 Open Access
Open Access
ARTICLE
A Secured and Continuously Developing Methodology for Breast Cancer Image Segmentation via U-Net Based Architecture and Distributed Data Training
1 Department of Computer Science and Engineering, Brac University, Dhaka, 1000, Bangladesh
2 Department of Computer Science and Engineering, George Mason University, Fairfax, VA 22030, USA
3 Department of Software Engineering, College of Computer and Information Sciences, King Saud University, Riyadh, 11543, Saudi Arabia
4 AI and Big Data Department, Endicott College, Woosong University, Daejeon, 34606, Republic of Korea
* Corresponding Author: Jia Uddin. Email:
Computer Modeling in Engineering & Sciences 2025, 142(3), 2617-2640. https://doi.org/10.32604/cmes.2025.060917
Received 12 November 2024; Accepted 06 February 2025; Issue published 03 March 2025
Abstract
This research introduces a unique approach to segmenting breast cancer images using a U-Net-based architecture. However, the computational demand for image processing is very high. Therefore, we have conducted this research to build a system that enables image segmentation training with low-power machines. To accomplish this, all data are divided into several segments, each being trained separately. In the case of prediction, the initial output is predicted from each trained model for an input, where the ultimate output is selected based on the pixel-wise majority voting of the expected outputs, which also ensures data privacy. In addition, this kind of distributed training system allows different computers to be used simultaneously. That is how the training process takes comparatively less time than typical training approaches. Even after completing the training, the proposed prediction system allows a newly trained model to be included in the system. Thus, the prediction is consistently more accurate. We evaluated the effectiveness of the ultimate output based on four performance matrices: average pixel accuracy, mean absolute error, average specificity, and average balanced accuracy. The experimental results show that the scores of average pixel accuracy, mean absolute error, average specificity, and average balanced accuracy are 0.9216, 0.0687, 0.9477, and 0.8674, respectively. In addition, the proposed method was compared with four other state-of-the-art models in terms of total training time and usage of computational resources. And it outperformed all of them in these aspects.Keywords
In today’s world, there are many cutting-edge technologies available in the field of healthcare. In this field, computer science plays a crucial role in advancing healthcare care, helping to save lives by preventing and controlling the spread of diseases. There are many diseases that are still visible that are difficult to diminish. And many people across the world are facing suffering due to these certain diseases. Breast cancer is one of the most alarming illnesses facing many women in the world today. According to WHO statistics, around 2.3 million women suffered from this particular disease while 670.000 died from breast cancer in the year 2022 [1]. Many women had to die because they were unable to diagnose this issue early and it is necessary to do so early. Otherwise, the possibility of losing your life will be high. To enhance the chance of staying alive, the patient should take steps as early as possible [2]. As a consequence, we decided to do research on breast cancer image segmentation to detect breast cancer in women.
Currently, many image segmentation architectures have been used for the task of segmenting breast cancer images, and UNet is one of them [3,4]. Anari et al. showed a way for the task of breast tumor segmentation via combining two algorithms called Vision Transformer and UNet [5]. Their approach is efficient and reduces the computational complexity. However, it is unable to ensure the data privacy of the users. Besides, they have not mentioned their required time for training data. So, we will not be able to fulfill the motive of ensuring data privacy and reducing the training time by following their proposed way. Additionally, another research project on biomedical image segmentation was conducted by Weng et al., and it is called INet [6]. Though their approach reduced the amount of parameters, it failed to provide confidentiality of the data. Besides, they have not disclosed anything regarding the required time for data training. Moreover, several machine learning techniques called Support Vector Machine, K-Nearest Neighbors, random forest, and Naive Bayes can be used for the breast cancer image analysis task [7].
In this research, the UNet-based architecture was used to predict output. Unlike the typical method of UNet, a unique way was followed for making the platform for breast cancer image segmentation. The objective of this research is to create a platform for breast cancer image segmentation that does not need high computational demand and takes comparatively less time to train. Besides, the data privacy will also be ensured. Because there are many instances where the privacy of data is required and people may not provide data for training purposes due to privacy issues. Because of this certain reason, the data will remain confidential to humans in the proposed system. Moreover, a phenomenal feature is also available here called Continuously Developing. By including this facility, the performance in terms of result prediction will be developed day by day. However, it is not possible to fulfill all these demands by following the typical prediction concept of the UNet. As a consequence, this research has been conducted to invent a new way of predicting breast cancer image segmentation.
Due to the ability to be trained faster and also train in low amounts of data, this system has a high potential to provide medical image segmentation. Clinically, this approach can be particularly beneficial in medical imaging, where accurate and timely segmentation of breast cancer images is critical for diagnosis and treatment planning. The ability to train models with smaller datasets and reduced computational demand allows for more widespread adoption in resource-constrained environments, improving diagnostic capabilities in underserved regions or smaller healthcare centers. Moreover, the ability of the method to consistently improve prediction accuracy with new datasets ensures that the system remains up-to-date and reliable over time, potentially leading to better clinical decision-making and improved patient outcomes.
The major objective is to invent a platform for breast cancer image segmentation via UNet architecture. However, the job of training data for image processing is complicated due to the need for powerful machines. Therefore, this process is introduced when the need for managing high-performance machines has diminished. To accomplish this motive, the data were separated into a few segments, and each segment was trained separately. In the case of predicting output, the output will be predicted from each trained model, and the ultimate output will be selected based on the pixel-wise majority voting of the predicted outputs [8]. Here, it is possible to train each model separately on different machines, which makes the process faster. That is how the objective of faster training was completed. Along with the mentioned purposes, this research had another objective of protecting data security. As the entire data will not stay in one place, the data will stay confidential. So, overall, by following the proposed algorithm, any person can train a model with a low-powerful machine in comparatively less time by using several computers at once, and data will also be secured. Along with these, the platform will be consistently developed in the sector of providing accurate output. In our research, we adopted a distributed training approach rather than training the entire dataset together. While this method performs well on the specific dataset used in the study, its effectiveness may vary when applied to other datasets, particularly those from different sources. The results could be less reliable in such cases due to variations in imaging conditions, demographics, or equipment. However, one of the key advantages of our proposed method is its ability to fix this challenge. With our approach, it is possible to integrate new training models based on new types of data without retraining the entire system. For example, if new varieties of ultrasound images are introduced, we can train models on these specific images separately and include this trained model in the existing system. That is how the proposed system adopts new data types without reprocessing all the previously trained data.
Image processing is too difficult for those people who do not have high-configuration computers. To train the model for image processing, a powerful Graphics Processing Unit (GPU) is needed, which is very expensive. Lots of people around the world hardly get such a GPU. U-Net performs phenomenally in medical image segmentation tasks. However, it has several limitations. For instance, a large amount of labeled data is needed to train any U-Net model. But there are some cases in which it is difficult to fulfill this task. Along with this, UNet is computationally expensive. As a result, the distribution method was followed to train data. That is how the necessity of managing the powerful machine to train the model for image processing is wiped out. On the other hand, medical data is so sensitive, and data privacy is required to ensure in such cases. As the models were trained with data separately, no one will access the entire data. Here, just trained models will be needed. Another problem may occur in this system. Since the data will not be trained at once and the training process will occur multiple times, it will take comparatively more time. In this case, it is possible to use multiple computers to eliminate this certain issue. Thus, it can train all the models faster. Let’s assume the data are divided into four segments and four computers are managed. If four models are trained with the four segments of data simultaneously, then it will not take much time and significantly less time than the traditional training method.
Motivated by the earlier work, Xu et al. [9], in this study, have reviewed federated learning technologies in the healthcare field. Because of having the ability to keep data secure, federated learning is being used in the healthcare sector. Unlike the typical machine learning approach, in federated learning, all the data is trained in a local model, and all the models are aggregated finally. In the global model, the average training weight is available instead of data. That is how it is feasible to keep data secure via federated learning. Moreover, Acar et al. presented a survey that provides an overview of homomorphic encryption [10]. By following the homomorphic encryption, it is possible to do any computational task without decrypting the data. Thus, data security is ensured by following this algorithm. By getting knowledge from the above two methods, this research for the breast cancer image segmentation task has been conducted. Here, a system of training via UNet has been proposed where data will stay secure. Below, the contributions of this research have been described:
• One of the most unique features of the proposed model is that it takes comparatively less time to train the model for the image segmentation task. This paper presents the comparison results of total training time with four other state-of-the-art models: UNet, Attention UNet, ParseNet, and Fully Convolutional Neural Network (FCNN). From the comparison chart, it is visible that the proposed approach takes 3.65 min to complete the training with the Tesla T4 GPU, whereas the other four models take from 7.20 to 12.81 min with the same data and the same machine.
• Additionally, the proposed method of image segmentation also needs fewer computational demands than typical state-of-the-art approaches. So, to prove that the experimental results of the minimum requirements of computational resources like Random Access Memory (RAM) and GPU of the proposed method and four other state-of-the-art models are presented in this paper. In this experimental research, it is proved that the invented approach of this research takes 3014.89 MB, whereas the rest need at least 3149.79 MB and at most 3382.66 MB. Like Random Access Memory (RAM), the four mentioned state-of-the-art models utilized more GPU than the proposed method. The proposed method utilized 88.00% in the Tesla T4 GPU of Google Colab, and the remaining four others need from 98.00% to 99.00%.
• Unlike the typical systems, the entire data is not trained at once, while it is feasible to use multiple machines to train all data. Thus, people will not access the entire data, and the data will stay secure. Because in this research only trained models are needed to make the prediction system for the breast cancer image segmentation task. This paper presents the effectiveness of the proposed method by evaluating it with some performance matrices: average pixel accuracy, mean absolute error, average specificity, and average balanced accuracy.
• Moreover, to explain the results from the trained model, the t-SNE (t-distributed Stochastic Neighbor Embedding) is used in this experiment. In this experiment, the entire data set was divided into a few parts, and each part was trained separately. This paper presents the t-SNE (t-distributed Stochastic Neighbor Embedding) of each trained model. The t-SNE (t-distributed Stochastic Neighbor Embedding) with the most data indicates the capability of separating classes appropriately. However, the model that has very little data has shown its potential to perform phenomenally with much data. Overall, the tSNE illustrates that the proposed architecture of UNet and the dataset are good enough to make a platform for breast cancer image segmentation.
The rest of the paper is organized as follows: Section 2 covers the background study of this research. In Section 3, the methodology of making the proposed breast cancer image segmentation system is explained. Following that, Section 4 covers the discussion of the experimental results, while Section 5 provides the conclusion of the paper.
In this section, we analyze relevant papers and research related to our study. Moreover, the architecture used in the proposed model is discussed.
In this research, the UNet was used to train the model [11]. This architecture is currently a highly used method for medical image segmentation tasks. This architecture consists of three segments, which are encoder, decoder, and bridge [12]. Here, the bridge works for connecting the encoder block with the decoder block via the skip connections. There are two types of fundamental operations visible in UNet. These are downsampling and upsampling. The downsampling is visible at the encoder part, while the upsampling works in the decoder part. There are typically an equal number of convolutional layers available in each segment. In the middle of the two parts, there is a layer visible named bottleneck. The encoder function works to fetch significant information from the input. The dimension of the feature map consistently decreases in the encoder by going through the downsampling. A method called max-pooling is used to reduce the dimension of the feature map. On the other hand, the feature became semantically rich in the deeper layer. While the opposite scenes are visible in the segment of the decoder. In the decoder phase, the spatial dimensions start to reconstruct from the deepest segment, and when it arrives at the top layer of the decoder, then the map gets its actual size with semantic information, which is obtained from the encoder level. The upsampling operation is responsible for accomplishing this task. The same layer of encoder and decoder is concatenated with each other via a bridge. It is helpful to keep a record of spatial details for each layer. Because after downsampling, the spatial details of the previous layer are removed. But due to the bridge between the same layer of encoder and decoder, it becomes possible to track such information [13].
Here’s a detailed breakdown of the key equations are given:
This equation represents the feature map of layer
Here,
This operation helps preserve the most important features while discarding spatial information, making the model more robust to variations in input.
Here,
Here,
Upsampling restores the spatial resolution of the feature maps, allowing the model to reconstruct the image dimensions to match the original input.
The concatenation operation (
After concatenation, the feature map undergoes a final convolution operation in the decoder, producing the final output of the U-Net architecture.
The final convolutional operation
Federated learning is one kind of machine learning approach [14]. Unlike typical machine learning methods, in federated learning, all data are not trained not once. Instead, all the data are divided into several segments, and each segment is trained separately. After completing the task of training all the segments, the training weights of these segments are averaged. Each segment is known as a local model, and the model where the average weight is set is known as a global model. After setting the average weight of local models in the global model, if the performance of accuracy and loss is poor, then this loop continues one more time. This loop continues until the performance is satisfactory. This loop is known as the communication round. Here, instead of data, the weight of the model will be passed in the central global model. That is how the data privacy is ensured. Along with data security, it plays essential roles in reducing the need for large data transfers. Currently, there are many sectors where federated learning is being used. However, people prefer to use this framework in medical data training. Because here the data is quite sensitive, it is needed to ensure privacy. Besides, in the finance sector, federated learning can be an amazing choice.
Transfer learning is one kind of machine learning technique where it is not needed to build a neural network from scratch to build a model [15]. Unlike typical machine learning algorithms, transfer learning allows a model to be partially trained using a pre-trained model. Therefore, by using transfer learning, we can reduce the time for training a model for any AI-related task. Since transfer learning is capable of being trained from the pre-trained model, it is not needed to collect much of the data that is required in typical machine learning methods. Along with this, the performance is quite phenomenal in transfer learning. Therefore, there are many instances where this method is being used. The two major sectors are computer vision and natural language processing, where transfer learning is being used much. Several deep learning models, such as VGG16, VGG19, Inception v3 (for computer vision), and GPT (for NLP), are frequently used in transfer learning workflows.
Michael et al. disclosed three types of image segmentation methods, where the first one is classical segmentation [16]. Machine learning segmentation is the second category out of three categories. While the last group is supervised, unsupervised, and deep-learning segmentation. According to their study, classical methods and machine learning methods are used for regional-based segmentation and machine learning segmentation, respectively. In the mammogram image segmentation, UNet is utilized.
Zhang et al. disclosed a way to identify breast cancer by following four steps and also explained the process to breast cancer diagnosis using some deep learning-based algorithms [17]. Moreover, some possible ways to do this task are also mentioned by the authors.
Abbas et al. effectively highlighted potential use cases in ISM band operations, which are critical for industrial automation, healthcare devices, and communication systems [18]. They use dilation and erosion for segmenting mammograms to detect breast cancer. By applying different structuring elements (3 × 3 to 15 × 15), the authors compute the morphological gradient. According to MSE and PSNR values, a 15 × 15 disk provides the best result by developing the image quality.
Debelee et al. have provided some statistics in the sector of breast cancer image analysis [19]. In their research, the authors have disclosed several current databases of breast cancer and reviewed some images of breast cancer. Along with this, some deep learning-based algorithms have been reviewed for analyzing medical images.
Zhang et al. have reviewed several facilities for using the concept of AI in breast cancer image segmentation [20]. They summarize the feasibility of using large-scale databases for robust training and diminish the problems that usually occur in AI-based software.
Liu et al. have proposed an idea of image segmentation of breast cancer mammography images using interval analysis [21]. They have developed the Laplacian of Gaussian (LoG) filter by adding interval-based modifications, effectively combining classic image processing with advanced tools to handle uncertainties. The authors have disclosed the issues that may arise due to growing selection ranges in interval analysis. They have not provided any solution for this problem. This lack of practical strategies reduces the feasibility of using the method, especially in situations where uncertainty is significant and needs careful management.
Hossam et al. have presented a novel approach to detect breast cancer based on ROI images of breast thermograms [22]. To train the model for detection purposes, they used SVM and ANN, while SVM performed better than ANN in terms of accuracy. The accuracy of SVM is 96.67%, and ANN provided 96.07%.
Huang et al. have summarized various ways of breast cancer ultrasound image segmentation [23]. They have highlighted issues like speckle noise and low contrast that are quite significant for computer-aided diag- nosis (CAD) systems. However, the datasets they have used are not well enough it limits the generalizability of their findings.
Xian et al. have reviewed various methods for breast ultrasound segmentation, including graph-based models, deformable models, learning-based methods, and classical techniques like thresholding [24]. The paper mentions the potential of deep learning but does not explore challenges like the need for large datasets or ensuring results are interpretable.
Almajalid et al. have shown a novel method for the image segmentation task using breast cancer ultrasound images and UNet architecture [25]. The authors have mentioned the issues of accurately identifying breast tumors in ultrasound images, essential for early cancer detection. However, the dataset that they have used is quite small. Such a limited dataset is insufficient for robust training and cannot be recommended for real-time applications. The results that they presented were on a single dataset with no external validation on independent datasets.
Mazouzi et al. have implemented a distributed system to train the model for image segmentation in a faster way [26]. They used a region-based approach for this kind of distributed training. In this research, they used two datasets, which are the ABW dataset and the Object Segmentation Database (OSD). Eventually, they proved their approach was faster by providing several evaluation performances.
Szénási et al. have provided a unique way to do the task of image processing via the region-growing method, which is implemented on CPUs and GPGPUs [27]. They compared traditional and distributed algorithms. They have found that the distributed approach is much faster with minimal loss in accuracy. However, the dataset used for testing (15 tissue images) is relatively small and lacks diversity. The results might not generalize well to other types of tissues or imaging modalities.
Bian et al. have introduced a modified UNet architecture called Blind UNet, which can train data for image segmentation while maintaining the confidentiality of the user’s data [28]. This design reduces the cryptographic workload and makes inference 14 times faster, with minimal impact on accuracy.
Deng et al. have provided an incredible method of image segmentation that programs better than the current image encryption algorithms [29]. It is very sensitive to key changes. That is why sometimes making decryption may not be feasible with small variations. Besides, the method has not been fully tested on RGB or multi-channel images, so it may not be suitable for modern multimedia use.
Marwan et al. have proposed a novel framework for medical image segmentation [30]. By maintaining the security of the medical data, it will be possible to do the task of image segmentation, and performance is also phenomenal.
Schutte et al. have presented an approach to detect breast cancer [31]. Besides, the authors have discussed 61 deep-learning methods for this certain task.
Tekin et al. have presented a new framework for breast cancer image segmentation, and they named it Tubule-U-Net [32]. In this framework, there is an encoder and a decoder part available. In the encoder, they experimented with EfficientNetB3, ResNet34, and DenseNet161, while UNet was used in the decoder segment with these three types of encoders separately. These three experiments provided promising results.
Wakili et al. have demonstrated an incredible way for the breast cancer image segmentation task by using a unique approach with comparatively lower computational demands than existing well-known approaches [33]. In their proposed approach, they used both transfer learning and DenseNet. Here, transfer learning was used to address the issues arising from feature extraction across the same distribution via DenseNet. In Table 1, a concise summary of the employed models, datasets, and limitations for several of the above-mentioned literatures is disclosed.
The Breast Ultrasound Images Dataset is used in this research to make the proposed breast cancer image segmentation system [34]. In this dataset, there are a total of 780 images, with their corresponding masks also available. These ultrasound images of the breast were collected from 600 female patients in the year 2018. The patients’ age range spans from 25 to 75 years, offering a diverse dataset. This diversity enables the model to effectively identify regions in ultrasound images across women of all ages. Here, a total of 780 data points were available for a total of 600 patients. The dataset is robust enough, and we use this dataset to justify the effectiveness of our proposed approach. The average image size of data is 500 × 500 pixels. The entire dataset is divided into three classes, which are benign, normal, and malignant. Due to this categorization, it will be quite uncomplicated for the model to predict accurate output for any image processing task. The total images of benign, normal, and malignant are 437, 133, and 210, respectively. The dataset represents real-world medical images, and so this dataset is highly relevant for developing machine- learning models for breast cancer image segmentation tasks. Preprocessing is a crucial part of training a model. This dataset is well suited to train any model easily due to its standard resolution and constant size of all images of the dataset. The dataset used in the research has been approved for research purposes. This dataset contains only ultrasound images. No patient information or any other confidential data is included. Therefore, any researcher can use this dataset for research purposes without hesitation. However, this dataset has a limitation. In image segmentation, the class imbalance (437 benign, 133 normal, 210 malignant) can lead to the model focusing on the majority class (benign) and underperforming on minority classes (normal, malignant). This can cause issues like over-segmentation of benign areas and under-segmentation of smaller classes.
This research has been done in several stages, where the first task was collecting data. After collecting data, the data have been preprocessed and trained, respectively. Following that, the prediction system was made eventually. In Fig. 1, the overview of this research is given.
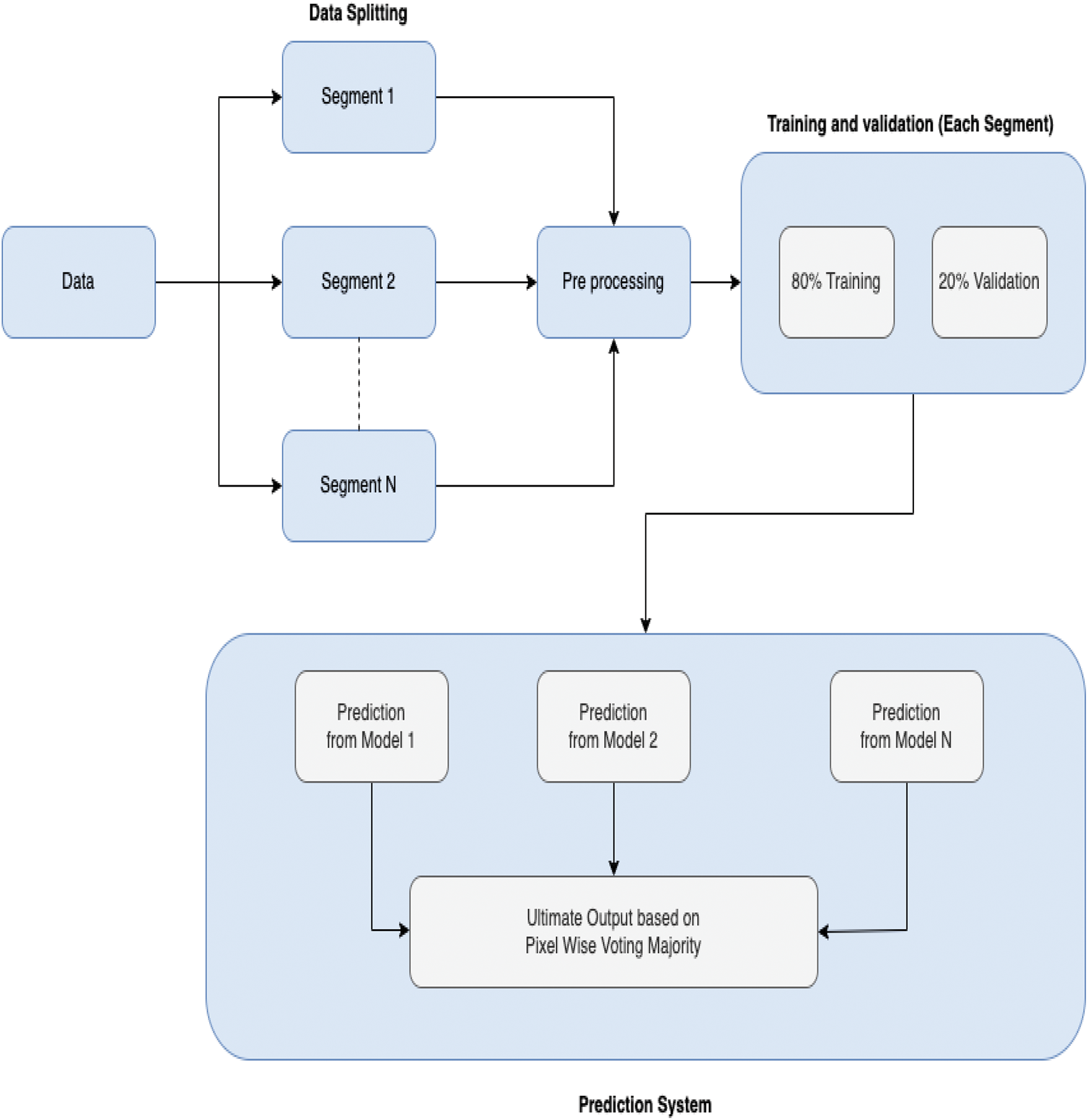
Figure 1: Overview of research
3.1 Data Collection and Splitting
The dataset of breast cancer ultrasound images has been collected from Kaggle. In this dataset, there are a total of 780 images, with their corresponding masks also available. These ultrasound images of the breast were collected from 600 female patients in the year 2018. The age range of patients is between 25 and 75 years. The data was acquired by the LOGIQ E9 ultrasound and LOGIQ E9 Agile ultrasound systems. Here, a total of 780 data points were available for the 600 patients. The dataset is robust enough, and we use this dataset to justify the effectiveness of our proposed approach. However, it is recommended to be concerned about the inconsistencies in patient numbers and potential data leakage in real-world projects. The average image size of data is 500 * 500 pixels. The entire dataset is divided into three classes, which are benign, normal, and malignant. The total images of bengin, normal, and malignant are 437, 133, and 210, respectively. The total of 780 data were divided into 3 segments, where each segment was allocated with 100, 300, and 380 data, respectively. In each segment, 20% of the entire data was allocated for the validation while the remaining 80% was allocated for training [35].
All the data of the dataset are divided into three classes: normal, benign, and malignant. Therefore, in Fig. 2, sample images of three classes are given. Along with input and output, the union of input and output is also provided for each data. The motive of providing this kind of union is to show the cancer-affected region of the input nicely. There are three samples given where the first sample is the data of the normal patient, and the remaining two samples have presented the ultrasound images of benign and malignant classes, respectively.
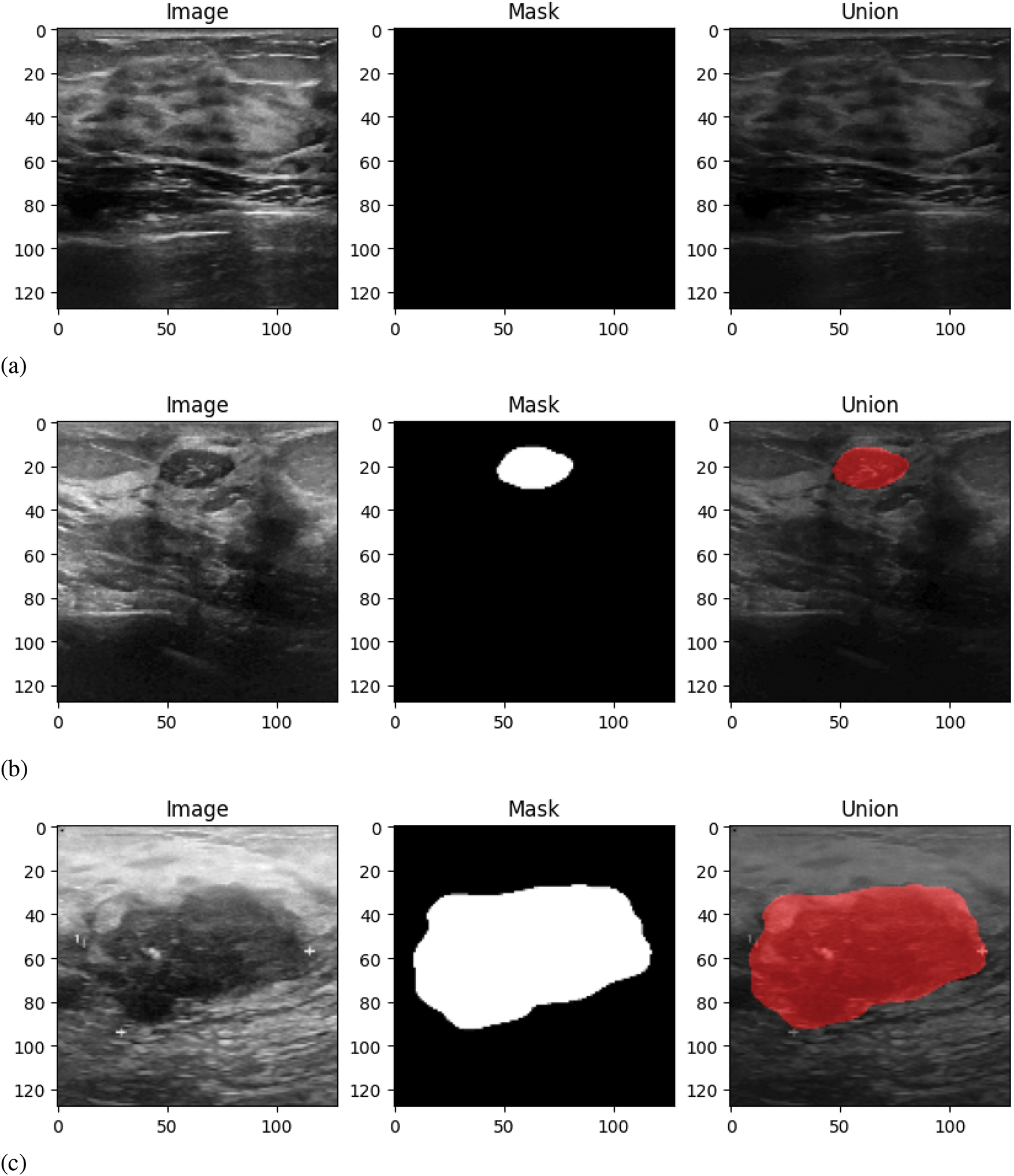
Figure 2: Sample data: (a) Normal class, (b) Benign class, (c) Malignant class
As in the proposed prediction system, the output will be predicted from each trained model, and the ultimate one will be selected using the proposed prediction algorithm of this research. As a result, it is not guaranteed that all trained models will have been trained with nearly equal amounts of data. To address this, we plan to train models using three different data sizes to evaluate the performance of the proposed system when models are trained on varying amounts of data. This approach provides insights into how the system works with different data.
Besides, for testing purposes, the data have been managed from external resources. From this external separate dataset, 210 images have been taken for testing purposes [36]. The 210 images of testing data are the modified training data. The testing data quality is modified by transforming it into various forms, like rotation in multiple angles. Due to this, the diversity and robustness of data are increased. Besides, each image is available in different sharpness levels that are beneficial for the model in any training and testing task.
After dividing the entire data into several segments, the data of each segment was preprocessed and trained separately by following the same methods.
First, all the inputs were resized into a uniform input with the dimension of 128 × 128 pixels, and the value of the channel was assigned as 1 [37]. Because the input images are converted to grayscale during preprocessing [38]. Because of the resizing of the input images, the training occurs more effectively.
Like inputs, it was needed to preprocess the outputs as well. To do this, the outputs were resized to be the same as the inputs, which are 128 × 128 pixels. Here, the number of channel is one. The shape of each output is (128, 128, 1). The single channel was used for the binary masks. This binary value was used to identify the region where a mask is available. In the region where the mask is available, it is assigned as 1, and except for this part, all the parts are assigned as 0.
Here, by resizing, all the inputs will have the same dimension, which is highly needed in any deep learning training. The smaller resolution can play a crucial role in faster computation. Along with this, grayscale conversion will reduce the data dimensionality. Eventually, the output mask resizing ensures the mask matches the input image size for accurate pixel-by-pixel comparison during training.
3.3 Model Training and Creating Prediction System
Like the preprocessing, the model architecture used for the training model via UNet is the same for each segment. In the U-Net model, there is an encoder path with four blocks. Each block consists of two convolutional layers and max pooling for downsampling. The bottleneck has two layers of convolutions, each with 1024 filters. On the other hand, the decoder path upsamples the feature maps using transposed convolutions and incorporates skip connections from the encoder. Finally, the final output layer uses a single filter with sigmoid activation for binary segmentation. Here Adam Optimizer, Binary Crossentropy Loss, and accuracy matrices were used. Binary Crossentropy Loss yielded excellent performance for our dataset. In this experiment, this loss function facilitated better convergence and performance in distinguishing the relevant features, leading to strong results in segmentation tasks. In Table 2, the model summary is given.
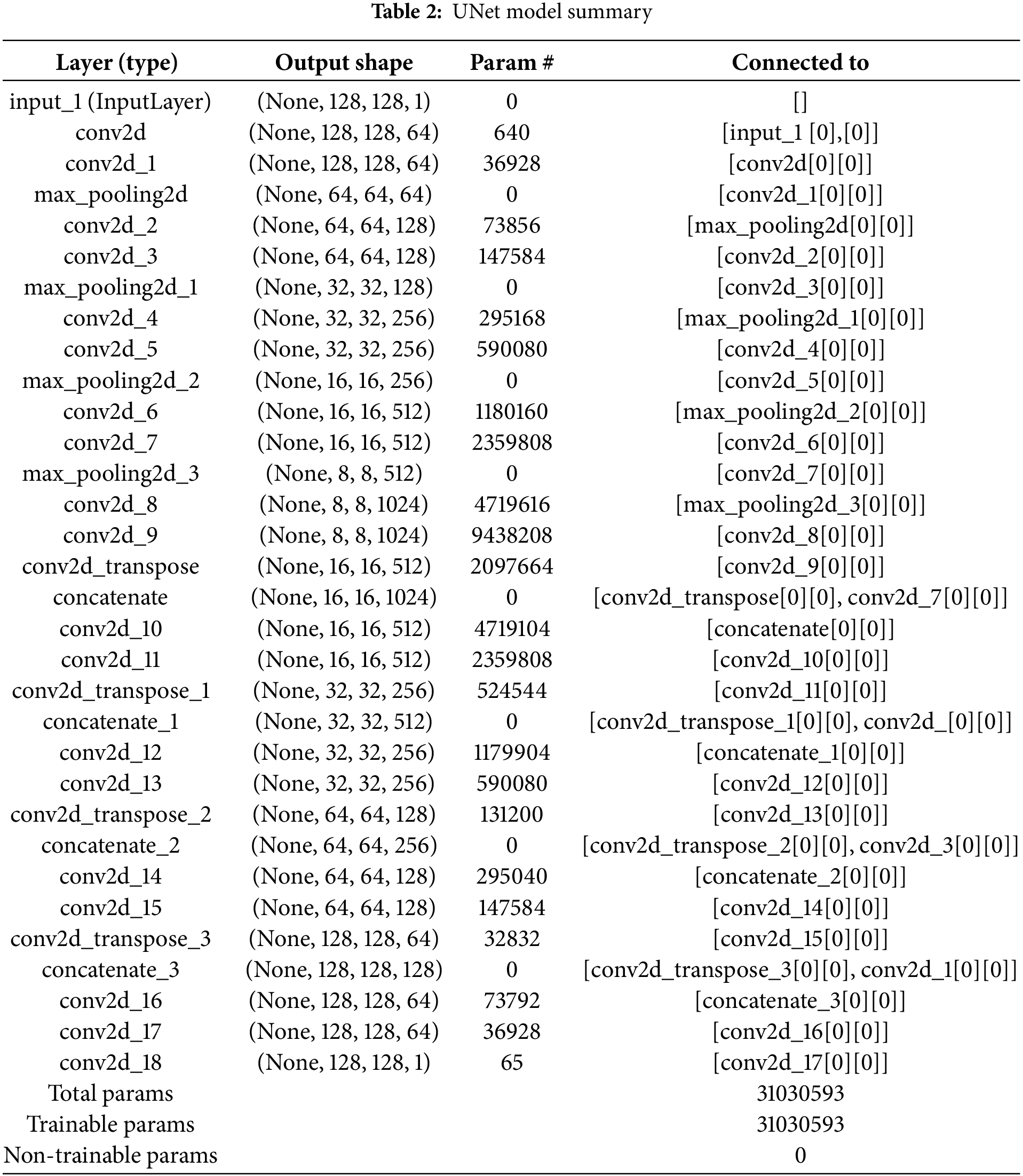
In this model summary, it is visible that there are mainly parts available in the UNet architecture are the encoder and decoder, respectively. And these two major parts are connected via a bottleneck. In this table, the first layer is the input layer, and it is visible that this model accepts the grayscale image with the 128 × 128 shape. After the input layer, the encoder layers consist of a series of convolutional and max-pooling layers. The max-pooling layers are used to perform downsampling. Due to downsampling, the spatial resolution of the input image is decreased and the depth of the feature maps is increased. Therefore, the amount of filters in the first encoder layer is 64, while 512 filters are visible in the last encoder layer. The bottleneck is the deepest layer of this UNet with a spatial resolution of (8, 8) and 1024 filters. Following the bottleneck, the layers of the decoder are started, where the opposite scenario of the encoders is visible. Here the spatial dimensions are increased and feature maps are decreased in each layer. In the initial decoder layer, the spatial dimension is (16, 16) and (128, 128) in the last layer. However, the number of filters is decreased in each layer of the decoder from 512 to 64. Due to the upsampling, these activities occur. Each encoder layer is connected to its corresponding decoder layer via skip connections. And that is why fine-grained details are preserved. The final layer is the output layer that outputs the final segmentation map with shape (128, 128, 1).
After completing the task of data training, the task of creating a prediction system was started. In Fig. 3, an overview of our prediction system is given. When the user gives an input, the output will be predicted from each model. To predict the output for an input, it is needed to preprocess the input. Initially, the input image is resized according to the trained data and converted to grayscale. Following that, the new dimension is added with the input image. That means it is assumed that the shape of the input image is (height, width, and color channels). After adding the batch dimension, the shape will be (batch size, height, width, and color channels). Then, the output will be predicted from each trained model. After obtaining predictions from each trained model, the predictions for each pixel position
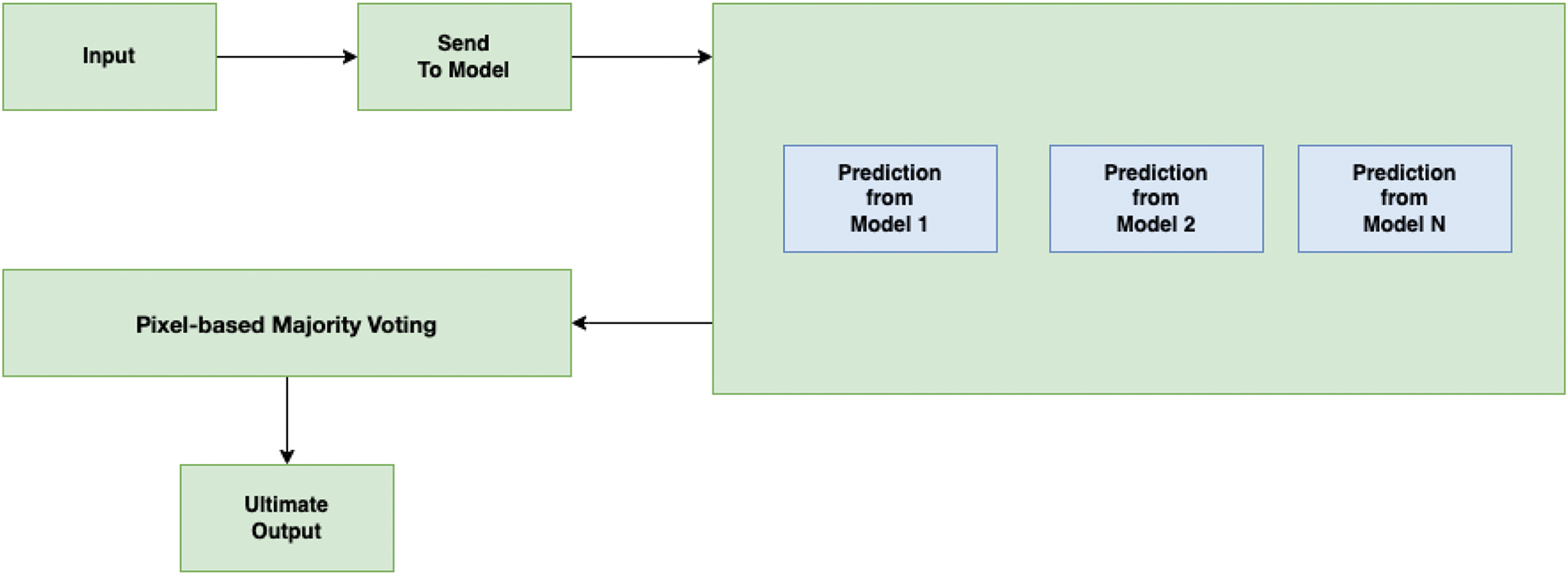
Figure 3: Overview of proposed output prediction system
The mode function returns the most frequent prediction among the N models for each pixel position
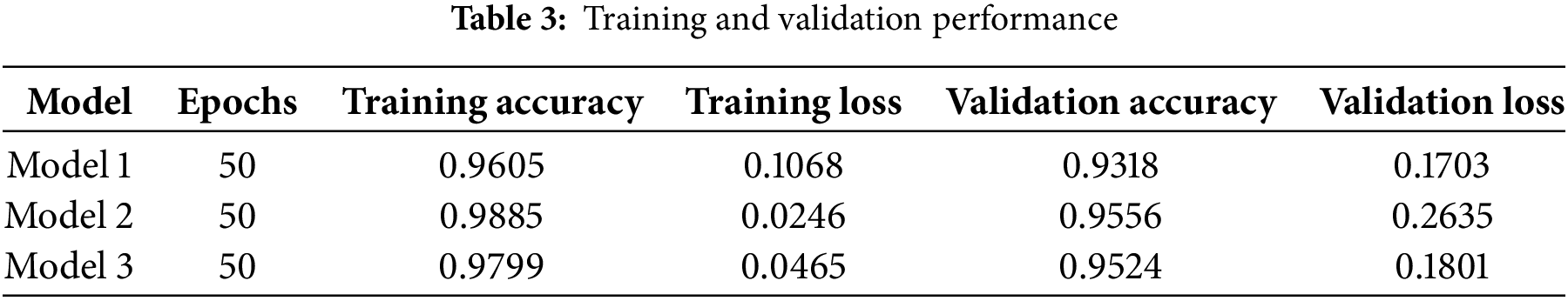
In this research, a unique way has been followed for the task of image segmentation. To evaluate the performance of this proposed methodology, it has not only focused on the testing performance but also has been concerned about the training performance. Each model has been run up to 50 epochs, and in Table 3, the training and validation results of the models have been provided. The performance metrics recorded include training accuracy, training loss, validation accuracy, and validation loss. Here, it is visible that each model provided outstanding performance.
Like training and validation, testing evaluation is also quite significant for measuring the capability of any model. In this proposed system, output will be predicted from multiple models, and the ultimate output will be selected based on the pixel-wise voting majority. Therefore, it is needed to follow a strategy, unlike most of the testing evaluation process. In the testing phase, the data from an external dataset was utilized. Due to this reason, it was not possible to employ cross-validation methods such as k-fold. Therefore, different approaches were used to measure the effectiveness of the proposed prediction system.
The perfectness of the ultimate outputs has been evaluated based on the four performance matrices. These are average pixel accuracy, mean absolute error, average specificity, and average balanced accuracy. In Table 4, the results of these matrices are provided.

Average pixel accuracy is one kind of performance matrix that is required to calculate the capability of predicting the pixels in the image correctly. It calculates the percentage of pixels that are correctly classified when comparing the model’s predicted output to the actual ground truth mask. We assumed the predicted output from the model of all data as the actual predicted output. Average pixel accuracy has been tested with 210 images of an external dataset and got a result of 0.9216.
Mean absolute error (MAE) is one kind of metric that calculates the perfectness of model prediction by averaging the differences between each predicted pixel value and its corresponding true pixel value [39]. By experimenting with the data of 210 images from the external dataset, we got an average MAE of 0.0687. The value is quite low. It indicates the difference is not so much. It can be said that our proposed prediction system performs well.
Average specificity is a metric that calculates the model´s ability to fetch negative pixels [40]. The proposed prediction system has got Average Specificity of 0.9477.
Average Balanced Accuracy means calculating the effectiveness of the model by averaging the predicting ability of negative and positive results [41]. We got 0.8674 in average balanced accuracy.
As shown in Table 3, it is visible that each model provided outstanding performance. Each model was able to secure more than 90% training and validation accuracy, while the amount of loss is quite low as well. It indicates effective learning. After having a look at Fig. 4, it is visible that the difference of training and validation results is not quite much in the three models. It can be disclosed that the training models are capable enough to predict accurately not only with the known data but also with new data. It also can be mentioned that these models do not have any issues like overfitting and underfitting. Because when the difference between training and validation is so high, then overfitting occurs. But this is not visible in this case. Therefore, it can be said that overfitting does not happen here. On the other hand, when the training and validation performance is poor, then underfitting happens. Such a situation is not also visible here. Therefore, it can be said that all the models are free from overfitting and underfitting.
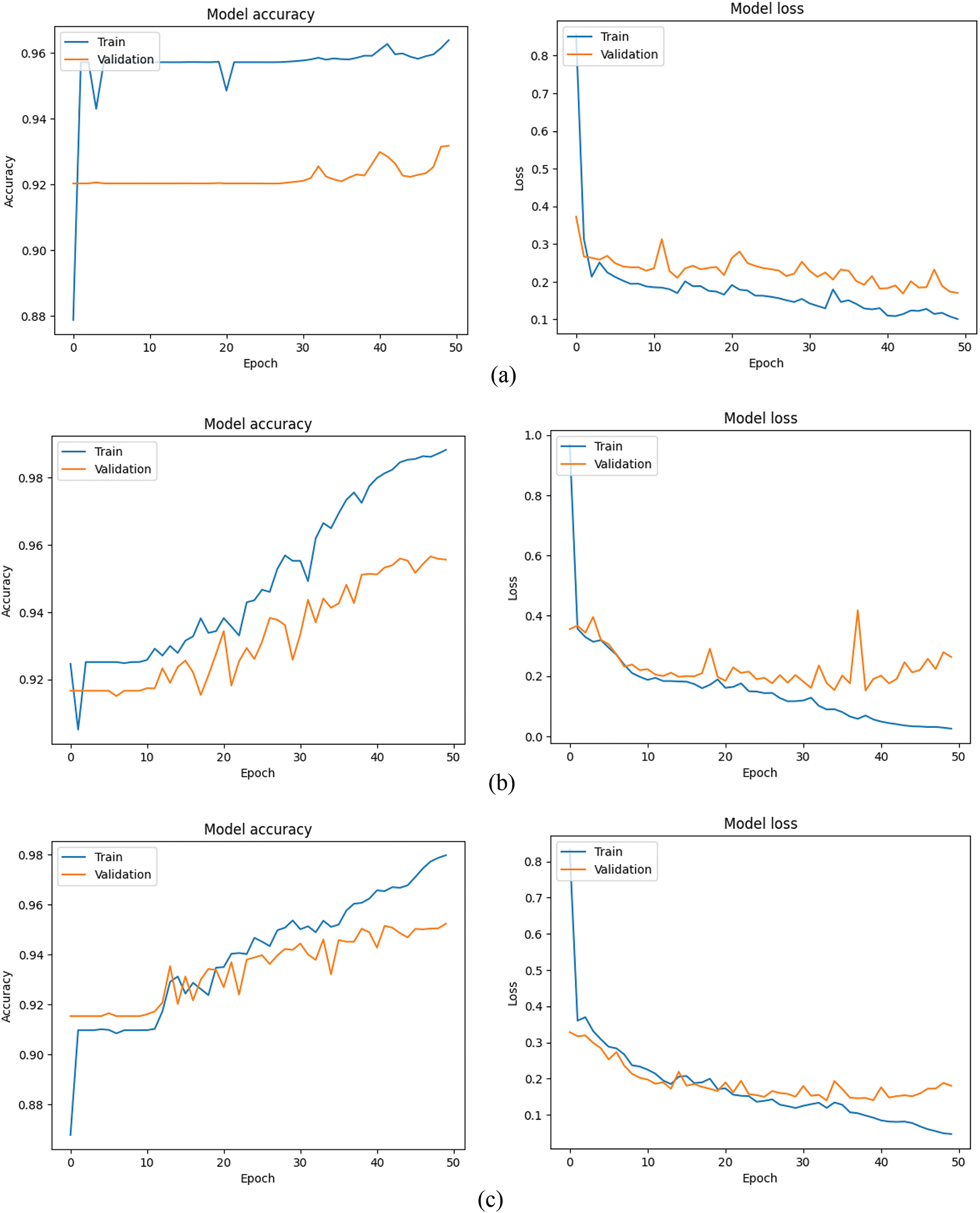
Figure 4: Training and validation performance of all models: (a) Model 1, (b) Model 2, (c) Model 3
In Fig. 5, the tSNE of three models has been given. In this figure, it is visible that the tSNE of the third model has the most of data among the three segments and provides the best performance among all. From Fig. 5, we can see a clear separation between different classes in the tSNE of the third model. Therefore, it can be disclosed that the third model can learn robust features. On the other hand, instead of having just 300 data, the second model is capable of learning effectively and generalizing from low data. Though the first model was trained with only 100 data, the Fig. 5 proves that this model has potential, and adding more data can improve its performance.

Figure 5: tSNE of models: (a) Model 1, (b) Model 2, (c) Model 3
In Table 4, the results of four performance matrices that are used for evaluating the testing performance are insane. The result of average pixel accuracy is 0.9216. That means 92.16% of the pixels of predicted output from the proposed method are similar to the predicted output of the trained model with all data. On the other hand, MAE is 0.0687. The ultimate predicted results are almost the same as the actual result. The remaining two metrics, average specificity and average balanced accuracy, also provided incredible performance. Therefore, it can be said that the proposed prediction method is good enough.
According to average pixel accuracy, 7.84% of predictions are errors in terms of the model that is trained with all data. The amount is quite low. This wrong prediction might happen due to subtle misclassifications in complex regions of the image, such as boundaries or areas with similar features. However, this performance can be developed more if it is possible to train in the dataset with balanced classes. Because in an imbalanced dataset, there is typically a bias toward the majority classes. Because during training, models tend to concentrate on the majority class. Therefore, the performance is comparatively better in the balanced classes dataset than imbalanced classes dataset. Besides, the MAE value indicates the low error availability in the predictions. However, it is also feasible to mitigate this low amount of error by working on a dataset. By reducing the presence of noise in the dataset, it is possible to decrease the value of error. On the other hand, the values of average specificity and average balanced accuracy are quite outstanding, which indicates the presence of low errors in the predictions. However, it is also possible to decrease the amount of error by working on the dataset. For instance, making balance on the dataset, reducing the noise of the dataset, and handling missing values more effectively.
In Fig. 6, sample outputs for 5 inputs are given. In Fig. 6, along with providing the ultimate output, the predicted output from each model is also given. In the proposed system, the output will be predicted from the trained models of each segment, and the ultimate output will be selected based on pixel-wise voting majority. By looking at Fig. 6, it is visible that the ultimate output is almost similar output with the predicted output from the trained model of all data. Therefore, it can be claimed that the proposed prediction system works appropriately.
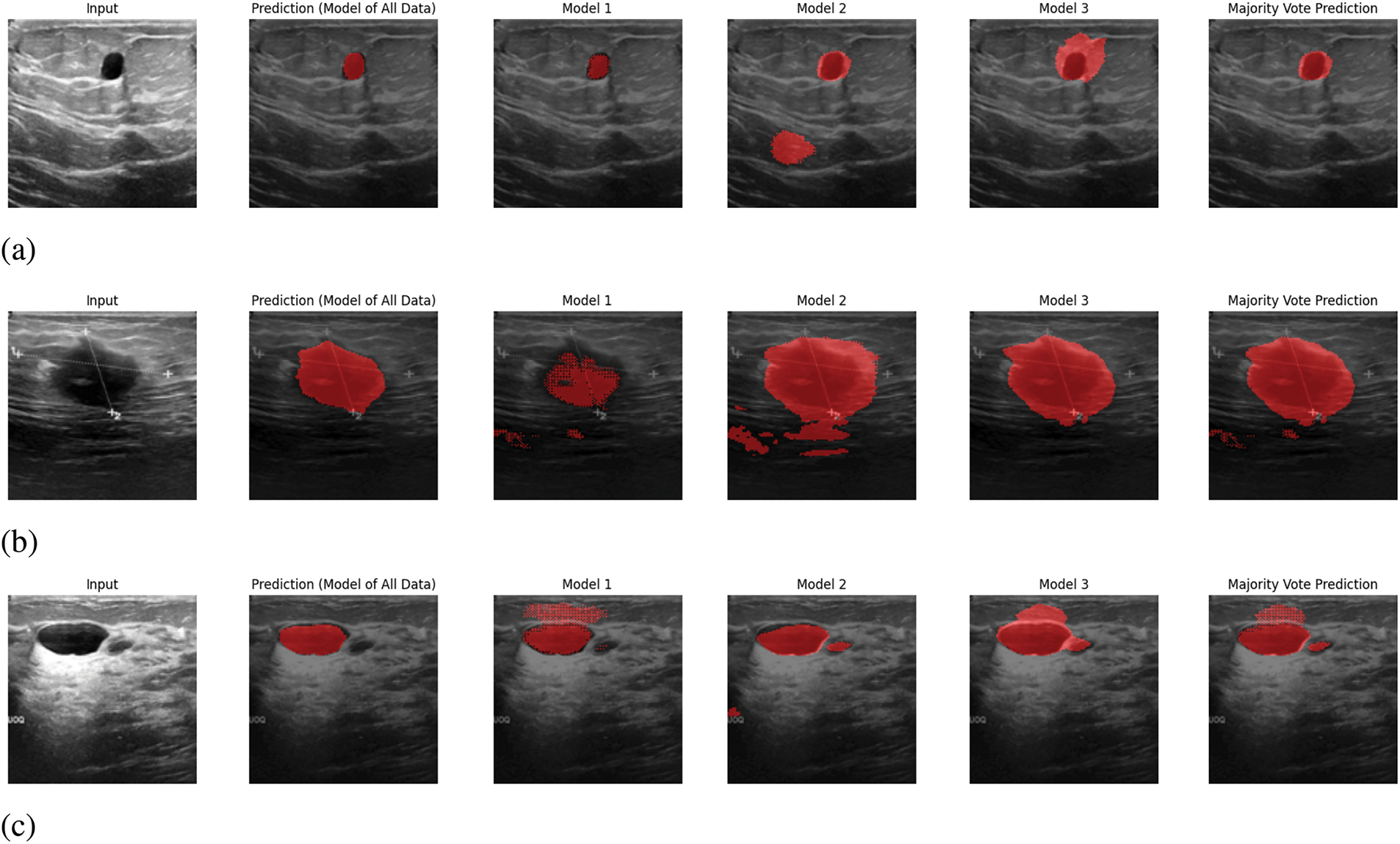
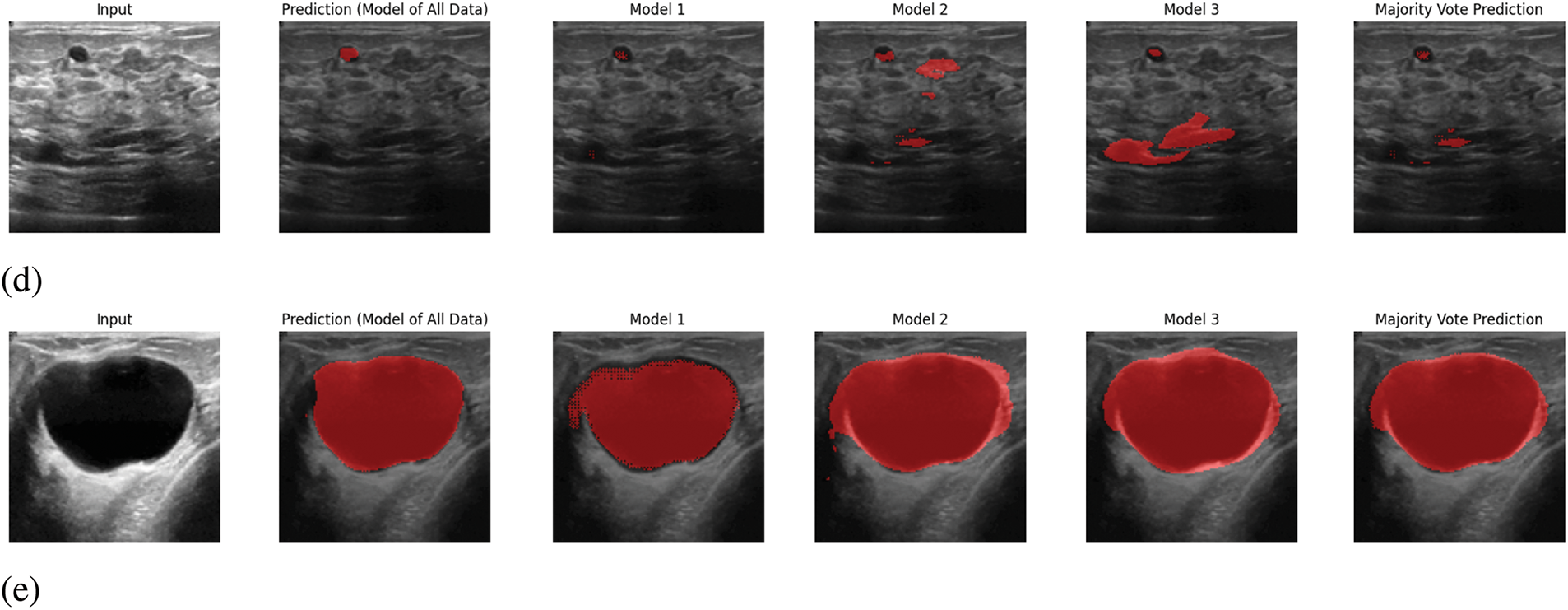
Figure 6: Sample output predictions: (a) Input 1, (b) Input 2, (c) Input 3, (d) Input 4, (e) Input 5
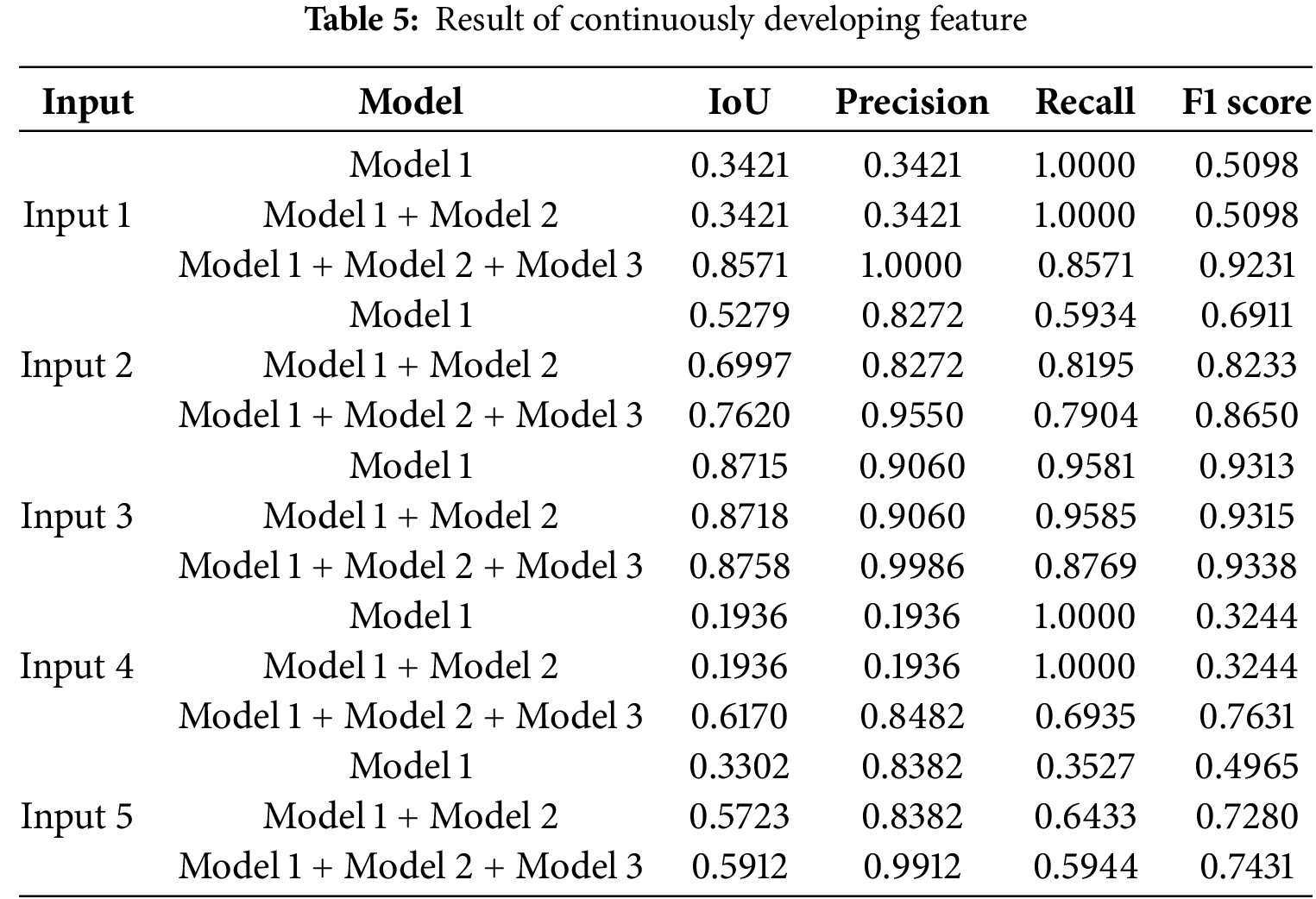
Besides, in Fig. 7, we have provided the voting majority output with Model 1, Model 1 + Model 2, and Model 1 + Model 2 + Model 3. The main objective of presenting this figure is to provide proof of the continuous development of the proposed system. As in the proposed system, it is feasible to add a new trained model to the prediction system that enhances the system’s ability to provide better performance. Whether this proposed system is being able to fulfil this certain objective or not, we have done the voting majority three times, initially with just Model 1. Later, we have done the voting majority with the predictions of Model 1 and Model 2. Finally, this pixel-based voting majority task was done with the predictions of Models 1, 2, and 3. Along with this figure, we have shown a table. In Table 5, we have provided the effectiveness of the ultimate predicted output of Model 1, Model 1+ Model 2, and Model 1+ Model 2+ Model 3 with five inputs based on IoU, Precision, Recall, and F1 score. In this table, it is visible that the voting majority output of model 1 and model 2 is comparatively more accurate than just Model 1. On the other hand, the voting majority output of Models 1, 2, and 3 is better than the combination of Model 1 and Model 2. So, based on the provided information Fig. 7 and Table 5, it can be said that the proposed way of continuous improvement is working successfully according to the plan.
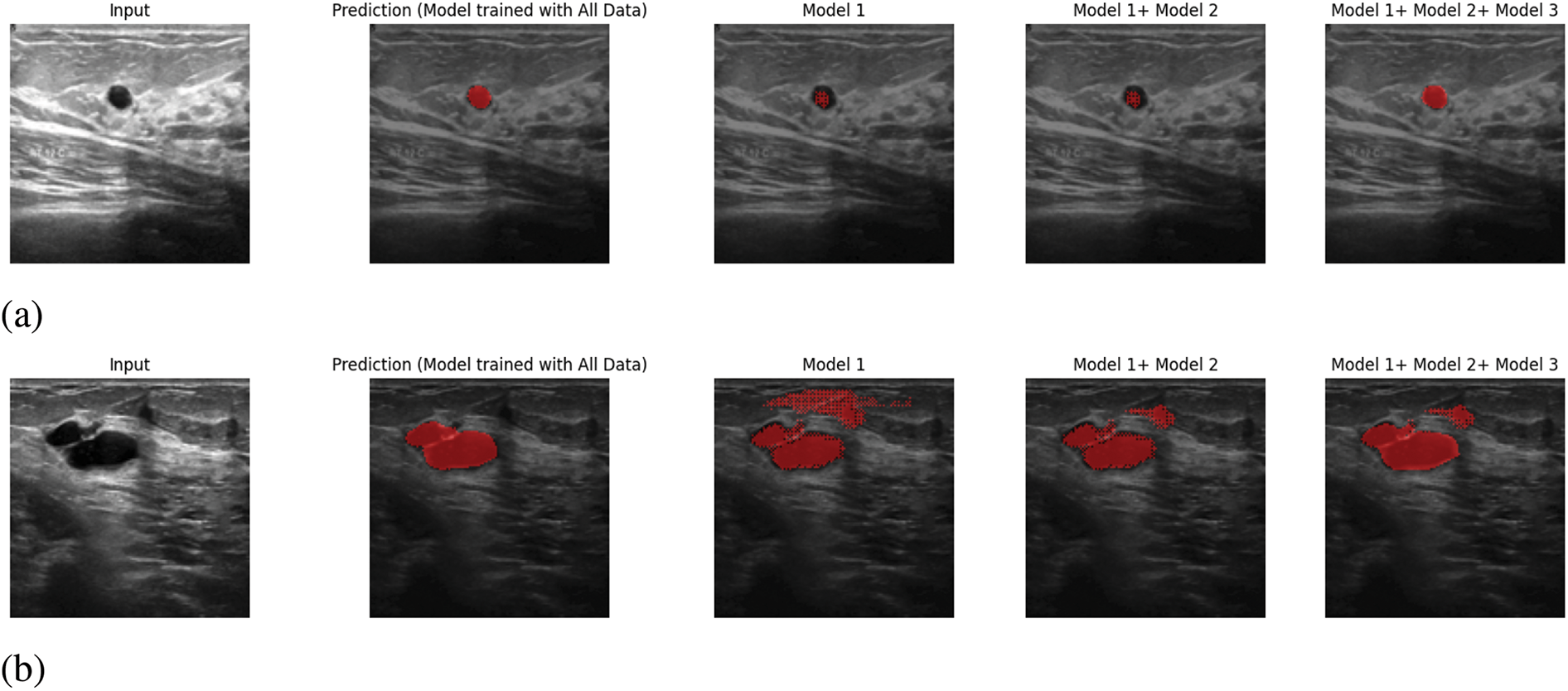

Figure 7: Continuously Developing: (a) Input 1, (b) Input 2, (c) Input 3, (d) Input 4, (e) Input 5
4.2 Comparison Results with State-of-Art Models
As it is already mentioned, the proposed method is invented to fulfil the purpose of training data in a comparatively less powerful machine. It is required to know whether the proposed system fulfils this objective or not. As a consequence, a comparison of the computational demands of four types of approaches with our proposed system was conducted. Initially, the comparison has been done based on the total training time among five approaches, including the proposed system of this research. In the proposed system, unlike typical methods, the entire data was divided into three segments, and each segment was trained separately. Thus, it can train the three segments simultaneously on three different machines at the same time. In this process, the total training time and most computational demand are for the segment with the most data. If data was trained simultaneously at the same time in three different machines using the proposed method, the total training time and computational resources required will be determined by the needs of the third segment, which has the most data. 380 data out of total 780 data are available in the third segment. Therefore, no other segment will take more time and more computational resources than the third segment. As a result, it is needed to compare only the third segment with UNet, PerseNet, Attention UNet, and FCNN [42–44]. In the case of computational demand, it is needed to focus on the usage of RAM and GPU utilization. In Table 6, the comparison among the five approaches is given, and it is visible that in terms of usages of RAM, GPU utilization, and total training time, the proposed system outperforms all the methods like FCNN, Typical UNet, Attention UNet, and Parse Net. In every case, the proposed method performs significantly better than the four other image segmentation methods. Eventually, it can be disclosed that the proposed system is capable of fulfilling the need of training in a less powerful machine and comparatively less time. For this comparison task, Google Colab with its Tesla T4 GPU was utilized for all five methods.
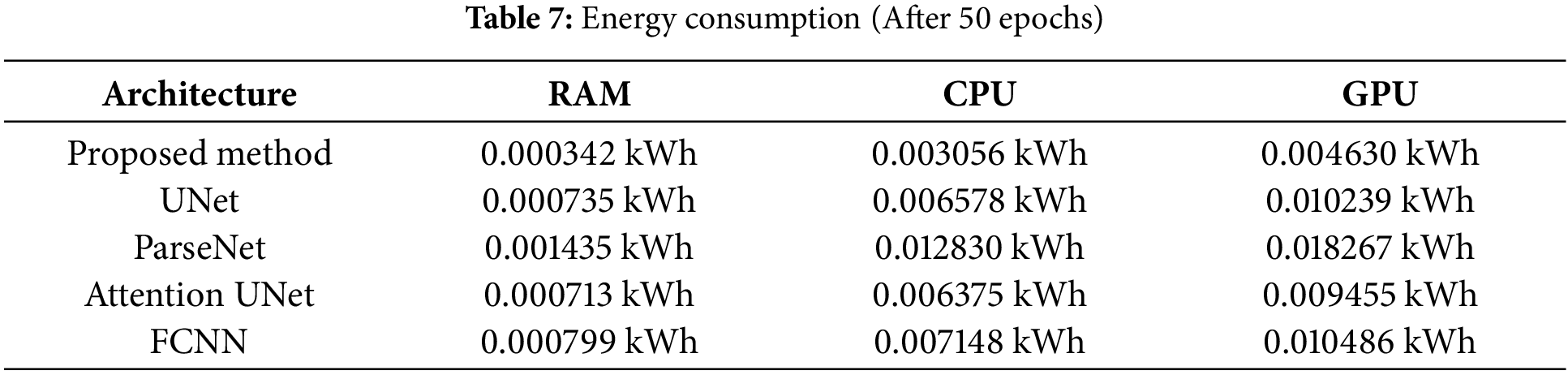
Besides, another statistical comparison has been conducted of the proposed system with these four state-of-the-art models. In Table 7, the comparison of energy consumption is provided. From this table, it is evident that the proposed system consumes the least energy among all these models. Therefore, it can be concluded that by following the proposed image segmentation system, it is possible to train the model on comparatively less powerful machines.
This paper introduces a new way for the image segmentation task by following the core concept of the UNet. Unlike typical prediction systems, our proposed prediction framework ensures data privacy by eliminating the need for high computational resources. This approach not only facilitates faster training with smaller datasets but also simplifies error detection. Besides, we have demonstrated the effectiveness of our system through evidence-based findings, showing that it outperforms four state-of-the-art models in terms of both computational resource usage and processing time.
In the future, we aim to optimise our prediction system to reduce the time complexity of predicting output. Mainly, we plan to merge all trained models into a unified framework, eliminating the need for sequential model output predictions. Because in our current approach, the output will be predicted from each trained model, and the ultimate one will be selected by following another process, which is time-consuming. This improvement will reduce processing time and enhance overall efficiency. This research is depending on U-Net, which is a popular and effective architecture for medical image segmentation, but it is not the only option. Therefore, in the future, we have planned to make a platform so that the trained model with any deep learning algorithm can be added to the system.
Acknowledgement: None.
Funding Statement: The authors extend their appreciation to the Researchers Supporting Project, King Saud University, Saudi Arabia, for funding this research work through Project No. RSPD2025R951.
Author Contributions: Conceptualization: Rifat Sarker Aoyon; methodology: Rifat Sarker Aoyon; validation and visualization: Ismail Hossain; formal analysis: Jia Uddin; data curation: Ismail Hossain; writing original draft preparation: Rifat Sarker Aoyon; writing review and editing: M. Abdullah-Al-Wadud; supervision: Jia Uddin; funding acquisition: M. Abdullah-Al-Wadud. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The dataset that we used in this research for training purposes is available at https://www.kaggle.com/datasets/aryashah2k/breast-ultrasound-images-dataset (accessed on 05 February 2025). And the dataset that we used in this research for testing purposes is available at https://www.kaggle.com/datasets/vuppalaadithyasairam/ultrasound-breast-images-for-breast-cancer (accessed on 05 February 2025).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. World Health Organization (WHO). Breast cancer [Internet]. [cited 2024 Mar 13]. Available from: https://www.who.int/news-room/fact-sheets/detail/breast-cancer. [Google Scholar]
2. Crivellari D, Aapro M, Leonard R, von Minckwitz G, Brain E, Goldhirsch A, et al. Breast cancer in the elderly. J Clini Oncol. 2007;25(14):1882–90. doi:10.1200/JCO.2006.10.2079. [Google Scholar] [PubMed] [CrossRef]
3. Iqbal A, Sharif M. UNet: a semi-supervised method for segmentation of breast tumor images using a U-shaped pyramid-dilated network. Expert Syst Appl. 2023;221(1):119718. doi:10.1016/j.eswa.2023.119718. [Google Scholar] [CrossRef]
4. Shinde VD, Surve J, Honade SJ, Naik K, Kuchibhotla S, Bangare SL, et al. Breast tumour segmentation using advanced UNet with saliency, channel, and spatial attention models. J Electr Syst. 2024;20(1s):488–97. doi:10.52783/jes.787. [Google Scholar] [CrossRef]
5. Anari S, de Oliveira GG, Ranjbarzadeh R, Alves AM, Vaz GC, Bendechache M. EfficientUNetViT: efficient breast tumor segmentation utilizing UNet architecture and pretrained vision transformer. Bioengineering. 2024;11(9):945. doi:10.3390/bioengineering11090945. [Google Scholar] [PubMed] [CrossRef]
6. Weng W, Zhu X. INet: convolutional networks for biomedical image segmentation. IEEE Access. 2021;9:16591–603. doi:10.1109/ACCESS.2021.3053408. [Google Scholar] [CrossRef]
7. Jasti VD, Zamani AS, Arumugam K, Naved M, Pallathadka H, Sammy F, et al. Computational technique based on machine learning and image processing for medical image analysis of breast cancer diagnosis. Secur Commun Netw. 2022;2022(1):1918379. doi:10.1155/2022/1918379. [Google Scholar] [CrossRef]
8. Lv X, Ming D, Lu T, Zhou K, Wang M, Bao H. A new method for region-based majority voting CNNs for very high resolution image classification. Remote Sens. 2018;10(12):1946. doi:10.3390/rs10121946. [Google Scholar] [CrossRef]
9. Xu J, Glicksberg BS, Su C, Walker P, Bian J, Wang F. Federated learning for healthcare informatics. J Healthc Inform Res. 2021;5(1):1–9. doi:10.1007/s41666-020-00082-4. [Google Scholar] [PubMed] [CrossRef]
10. Acar A, Aksu H, Uluagac AS, Conti M. A survey on homomorphic encryption schemes: theory and implementation. ACM Comput Surv. 2018;51(4):1–35. doi:10.1145/3214303. [Google Scholar] [CrossRef]
11. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015: 18th International Conference; 2015 Oct 5–9; Munich, Germany: Springer International Publishing. p. 234–41. [Google Scholar]
12. Alharbi AH, Aravinda CV, Lin M, Venugopala PS, Reddicherla P, Shah MA. Segmentation and classification of white blood cells using the UNet. Contr Media Molecul Imag. 2022;2022(1):5913905. doi:10.1155/2022/5913905. [Google Scholar] [PubMed] [CrossRef]
13. Soulami KB, Kaabouch N, Saidi MN, Tamtaoui A. Breast cancer: one-stage automated detection, segmentation, and classification of digital mammograms using UNet model based-semantic segmentation. Biomed Signal Process Control. 2021;66(24):102481. doi:10.1016/j.bspc.2021.102481. [Google Scholar] [CrossRef]
14. Li T, Sahu AK, Talwalkar A, Smith V. Federated learning: challenges, methods, and future directions. IEEE Signal Processing Magazine. 2020;37(3):50–60. doi:10.1109/MSP.2020.2975749. [Google Scholar] [CrossRef]
15. Torrey L, Shavlik J. Transfer learning. In: Handbook of research on machine learning applications and trends: Algorithms, methods, and techniques. Hershey, PA, USA: IGI Global; 2010. p. 242–64. [Google Scholar]
16. Michael E, Ma H, Li H, Kulwa F, Li J. Breast cancer segmentation methods: current status and future potentials. Biomed Res Int. 2021;2021(1):9962109. doi:10.1155/2021/9962109. [Google Scholar] [PubMed] [CrossRef]
17. Zhang YN, Xia KR, Li CY, Wei BL, Zhang B. Review of breast cancer pathologigcal image processing. Biomed Res Int. 2021;2021(1):1994764. doi:10.1155/2021/1994764. [Google Scholar] [PubMed] [CrossRef]
18. Abbas AH, Kareem AA, Kamil MY. Breast cancer image segmentation using morphological operations. Int J Electr Commun Eng Technol. 2015;6(4):8–14. [Google Scholar]
19. Debelee TG, Schwenker F, Ibenthal A, Yohannes D. Survey of deep learning in breast cancer image analysis. Evolv Syst. 2020;11(1):143–63. doi:10.1007/s12530-019-09297-2. [Google Scholar] [CrossRef]
20. Zhang J, Wu J, Zhou XS, Shi F, Shen D. Recent advancements in artificial intelligence for breast cancer: image augmentation, segmentation, diagnosis, and prognosis approaches. In: Seminars in cancer biology. London, UK: Academic Press; 2023. [Google Scholar]
21. Liu Q, Liu Z, Yong S, Jia K, Razmjooy N. Computer-aided breast cancer diagnosis based on image segmentation and interval analysis. Automatika. 2020;61(3):496–506. doi:10.1080/00051144.2020.1785784. [Google Scholar] [CrossRef]
22. Hossam A, Harb HM, Abd El Kader HM. Automatic image segmentation method for breast cancer analysis using thermography. JES J Eng Sci. 2018;46(1):12–32. doi:10.21608/jesaun.2017.114377. [Google Scholar] [CrossRef]
23. Huang Q, Luo Y, Zhang Q. Breast ultrasound image segmentation: a survey. Int J Comput Assist Radiol Surg. 2017;12(3):493–507. doi:10.1007/s11548-016-1513-1. [Google Scholar] [PubMed] [CrossRef]
24. Xian M, Zhang Y, Cheng HD, Xu F, Zhang B, Ding J. Automatic breast ultrasound image segmentation: a survey. Pattern Recognit. 2018;79(3):340–55. doi:10.1016/j.patcog.2018.02.012. [Google Scholar] [CrossRef]
25. Almajalid R, Shan J, Du Y, Zhang M. Development of a deep-learning-based method for breast ultrasound image segmentation. In: 17th IEEE International Conference on Machine Learning and Applications (ICMLA); 2018; Piscataway, NJ, USA: IEEE. p. 1103–8. [Google Scholar]
26. Mazouzi S, Guessoum Z. A fast and fully distributed method for region-based image segmentation: fast distributed region-based image segmentation. J Real-Time Image Process. 2021;18(3):793–806. doi:10.1007/s11554-020-01021-7. [Google Scholar] [CrossRef]
27. Szénási S. Distributed region growing algorithm for medical image segmentation. Int J Circ Syst Signal Process. 2014;8(1):173–81. [Google Scholar]
28. Bian S, Xu X, Jiang W, Shi Y, Sato T. BUNET: blind medical image segmentation based on secure UNET. In: Medical Image Computing and Computer Assisted Intervention-MICCAI 2020: 23rd International Conference; 2020 Oct 4–8; Lima, Peru: Springer International Publishing. p. 612–22. [Google Scholar]
29. Deng J, Zhou M, Wang C, Wang S, Xu C. Image segmentation encryption algorithm with chaotic sequence generation participated by cipher and multi-feedback loops. Multim Tools Appl. 2021;80(9):13821–40. doi:10.1007/s11042-020-10429-z. [Google Scholar] [CrossRef]
30. Marwan M, Kartit A, Ouahmane H. A novel approach based on segmentation for securing medical image processing over cloud. J Data Min Digital Human. 2018. doi:10.46298/jdmdh.3154. [Google Scholar] [CrossRef]
31. Schutte S, Uddin J. Deep segmentation techniques for breast cancer diagnosis. BioMedInformatics. 2024;4(2):921–45. doi:10.3390/biomedinformatics4020052. [Google Scholar] [CrossRef]
32. Tekin E, Yazıcı Ç, Kusetogullari H, Tokat F, Yavariabdi A, Iheme LO, et al. Tubule-U-Net: a novel dataset and deep learning-based tubule segmentation framework in whole slide images of breast cancer. Sci Rep. 2023;13(1):128. doi:10.1038/s41598-022-27331-3. [Google Scholar] [PubMed] [CrossRef]
33. Wakili MA, Shehu HA, Sharif MH, Sharif MH, Umar A, Kusetogullari H, et al. Classification of breast cancer histopathological images using DenseNet and transfer learning. Comput Intell Neurosci. 2022;2022(1):8904768. doi:10.1155/2022/8904768. [Google Scholar] [PubMed] [CrossRef]
34. Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Dataset of breast ultrasound images. Data Brief. 2020;28(5):104863. doi:10.1016/j.dib.2019.104863. [Google Scholar] [PubMed] [CrossRef]
35. Polyzotis N, Zinkevich M, Roy S, Breck E, Whang S. Data validation for machine learning. Proc Mach Learn Syst. 2019;1:334–47. [Google Scholar]
36. Ultrasound breast images for breast cancer detection: data pre-processing [Internet]. [cited 2025 Feb 5]. Available from: https://www.kaggle.com/datasets/vuppalaadithyasairam/ultrasound-breast-images-for-breast-cancer. [Google Scholar]
37. Blinn JF. What is a pixel? IEEE Comput Graph Appl. 2005;25(5):82–7. doi:10.1109/MCG.2005.119. [Google Scholar] [PubMed] [CrossRef]
38. Saravanan C. Color image to grayscale image conversion. In: 2010 Second International Conference on Computer Engineering and Applications; 2010; Piscataway, NJ, USA: IEEE. Vol. 2, p. 196–9. [Google Scholar]
39. Nečasová T, Burgos N, Svoboda D. Validation and evaluation metrics for medical and biomedical image synthesis. In: Biomedical image synthesis and simulation. London, UK: Academic Press; 2022. p. 573–600. [Google Scholar]
40. McKay KM, Lim LL, Van Gelder RN. Rational laboratory testing in uveitis: a Bayesian analysis. Surv Ophthalmol. 2021;66(5):802–25. doi:10.1016/j.survophthal.2021.02.002. [Google Scholar] [PubMed] [CrossRef]
41. Brodersen KH, Ong CS, Stephan KE, Buhmann JM. The balanced accuracy and its posterior distribution. In: 2010 20th International Conference on Pattern Recognition; 2010; Piscataway, NJ, USA: IEEE. p. 3121–4. [Google Scholar]
42. Liu W. Parsenet: looking wider to see better. arXiv:1506.04579. 2015. [Google Scholar]
43. Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, et al. Attention u-net: learning where to look for the pancreas. arXiv:1804.03999. 2018. [Google Scholar]
44. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015; Piscataway, NJ, USA: IEEE; p. 3431–40. [Google Scholar]
45. Saood A, Hatem I. COVID-19 lung CT image segmentation using deep learning methods: U-Net versus SegNet. BMC Med Imaging. 2021;21(1):19. doi:10.1186/s12880-020-00529-5. [Google Scholar] [PubMed] [CrossRef]
46. Zubair M. ParseNet Overview. Medium [Internet]. [cited 2024 Aug 25]. Available from: https://medium.com/@zakhtar2020/parsenet-overview-23432ac425cc. [Google Scholar]
47. Sun Y, Bi F, Gao Y, Chen L, Feng S. A multi-attention UNet for semantic segmentation in remote sensing images. Symmetry. 2022;14(5):906. doi:10.3390/sym14050906. [Google Scholar] [CrossRef]
48. Xu Y, Quan R, Xu W, Huang Y, Chen X, Liu F. Advances in medical image segmentation: a comprehensive review of traditional, deep learning and hybrid approaches. Bioengineering. 2024;11(10):1034. doi:10.3390/bioengineering11101034. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF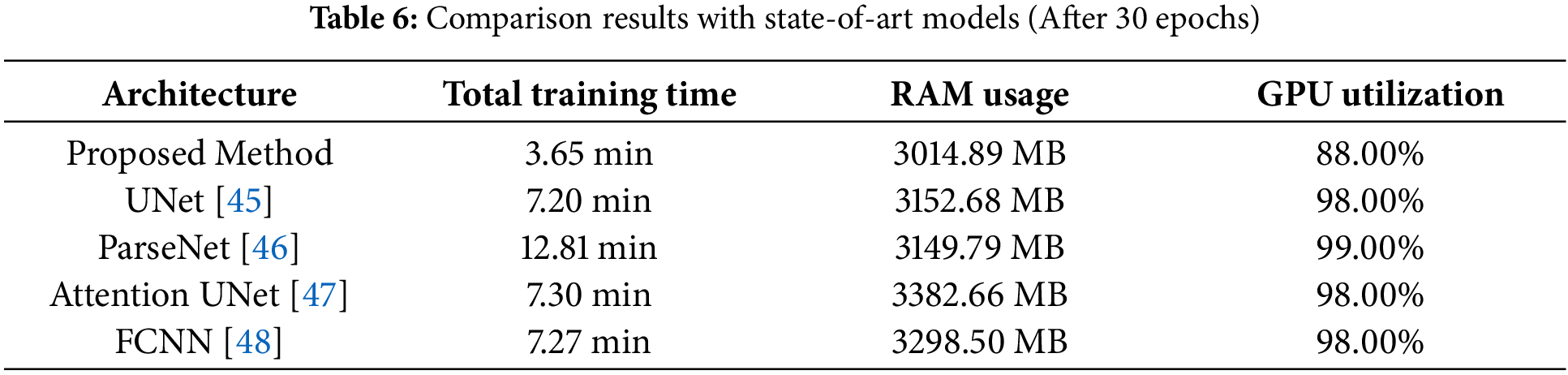
 Downloads
Downloads
 Citation Tools
Citation Tools