A. Expected Bayesian estimation for exponential model based on simple step stress with Type-I hybrid censored data. Math Biosci Eng. 2022;19(10):9773–979. doi:10.3934/mbe.2022455. [Google Scholar] [PubMed] [CrossRef]
22. Balakrishnan N, Sandhu RA. Best linear unbiased and maximum likelihood estimation for exponential distributions under general progressive type-II censored samples. Sankhyã: Indian J Stat, Ser B. 1996;58(1):1–9. [Google Scholar]
23. Mohie El-Din MM, Sharawy A, Abu-Moussa MH. E-Bayesian estimation for the parameters and hazard function of Gompertz distribution based on progressively type-II right censoring with application. Qual Reliab Eng Int. 2023;39(4):1299–317. doi:10.1002/qre.3292. [Google Scholar] [CrossRef]
24. Dutta S, Kayal S. Estimation and prediction for Burr type III distribution based on unified progressive hybrid censoring scheme. J Appl Stat. 2024;51(1):1–33. doi:10.1080/02664763.2022.2113865. [Google Scholar] [PubMed] [CrossRef]
25. Dey D, Gosh M, Srinivasan C. Simultaneous estimation of parameters under entropy loss. J Stat Plan Inference. 1986;15:347–63. doi:10.1016/0378-3758(86)90108-4. [Google Scholar] [CrossRef]
26. Nasir W, Aslam M. Bayes approach to study shape parameter of Frechet distribution. Int J Basic Appl Sci. 2015;4(3):246. doi:10.14419/ijbas.v4i3.4644. [Google Scholar] [CrossRef]
27. Tummala VMR, Sathe PT. Minimum expected loss estimators of reliability and parameters of certain lifetime distributions. IEEE Trans Reliab. 1978;27(4):283–5. doi:10.1109/TR.1978.5220373. [Google Scholar] [CrossRef]
28. Han M. The structure of hierarchical prior distribution and its applications. Chin Oper Res Manag Sci. 1997;6(3):31–40. [Google Scholar]
29. Lindley DV, Smith AF. Bayes estimates for the linear model. J Royal Statist Soc Series B: Statist Methodol. 1972;34(1):1–18. doi:10.1111/j.2517-6161.1972.tb00885.x. [Google Scholar] [CrossRef]
30. Kohansal A. On estimation of reliability in a multicomponent stress-strength model for a Kumaraswamy distribution based on progressively censored sample. Stat Pap. 2019;60(6):2185–224. doi:10.1007/s00362-017-0916-6. [Google Scholar] [CrossRef]
31. Tu J, Gui W. Bayesian inference for the Kumaraswamy distribution under generalized progressive hybrid censoring. Entropy. 2020;22(9):1032. doi:10.3390/e22091032. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools