 Open Access
Open Access
ARTICLE
An Enhanced Lung Cancer Detection Approach Using Dual-Model Deep Learning Technique
1 Department of Information Technology, Institute of Graduate Studies and Research, Alexandria University, Alexandria, 21526, Egypt
2 College of Computing and Information Technology, Arab Academy for Science, Technology and Maritime Transport, Alexandria, P.O. Box 1029, Egypt
* Corresponding Author: Saad Mohamed Darwish. Email:
(This article belongs to the Special Issue: Advances in AI-Driven Computational Modeling for Image Processing)
Computer Modeling in Engineering & Sciences 2025, 142(1), 835-867. https://doi.org/10.32604/cmes.2024.058770
Received 20 September 2024; Accepted 15 November 2024; Issue published 17 December 2024
Abstract
Lung cancer continues to be a leading cause of cancer-related deaths worldwide, emphasizing the critical need for improved diagnostic techniques. Early detection of lung tumors significantly increases the chances of successful treatment and survival. However, current diagnostic methods often fail to detect tumors at an early stage or to accurately pinpoint their location within the lung tissue. Single-model deep learning technologies for lung cancer detection, while beneficial, cannot capture the full range of features present in medical imaging data, leading to incomplete or inaccurate detection. Furthermore, it may not be robust enough to handle the wide variability in medical images due to different imaging conditions, patient anatomy, and tumor characteristics. To overcome these disadvantages, dual-model or multi-model approaches can be employed. This research focuses on enhancing the detection of lung cancer by utilizing a combination of two learning models: a Convolutional Neural Network (CNN) for categorization and the You Only Look Once (YOLOv8) architecture for real-time identification and pinpointing of tumors. CNNs automatically learn to extract hierarchical features from raw image data, capturing patterns such as edges, textures, and complex structures that are crucial for identifying lung cancer. YOLOv8 incorporates multi-scale feature extraction, enabling the detection of tumors of varying sizes and scales within a single image. This is particularly beneficial for identifying small or irregularly shaped tumors that may be challenging to detect. Furthermore, through the utilization of cutting-edge data augmentation methods, such as Deep Convolutional Generative Adversarial Networks (DCGAN), the suggested approach can handle the issue of limited data and boost the models’ ability to learn from diverse and comprehensive datasets. The combined method not only improved accuracy and localization but also ensured efficient real-time processing, which is crucial for practical clinical applications. The CNN achieved an accuracy of 97.67% in classifying lung tissues into healthy and cancerous categories. The YOLOv8 model achieved an Intersection over Union (IoU) score of 0.85 for tumor localization, reflecting high precision in detecting and marking tumor boundaries within the images. Finally, the incorporation of synthetic images generated by DCGAN led to a 10% improvement in both the CNN classification accuracy and YOLOv8 detection performance.Keywords
Lung cancer is one of the leading causes of cancer-related deaths worldwide, accounting for a significant percentage of all cancer diagnoses. It is a malignant tumor that originates in the tissues of the lungs, typically in the cells lining the air passages [1]. Early diagnosis of lung cancer is crucial for improving patient outcomes, as the disease is often asymptomatic in its early stages and is frequently diagnosed at an advanced stage. The most common methods of diagnosing lung cancer include imaging techniques such as chest X-rays, Computed Tomography (CT scans), Positron Emission Tomography (PET) scans, and tissue biopsy. These methods, while effective, have their limitations and challenges [2]. While imaging techniques like CT and PET scans are powerful tools, they may not always accurately distinguish between cancerous and non-cancerous conditions. This can lead to either missed diagnoses or unnecessary interventions. Biopsies, although definitive, involve risks associated with invasive procedures. This is particularly concerning in patients with compromised lung function or other health issues. Yet, advanced imaging and biopsy techniques can be expensive and may not be readily accessible to all patients, especially in regions with limited healthcare infrastructure. Furthermore, repeated use of imaging techniques like CT scans involves exposure to radiation, which can be a concern, particularly in monitoring patients over time. These methods are generally more effective at detecting lung cancer once it has reached a certain size or stage. Detecting lung cancer in its very early stages, when it is most treatable, remains a significant challenge [2,3].
Lung cancer detection and localization are critical components of diagnosing and effectively treating the disease. Detection refers to the identification of the presence of lung cancer, while localization involves pinpointing the exact location and extent of the tumor within the lungs. Accurate detection and precise localization are essential for determining the appropriate treatment plan. Deep learning, a subset of machine learning, involves the use of artificial neural networks to automatically learn and identify patterns in large datasets. In the context of lung cancer detection and localization, deep learning has shown great promise, particularly with imaging data from modalities like CT scans, PET scans, and X-rays [4,5]. Yet, training deep learning models requires large amounts of annotated data. Inadequate or biased datasets can lead to poor model performance, particularly in detecting rare or atypical cases.
Deep learning techniques for lung cancer detection and localization can be broadly categorized into single-model and dual-model approaches. Each has its advantages and disadvantages [5,6]. Single-model approaches involve using a single neural network model to perform tasks such as detection, segmentation, or classification of lung cancer. These techniques typically focus on leveraging a single type of deep learning architecture to handle the task. Dual-model approaches involve using two complementary deep learning models to leverage the strengths of different architectures or modalities. These methods often integrate information from multiple sources or combine different types of neural networks to improve performance. The choice between single-model and dual-model techniques depends on the specific requirements of the lung cancer detection and localization task, including the complexity of the data, available resources, and desired performance outcomes. In general, by combining models trained on different types of data or using different architectures, dual-model approaches can handle variability in imaging conditions, patient demographics, and tumor presentations more effectively. However, it requires careful consideration of how different models or data types contribute to the final decision.
In lung cancer detection, one of the major combinations in dual-model deep learning approaches involves using CNNs (Convolutional Neural Networks) combined with RNNs (Recurrent Neural Networks), specifically LSTMs (Long Short-Term Memory networks), or CNNs combined with object detection models like YOLO (You Only Look Once). These combinations are frequently used because they can capture both spatial and temporal features or integrate fast object detection with detailed feature extraction [7,8]. Both CNN + RNN and CNN + YOLO combinations are prone to overfitting when trained on small datasets, leading to models that do not generalize well to new patients or different imaging conditions. Small datasets require high-quality, expertly annotated data to mitigate some of the disadvantages. However, obtaining such annotations is often costly and time-consuming, particularly in medical fields like lung cancer detection [9]. To address these disadvantages, researchers often employ strategies such as: transfer learning, data augmentation, regularization techniques, and cross-validation. See [10] for more details.
Dual-model architectures, by nature, are complex, with a large number of parameters that require extensive data to train effectively. With small datasets, these models are prone to overfitting, where they learn to memorize the training data rather than generalize from it. This leads to poor performance on unseen data, which is critical in medical applications like lung cancer detection. Lung cancer detection datasets often suffer from class imbalance, with many more examples of healthy tissue than cancerous lesions. In small datasets, this imbalance is more pronounced, leading to biased models that are more likely to predict the majority class (healthy) and miss cancerous regions. While data augmentation can help increase the effective size of the training set, it may not fully capture the variability and complexity of real-world lung cancer cases. For example, simple transformations like rotation or flipping may not add significant new information to the dataset, limiting the benefits of augmentation. The novelty of our approach is refining data augmentation methods that go beyond standard techniques, potentially incorporating generative models (like generative adversarial networks) to create realistic synthetic data that mimics the diversity of real-world medical images. Such technique could effectively increase the size and diversity of the dataset, allowing dual-model architectures to perform better even when the original dataset is small.
The remainder of this paper consists of the following sections: Section 2 provides a literature review of relevant publications regarding the lung cancer detection frameworks. The suggested approach is presented in Section 3. The assessment of the suggested approach, including results and discussion, is presented in Section 4. The study is concluded, and possible future directions are discussed, in Section 5.
Lung cancer detection has been a major focus of research due to its high mortality rate, and numerous approaches have been developed over the years. The majority of lung cancer detection approaches rely on deep learning concepts due to several key advantages that deep learning offers, particularly in the context of medical imaging and data analysis [1,2]. Deep learning models, particularly Convolutional Neural Networks (CNNs), can automatically learn and extract complex features from medical images, such as CT scans and X-rays. These features are often more informative and nuanced than those identified through traditional image processing methods, leading to improved detection accuracy [4]. Furthermore, deep learning can effectively integrate different types of data (e.g., imaging, clinical, and genomic) into a single predictive model. This multimodal approach enhances the overall accuracy and reliability of lung cancer detection systems.
In a study by Toğaçar et al. [6], a CNN was employed to analyze CT scans for lung cancer detection. The researchers enhanced classification accuracy by zooming in on the CT images and carefully selecting specific features. While this approach led to improved performance, it also highlighted the requirement for high-quality images and substantial computing power, which may limit the method’s accessibility, particularly in regions with limited resources. Similarly, Alakwaa et al. [7] improved lung cancer diagnosis by integrating 3D CNNs with feature-based classifiers, which were optimized using metaheuristics methods. Although this approach shows significant potential, its complexity and the need for in-depth validation before clinical application should be carefully considered.
Transfer learning has been explored as a means to enhance diagnostic accuracy. In a study by Mehmood et al. [8], they demonstrated that using a pre-trained AlexNet model could efficiently diagnose lung and colon cancer from histopathology images. This approach showed both high accuracy and efficiency in diagnosis. However, its success is heavily dependent on the diversity and quality of the training data, which could limit its effectiveness in certain scenarios. In another study, Zhao et al. [9] introduced a technique that integrates scale feature fusion with CNN for classifying pulmonary nodules, achieving remarkable diagnostic results. Despite these advancements, further research is needed to validate the model’s effectiveness, as its performance may vary depending on specific characteristics and image quality.
Khanmohammadi et al. [10] explored the use of biosensors as a cost-effective and non-invasive method for detecting lung cancer biomarkers. Although this approach is promising, its success is largely dependent on the specific biomarkers and their concentrations, necessitating thorough clinical validation. Salama et al. [11] utilized sampling models to enhance small and imbalanced datasets of chest X-ray images. Their work showed that leveraging these models could significantly improve the performance of the Residual Convolutional Neural Network (ResNet)-50 classifier. However, there are concerns that this method could introduce biases, potentially affecting the model’s ability to generalize effectively in real-world scenarios.
In a study by Naseer et al. [12], the benefits of integrating deep learning with traditional machine learning methods for lung cancer diagnosis were highlighted. The researchers used a modified version of AlexNet combined with Support Vector Machines (SVM). However, this approach requires domain expertise to fine-tune and optimize different configuration parameters for both AlexNet and SVM effectively. Integrating gene expression data with deep learning has proven effective in detecting lung cancer, as demonstrated by Liu et al. [13]. The model effectively handles complex data, but its utility may be limited by the availability and quality of gene expression datasets. Reddy et al. [14] highlighted the use of different CNN models to improve pulmonary nodule detection, underscoring the critical role of data processing methods. However, the effectiveness of these models is highly dependent on the availability of comprehensive datasets.
Transfer learning has also been employed to address the challenge of imbalanced data in lung cancer classification. For instance, Zhan et al. [15] improved the accuracy of identifying small cell lung cancer (NSCLC) patients using transfer learning. While this approach effectively mitigates data imbalances, there remains a concern about overfitting if significant differences exist between the source and target datasets. In a study by Nageswaran et al. [16], it was demonstrated that Artificial Neural Network (ANN) models can achieve high accuracy in classifying lung cancer using CT scans. However, these models require substantial computational resources and extensive data preparation. The DenseNet architecture has also been applied to lung cancer classification. Zhang et al. [17] used DenseNet to achieve impressive results, highlighting the effectiveness of deep learning structures. However, the high computational demands of these models may hinder their real-time application. In a related context, Qiao et al. [18] developed a deep neural network for identifying GAN (Generative Adversarial Networks)-generated faces, suggesting potential adaptations of this approach for direct application in lung cancer detection.
Advancements in gene selection for cancer prediction have progressed with the use of classifiers. Swathi et al. [19] improved the effectiveness of the XGBoost classifier for gene selection in cancer prediction by employing Hyperband optimization. However, managing the complexities and computational demands of this optimization process can be quite challenging. Research studies have emphasized the significance of using image processing for lung cancer detection. In a study by Bhalerao et al. [20], they combined image processing with CNNs to achieve high accuracy in lung cancer identification, showcasing the effectiveness of merging traditional image processing techniques with deep learning. However, the success of this approach is highly dependent on the quality of the images and the preprocessing steps, which can be challenging when working with diverse datasets. Moreover, Nadkarni et al. [21] and Sori et al. [22] demonstrated promising results using CT image analysis techniques like Detail-Fidelity Deep Network (DFDNet). However, the effectiveness of these approaches may be constrained when applied to unmodified imaging datasets.
Forte et al. [23] demonstrated that Convolutional Neural Networks (CNNs) are highly accurate in analyzing medical images, which underscores their reliability in various imaging tasks. CNNs are particularly effective in recognizing patterns and features within complex image data, making them ideal for applications like detecting abnormalities in medical scans. The study likely highlighted the CNNs’ ability to deliver consistent, precise results, reinforcing their role as a dependable tool in medical imaging. However, despite their accuracy, the study may also have noted the need for extensive computational resources and large datasets to train these models effectively. In a related research effort, Behara et al. [20] employed Deep Convolutional Generative Adversarial Networks (DCGAN) to classify skin lesions, demonstrating the flexibility and potential of GANs in dermatological imaging. The study highlighted the adaptability of GANs in producing realistic images that can be used to train models for various medical imaging tasks. However, it also pointed out significant challenges, particularly in fine-tuning these models, which can be a delicate and time-consuming process. Additionally, the study noted the lengthy training process required for GANs. Training these models often involves multiple iterations and requires considerable computational power and time, making it a resource-intensive process.
A literature survey on dual-model deep learning focuses on exploring the development and application of dual-model architectures, where two distinct models or networks work together synergistically. These architectures are employed across various domains, including computer vision, natural language processing, and other machine learning tasks [24]. Rathod et al. [25] suggested DLCTLungDetectNet, a novel CNN framework tailored for early lung cancer detection through comprehensive CT scan analysis. The core innovation of DLCTLungDetectNet is its hybrid model, FusionNet, which merges the complementary strengths of two advanced architectures: ResNet-50 and InceptionV3. ResNet-50 contributes its deep residual learning capabilities, allowing the network to retain essential features across numerous layers, thereby enhancing the model’s ability to capture subtle and complex characteristics in CT images. InceptionV3, known for its multi-scale feature extraction, adds further robustness by enabling the model to detect varied spatial patterns associated with lung tumors. Together, these architectures form a highly efficient system designed for accurate detection. Comparative analyses highlight DLCTLungDetectNet’s superior performance, consistently outperforming other established architectures, such as Visual Geometry Group models (VGG16 and VGG19) and standalone InceptionV3. This performance is reflected in high scores across multiple evaluation metrics, including accuracy, precision, Area Under Curve (AUC), and F1-score, confirming its effectiveness in identifying and localizing lung tumors with remarkable reliability. This research not only exemplifies the power of deep learning in advancing diagnostic methods for lung cancer but also establishes a new benchmark. FusionNet’s integrated design proves instrumental in achieving this.
Narotamo et al. [26] suggested a dual-model deep learning approach designed to enhance representation learning and introduce long-term temporal dependence in the analysis of electrocardiogram (ECG) data. Their method specifically addresses the challenges of imbalanced data and the need for accurate temporal modeling in medical diagnostics. To address the common issue of imbalanced datasets—where one class (e.g., abnormal heart rhythms) is underrepresented—the adaptive synthetic (ADASYN) sampling method is used. ADASYN generates synthetic examples of the minority class, thereby balancing the dataset and improving the model’s ability to learn from all classes effectively. Following this preprocessing, the researchers developed a representation learning model based on a one-dimensional convolutional neural network (1DCNN-RLM). This model is responsible for extracting feature vectors from the RRI (R-R Interval) series, effectively capturing the most relevant information from the time-series data. The extracted features serve as a condensed representation of the ECG segment, facilitating efficient and accurate analysis. The second component of the dual-model framework is a temporal dependence model built on the bidirectional gated recurrent unit (BiGRU-TDM). The BiGRU is particularly suited for capturing long-term dependencies in sequential data, making it ideal for understanding the transition patterns between different states—such as abnormal (SA) and normal heart rhythms—across the ECG segments. The complexity of the dual-model architecture, particularly the combination of 1DCNN and BiGRU, may result in higher computational demands, which could be a challenge in real-time or resource-limited settings.
In their research, Mirza et al. [27] introduced a fully automated approach for classifying the presence of lower-grade gliomas (a type of brain tumor) using a deep-learning-based segmentation algorithm. The study focuses on developing two distinct deep-learning models that work together to achieve accurate tumor detection and localization. The first model is designed to determine whether a tumor is present in the brain. This initial step is crucial for identifying individuals who may require further medical intervention. Once a tumor is detected, the second model comes into play, which focuses on generalizing the tumor’s location within the brain. This model provides a detailed segmentation of the tumor, mapping its exact position, which is essential for planning treatment strategies. Furthermore, the researchers employed the Tversky loss function, which is particularly effective in handling imbalanced datasets by focusing on minimizing false negatives and false positives. The fully automated nature of the approach can significantly reduce the workload of radiologists, allowing for faster diagnosis and more consistent results. Yet, the reliance on the Tversky loss function, while effective for handling class imbalance, may need careful tuning to ensure optimal performance, particularly in cases where the degree of imbalance varies significantly.
Kiran et al. [28] introduced a new hybrid deep learning approach that merges the powerful feature extraction capabilities of EfficientNetB3 with the simplicity and effectiveness of the k-Nearest Neighbors (k-NN) algorithm. This approach aims to leverage the strengths of both techniques to improve performance on complex classification tasks. The process begins with EfficientNetB3, a state-of-the-art convolutional neural network architecture that is pre-trained on the extensive ImageNet dataset. This model is repurposed as a feature extractor, capitalizing on its ability to capture intricate patterns and features from input images. To further refine the feature set, a GlobalAveragePooling2D layer is applied, which effectively reduces the dimensionality of the feature maps while retaining the most important information. Data augmentation techniques are also a key part of the process. By applying various transformations such as rotations, shifts, flips, and zooms, the researchers aimed to make the model more robust and capable of generalizing well to new, unseen data. These augmentations help the model learn to recognize objects and patterns in a variety of contexts, enhancing its overall performance. However, the reliance on EfficientNetB3, a deep neural network, requires significant computational resources for feature extraction, which might limit the model’s accessibility in resource-constrained environments.
To advance dual-model deep learning approaches for lung cancer detection, it is crucial to extend the related work in several key areas. Addressing these needs will enhance the effectiveness, applicability, and reliability of these models in clinical settings. The integration of real-time capabilities into tumor identification models is essential, as current models often face challenges related to high computational demands. Incorporating YOLOv8 can address this need, providing fast and accurate detection suitable for real-time analysis. Additionally, current models sometimes struggle with data quality and imbalance, which can be mitigated by combining CNN with advanced data augmentation techniques or generative models like DCGAN to create more robust datasets. To reduce complexity and enhance model interpretability, using CNN for categorization in conjunction with YOLOv8 offers a streamlined approach that balances accuracy with efficiency. Finally, improving generalizability and adaptability can be achieved by employing a hybrid model that leverages CNN’s feature extraction capabilities and YOLOv8’s real-time detection, ensuring robust performance across diverse datasets and conditions.
The suggested approach combines the strengths of CNN and YOLOv8 to create an efficient and robust model for tumor identification. CNN is utilized for categorization, leveraging its ability to extract hierarchical features from images for strong feature extraction. YOLOv8 is integrated for real-time identification and localization, ensuring timely and accurate detection of tumors. To enhance model robustness, advanced data augmentation techniques like DCGAN are employed, improving the model’s ability to handle diverse data scenarios. The proposed model offers a more efficient, adaptable, and accurate solution for real-time tumor detection and categorization.
The enhanced lung cancer detection approach utilizing CNNs for categorization and YOLOv8 for real-time tumor identification and pinpointing combines the strengths of both models. CNN excels in feature extraction and classification, while YOLOv8 provides efficient and accurate tumor localization. This dual-model strategy improves detection accuracy, enhances real-time capabilities, and offers a robust solution for lung cancer diagnosis. For improving the performance of lung cancer recognition models, the suggested approach uses DCGANs as an effective method in augmenting data by generating synthetic images. Data augmentation is crucial, especially when dealing with limited medical imaging data. Data augmentation techniques enhance the diversity of the training dataset by applying various transformations to existing images, which helps the model generalize better and prevents overfitting. Here’s an in-depth overview of how these models work together and Fig. 1 shows how it works.

Figure 1: The suggested lung cancer detection approach using dual-model deep learning technique
Data collection is a critical first step in developing a robust lung cancer detection system. It involves gathering a diverse and high-quality dataset that accurately represents the variations in lung cancer images and associated clinical information [28]. Two meticulously selected datasets were utilized to advance lung cancer research: the Comprehensive CT and PET/CT Dataset (Lung PET-CT-Dx), https://www.cancerimagingarchive.net/collection/lung-pet-ct-dx/ (accessed on 14 November 2024) aimed at lung cancer identification, and the IQ-OTH/NCCD Lung Cancer Dataset https://www.kaggle.com/datasets/hamdallak/the-iqothnccd-lung-cancer-dataset (accessed on 14 November 2024), focused on categorization. The IQ-OTH/NCCD dataset (see Fig. 2), gathered from the Iraq Oncology Teaching Hospital and the National Center for Cancer Diseases over three months in Autumn 2019, comprises 1190 CT images from 110 patients. These images, categorized into 40 malignant, 15 benign, and 55 normal cases, were captured in Digital Imaging and Communications in Medicine (DICOM) format using a Siemens SOMATOM scanner with precise settings (120 kV, 1 mm slice thickness, and window widths between 350 to 1200 HU). The scans were performed while patients held their breath at inhalation to ensure high detail, with each scan containing between 80 to 200 slices providing various perspectives of the chest area. Detailed annotations accompany each image, enhancing the dataset’s utility for training robust models capable of accurately distinguishing between different lung conditions. This comprehensive dataset is crucial for improving diagnostic precision by enabling the model to handle a diverse range of lung cancer presentations effectively [29].

Figure 2: Example CT scan images from the IQ-OTH/NCCD dataset
To enhance our model’s ability to detect and differentiate between lung cancer conditions, we selected a subset of 352 images from the Lung-PET-CT-Dx dataset. This subset was carefully chosen to ensure it includes a diverse mix of malignant cases, which is crucial for training models to accurately identify and localize tumors. The Lung-PET-CT-Dx dataset (see Fig. 3), featuring both CT and PET/CT scans, provides comprehensive imaging data essential for detecting and pinpointing lung tumors. By utilizing this diverse subset, we ensure that our YOLOv8 model is trained on a representative sample of various tumor types, enhancing its accuracy and reliability in clinical settings. The combination of high-quality images and detailed annotations from this dataset forms a robust foundation for developing deep learning models capable of precise lung cancer identification and categorization. The images were analyzed on the mediastinum (window width, 350 HU; level, 40 HU) and lung (window width, 1400 HU; level, –700 HU) settings. The reconstructions were made in 2 mm-slice-thick and lung settings. The CT slice interval varies from 0.625 to 5 mm. Scanning mode includes plain contrast and 3D reconstruction [30].

Figure 3: Examples of CT and PET-CT DICOM images of lung cancer form Lung-PET-CT-Dx dataset subjects with XML Annotation files that indicate tumor location with bounding boxes
To address the class imbalance in the IQ-OTH/NCCD Lung Cancer Dataset, where the benign class was significantly underrepresented with only 120 images compared to 561 malignant images and 416 normal images, we implemented a method using DCGAN. This approach was specifically chosen to generate high-quality synthetic images for the minority class of benign lung tumors, thereby expanding the dataset and achieving a balanced distribution with 561 images for each class. DCGAN operates through a unique adversarial system comprising two neural networks (see Fig. 4): the generator and the discriminator. The generator’s objective is to create images that closely mimic real ones, while the discriminator’s role is to differentiate between genuine and synthetic images. During training, these two networks engage in a competitive process, where the generator continually improves its ability to produce realistic images, and the discriminator becomes more adept at identifying fake images. This iterative adversarial process continues until the generator can produce images that are so realistic that the discriminator struggles to distinguish them from real images. This process not only balanced the dataset but also enhanced the model’s ability to generalize across different tumor types [31,32].

Figure 4: DCGAN generator structure
3.2.2 Residual CNN Model for Lung Image Classification
The Residual Convolutional Neural Network (ResNet) model has emerged as a powerful tool for lung image classification due to its ability to overcome the vanishing gradient problem, which can hinder the performance of deep learning models. By introducing skip (residual) connections, ResNet enables efficient training of very deep networks, allowing for the extraction of complex features in lung images, such as small tumors and lesions that might be missed by shallower networks. This architecture also preserves both low- and high-level features, making it highly effective in detecting subtle abnormalities in lung CT scans or X-rays. Additionally, ResNet reduces the complexity of training deep networks by focusing on learning residuals, leading to faster convergence times and lower computational overhead, which is especially beneficial when working with large medical imaging datasets [33].
Furthermore, ResNet’s design offers enhanced robustness and generalization across diverse lung image datasets, reducing the risk of overfitting even when data is limited. Its flexibility allows it to be scaled according to task complexity, from large-scale lung cancer detection using CT scans to more focused tasks like pneumonia detection in X-rays. The model’s capacity for transfer learning makes it easy to fine-tune pre-trained ResNet models on specific medical imaging tasks, improving diagnostic accuracy with less data. With its ability to handle complex classifications, ResNet has consistently delivered state-of-the-art performance in medical image analysis, making it a reliable and efficient choice for lung disease detection and classification [34,35].
The goal is to develop a ResNet model for the classification of lung images into categories such as normal, benign, or malignant. Lung diseases such as lung cancer, pneumonia, and fibrosis can be detected from medical images like CT scans or X-rays. The model should be capable of learning both low-level and high-level features from these images, dealing with the vanishing gradient problem in deep neural networks, and handling the complexity of multi-class classification. Let the input lung image be represented by a 3D matrix
The first layer applies convolutional filters to the input image to extract local features like edges, textures, and small structures such as nodules in lung scan. Let the convolution operation be denoted as:
After several residual blocks and pooling layers, the output is passed through a fully connected layer for classification. The fully connected layer combines the features learned by the network and produces the raw scores (logits) for each class:
The output of this training process is saved as Model.H5, which a file format is commonly used to store trained models in Keras, a deep learning library.
Data annotation for the Lung-PET-CT-Dx dataset is essential for developing accurate AI models for lung cancer detection, diagnosis, and treatment planning. By providing well-labeled data, it enables machine learning models like YOLOv8 to learn how to detect and localize cancerous regions or abnormalities in both PET and CT scans. Annotating this multimodal dataset helps integrate functional information from PET scans with structural data from CT scans, enhancing the model’s ability to identify abnormalities based on both tumor activity and physical structure. High-quality annotations improve diagnostic accuracy by enabling the model to distinguish between benign, malignant, and normal findings, while also helping it handle subtle and complex data, such as early-stage tumors. Additionally, annotating multiple slices in CT scans provides a 3D understanding of abnormalities, which is critical for accurate tumor localization and size estimation [36,37].
The ultimate goal is to create a highly accurate dataset that enables YOLOv8 to effectively detect and localize lung cancer in real time, supporting faster and more accurate diagnosis. The Roboflow toolbox [38] is highly beneficial for data annotation in lung cancer detection, offering a range of tools to streamline the preparation of CT and PET scan datasets. It enables users to draw and edit bounding boxes around tumors or lesions, as well as perform pixel-level segmentation for detailed annotation of irregularly shaped abnormalities, crucial for improving diagnosis and treatment planning. Roboflow supports multiclass labeling, allowing the classification of regions as benign, malignant, or normal, and provides multi-slice support for volumetric data, offering a 3D context for abnormalities. Additionally, the toolbox includes preprocessing options like normalization, cropping, and resizing, ensuring image consistency. With dataset versioning and seamless export to formats compatible with YOLOv8, Roboflow ensures easy integration into the model training pipeline. It also offers a model training environment, allowing users to train their lung cancer detection models directly within the platform.
3.3.2 YOLOv8 for Lung Tumor Detection
YOLOv8 is a real-time object detection algorithm that can be applied to lung tumor detection by localizing and classifying abnormal regions (e.g., tumors, lesions) in medical images such as CT or PET scans. The formal definition of YOLOv8 for lung tumor detection involves the detection and classification of tumors within an image using CNNs and an efficient single-shot detection architecture [39,40]. Given an input image
In the detection process, YOLOv8 first passes the input image III through a CNN backbone, such as CSPDarknet (Cross Stage Partial Darknet), to extract feature maps that capture high-level patterns. Based on these features, the network predicts bounding boxes at multiple scales using anchor boxes, adjusting them to match potential tumor locations. For each predicted bounding box, YOLOv8 assigns a confidence score
In summary, the dual-model deep learning approach for lung cancer detection integrates CNNs for tumor classification with YOLOv8 for real-time localization, enhancing diagnostic accuracy and efficiency by utilizing DCGAN to tackle class imbalance, harnessing the strengths of both models and providing scalability for other medical imaging applications. By automating detection and classification, it minimizes human error and delivers more consistent results. The dual-model system offers high sensitivity and specificity by reducing false positives and negatives as confirmed in the next section, leading to more accurate cancer detection.
4 Experimental Results and Discussions
4.1 Datasets, Evaluation Metrics, and Implementation
In this section, the proposed model was validated using two distinct datasets: Lung-PET-CT-Dx and IQ-OTH/NCCD, both containing images of lung cancer. Each image in these datasets represents a case of lung cancer with unique characteristics, including variations in cancer type, tumor location, size, and intensity. The use of these two well-established datasets ensured that the validation process covered a comprehensive range of lung cancer manifestations, contributing to the model’s ability to generalize across diverse patient cases (see Section 3.1 for further details on the characteristics of the datasets used).
The Comprehensive CT and PET/CT Dataset is thoughtfully structured to reflect a diverse patient population, offering valuable insights into factors that influence lung cancer detection and diagnosis. It encompasses various age groups, including children (0–17 years), young adults (18–34 years), middle-aged adults (35–54 years), and older adults (55 years and above), with a particular focus on older adults, who constitute a significant portion of lung cancer cases due to age-related risk factors. The dataset also maintains a balanced gender distribution, capturing the historically higher prevalence of lung cancer in males linked to smoking and occupational exposures, while also recognizing the rising incidence among females due to changing smoking trends and environmental influences. Additionally, it incorporates a wide range of medical histories, including smoking status (current, former, or non-smoker) with details on duration and intensity, occupational exposures to carcinogens, pre-existing conditions such as chronic obstructive pulmonary disease (COPD) or tuberculosis, and family history of lung cancer or other cancers, thereby enhancing the dataset’s richness and the robustness of machine learning models for lung cancer detection [28,29].
The IQ-OTHNCCD Lung Cancer Dataset is meticulously curated to ensure a comprehensive representation of lung cancer cases, showcasing significant diversity in patient demographics and imaging modalities. It encompasses a broad spectrum of age groups, including children (0–17 years), young adults (18–34 years) with potential risk factors, middle-aged adults (35–54 years), and older adults (55 years and above), particularly emphasizing the importance of age as a risk factor. The dataset also maintains a balanced gender distribution, reflecting the historical prevalence of lung cancer in males and the rising incidence among females, allowing for an in-depth understanding of gender-specific risk factors. Additionally, it includes patients from various ethnic backgrounds and socioeconomic statuses, which can influence lung cancer risks and healthcare access. In terms of imaging modalities, the dataset primarily features detailed CT scans for identifying tumors, supplemented by PET scans to assess metabolic activity, enhancing early detection and differentiation between benign and malignant nodules. It also incorporates varied imaging protocols that reflect different healthcare practices. Furthermore, the dataset includes comprehensive medical histories detailing smoking status, exposure to environmental carcinogens, and pre-existing conditions, as well as treatment outcomes related to various therapeutic strategies, thus providing valuable insights into the effectiveness of treatments across diverse patient demographics [30].
The experiment was carried out on a system equipped with an Intel(R) Core(TM) i3 processor and 8 GB of RAM. This hardware configuration, though modest, provided sufficient computational power to perform the necessary image processing and analysis tasks effectively. We conducted our model testing using Google Colab, a cloud-based platform that is not only user-friendly but also offers GPU support. The integration of GPU acceleration in Colab significantly enhanced the speed of our model training, allowing us to handle the computational demands of deep learning tasks with greater efficiency. This acceleration was particularly beneficial given the complexity and high processing power required for medical imaging data, where large datasets and intricate image details must be processed.
The effectiveness of the proposed dual-model deep learning approach for lung cancer detection is assessed using a range of key performance metrics, including precision, recall, accuracy, specificity, sensitivity, and the confusion matrix. Precision measures the proportion of true positive predictions among all positive predictions, indicating the model’s accuracy in detecting lung cancer without misclassifying healthy lungs. Recall (or sensitivity) evaluates the model’s ability to correctly identify actual lung cancer cases, minimizing false negatives. Accuracy provides an overall measure of correctness by showing the percentage of all correctly classified cases, both positive and negative. Specificity reflects how well the model identifies non-cancerous cases, reducing the risk of false positives. Sensitivity and specificity are critical in ensuring that both cancer and non-cancerous cases are properly distinguished. Finally, the confusion matrix gives a detailed breakdown of the model’s performance by showing the true positives, true negatives, false positives, and false negatives, offering deeper insights into areas for improvement. These metrics together provide a comprehensive evaluation of how well the dual-model approach handles the complexities of medical image classification [1,2].
Our dataset was divided into two portions: 75% was allocated for training, while the remaining 25% was reserved for validation. By maintaining this separation, we could assess how well the model generalized beyond the training set. During the training phase, we used a batch size of 32, which enabled efficient processing of the data while ensuring the model could handle the complexity of the task. The model was trained over 80 epochs, striking a balance between computational efficiency and model stability. Running it through these many epochs helped ensure that the model had ample opportunity to learn patterns in the data, but without overfitting. To optimize the training process further, we incorporated an early stopping mechanism. This feature automatically halted training if no improvements in performance were observed after 20 consecutive epochs. This strategy allowed us to maximize computational resources, preventing unnecessary training and saving time when the model had already reached its peak performance.
4.2 Learning Rate Sensitivity Analysis
In Table 1, we evaluate the impact of different learning rates on the model’s performance. We chose the Adam optimizer to ensure an efficient and stable optimization process for both the Residual CNN model and the YOLOv8 architecture. Adam (Adaptive Moment Estimation) is widely favored in deep learning due to its adaptive learning rate capabilities, which combine the advantages of both the Root Mean Square Propagation (RMSProp) and Stochastic Gradient Descent (SGD) optimizers. It adjusts the learning rate for each parameter dynamically, based on first order and second-order moments of the gradient, allowing the optimizer to navigate complex loss surfaces more effectively [42]. For both models, learning rates significantly influence key metrics like accuracy, specificity, and sensitivity, which are crucial in medical diagnostics. While lower learning rates can lead to slower convergence, they often yield more precise and stable results. In contrast, higher learning rates may speed up the training process but risk overshooting the global minimum, leading to suboptimal detection of lung cancer cases. This balance between learning rate and performance is critical to achieving high sensitivity in detecting cancer cases and maintaining specificity to avoid false positives.

The results revealed in the table can be interpreted as follows, with a learning rate of 0.01 the model achieved an accuracy of 93.21%, specificity of 92.25%, and sensitivity of 91.24%. While these results are acceptable, the higher learning rate may have caused the optimizer to overshoot the optimal solution, resulting in slightly lower performance and less stable convergence. Reducing the learning rate to 0.001 significantly improved performance, with accuracy rising to 96.74%, specificity to 96.45%, and sensitivity to 95.98%, as the optimizer’s adjustments became more refined and balanced. At a learning rate of 0.0001, the model reached the highest accuracy of 97.82%, with specificity at 98.34% and sensitivity at 95.82%. This lower rate facilitated more precise weight updates, enhancing overall accuracy and specificity, though sensitivity decreased slightly compared to 0.001. Finally, with a very low learning rate of 0.00001, accuracy decreased to 97.02%, specificity to 97.29%, and sensitivity improved to 96.63%. While this rate enabled finer model tuning and better cancer detection, the slower convergence and potential for getting trapped in local minima resulted in reduced accuracy and specificity.
The results show that while a learning rate of 0.001 provides a good balance between performance and stability, a rate of 0.0001 delivers the best overall accuracy and specificity, though with a slight decrease in sensitivity. The very low rate of 0.00001 improves sensitivity but at the cost of slight reductions in other metrics, highlighting the delicate balance required in choosing the optimal learning rate. Furthermore, higher learning rates can speed up convergence but may risk overshooting, refers to a situation where an optimization algorithm makes updates that are too large, causing the model’s parameter values to move beyond the optimal solution, and instability; whereas lower rates improve precision and stability but may slow down convergence. The optimal learning rate of 0.0001 achieves high accuracy and specificity while maintaining reasonable sensitivity, indicating it provides a good trade-off for stable and effective training in this context. For practical applications, particularly in medical imaging where accuracy and specificity are crucial, a learning rate of 0.0001 appears to be the most effective choice. It ensures that the model achieves high performance in identifying both cancerous and non-cancerous cases while balancing the computational efficiency of training.
4.3 Lung Cancer Classification Accuracy Evaluation Using ResNet
The second set of experiments was designed to evaluate the accuracy of the ResNet model in classifying lung cancer into benign, malignant, and normal categories. By utilizing a confusion matrix, we can gain deeper insights into the model’s performance. The matrix will display the number of correct and incorrect predictions for each class, helping to identify where the ResNet model performs well and where it encounters difficulties. This detailed breakdown allows us to assess the model’s strengths in detecting specific tumor types, as well as any misclassification patterns, providing a clearer picture of its overall classification effectiveness. Herein, we utilize a ResNet-18, a standard variant of the ResNet architecture, consisting of 18 layers with Basic Blocks, each containing two convolutional layers. The model starts with a 7 × 7 convolution layer with 64 filters and a stride of 2, followed by a 3 × 3 max pooling layer. Each residual block contains two 3 × 3 convolutions, batch normalization, and ReLU activation, with skip connections allowing the input to bypass certain layers. The input image size is typically 224 × 224 pixels, and the final fully connected layer maps to 3 classes by default. Adam optimizer is the default optimizer, the initial learning rate is 0.01, and the weight decay is set to 1 × 10−4. The model uses cross-entropy loss for classification and typically trains 80 epochs with a batch size of 32.
As confirmed by Table 2, the confusion matrix for the residual CNN model shows strong classification performance across the three categories: normal, benign, and malignant. The model correctly classified 90% of normal cases (True Normal) and misclassified 10% as malignant. It achieved perfect classification for benign cases, with 100% accuracy. For malignant cases, the model correctly identified 97% (True Malignant) but misclassified 3% as benign. The overall performance indicates that the model handles benign and malignant cases particularly well, with slight misclassification between normal and malignant, yielding an accuracy rate of approximately 95.67%.

4.4 ResNet Depth Impact Analysis on Lung Cancer Classification
The objective of these experiments is to assess the impact of different ResNet model depths (ResNet-18, ResNet-34, ResNet-50, and ResNet-101) on the accuracy of lung cancer classification into benign, malignant, and normal categories. Using the balanced IQ-OTHNCCD Lung Cancer Dataset, standard preprocessing steps such as resizing images to 224 × 224 pixels, normalization, and augmentation (if necessary) will be applied. ResNet-18 and ResNet-34 use the Basic Block architecture, while ResNet-50 and ResNet-101 employ Bottleneck blocks. Each model will be trained with the same optimizer, learning rate, and cross-entropy loss for 80 epochs. After training, evaluation metrics like accuracy, precision, recall, and F1-score will be analyzed to understand misclassification patterns. The hypothesis suggests ResNet-18 might struggle with complex patterns, while deeper models like ResNet-34 and ResNet-50 should achieve better accuracy. ResNet-101 may offer the highest accuracy but risks overfitting and increased computational costs, especially with smaller datasets. The results in Table 3 confirm that while deeper models such as ResNet-101 can provide the highest accuracy, ResNet-50 achieves a nearly comparable performance with fewer computational resources and a reduced risk of overfitting. Thus, for lung cancer classification, ResNet-50 offers the most practical and effective balance between accuracy and complexity. The observation that ResNet-34 has lower accuracy than ResNet-18, despite its greater depth, might seem counterintuitive. One possible justification for this results is that if the dataset is not sufficiently large, the model might be overfit to the training data, leading to reduced generalization on the test set. This overfitting can manifest as a lower accuracy on unseen data compared to a shallower model like ResNet-18.

Fig. 5 provides a detailed graphical representation of the performance comparison between four ResNet models: ResNet-18, ResNet-34, ResNet-50, and ResNet-101. The graph illustrates how each model performs across four key evaluation metrics: Accuracy, Precision, Recall, and F1-score. The performance of the models is displayed as percentages, allowing for a clear visual comparison. ResNet-101 consistently achieves the highest scores across all metrics, followed by ResNet-50. ResNet-18 and ResNet-34 show lower performance, with ResNet-34 particularly underperforming in Precision and Recall. This figure visually confirms the trend observed in the numerical results, highlighting the superior performance of deeper ResNet models, especially ResNet-101, in terms of overall accuracy and balanced evaluation metrics.

Figure 5: The performance comparison of various ResNet models (ResNet-18, ResNet-34, ResNet-50, and ResNet-101) across different metrics such as accuracy, precision, recall, and F1-score
4.5 Epoch Variation Impact on ResNet-50 Classification Accuracy and Loss in Lung Cancer Detection
The number of epochs in a ResNet model refers to the number of times the entire training dataset is passed through the model during training. The choice of the number of epochs has a significant impact on the model’s performance, and finding the optimal number is essential. The number of epochs in training a ResNet model significantly impacts performance, requiring a careful balance to avoid underfitting or overfitting. Training for too few epochs can result in underfitting, where the model fails to capture enough patterns from the data, leading to poor performance on both training and unseen data. On the other hand, training for too many epochs can cause overfitting, where the model learns not just the relevant patterns but also noise, reducing its generalization ability and causing it to perform well only on the training set. Accuracy typically improves quickly in early epochs before reaching a plateau, where further training yields diminishing returns or worsens performance. To prevent unnecessary training and resource usage, early stopping mechanisms are often employed once validation performance no longer improves. Learning rate schedules also help ensure optimal learning, as the rate is usually reduced over time to aid convergence. Monitoring validation loss and accuracy allows for determining the optimal number of epochs, often aided by methods like cross-validation or early stopping with patience to ensure the model stops training at the right time [14,35].
To explore how varying the number of epochs affects the classification accuracy and loss rate of ResNet models (e.g., ResNet-50) in lung cancer classification. This experiment aims to identify optimal training conditions that maximize model performance. The results shown in Table 4 illustrate the training and validation performance of a model over 50 epochs. Initially, the model shows modest performance improvements, with train accuracy rising from 53.09% to 82.55% and validation accuracy increasing from 50.55% to 65.45% by Epoch 10. However, the key insights emerge in the later epochs. By Epoch 40, the model achieves a high train accuracy of 97.45% and a low train loss of 0.0702, alongside a substantial improvement in validation accuracy to 94.91% and a validation loss of 0.1877. The optimal performance is seen in Epoch 50, with a peak validation accuracy of 97.09% and a low validation loss of 0.1450, indicating excellent generalization. This suggests that Epoch 50 is the optimal point, balancing high accuracy and low loss, demonstrating effective learning and generalization without significant overfitting. Both Figs. 6 and 7 confirm the above hypotheses by illustrating the effects of epoch count on model performance across different large-scale ranges.

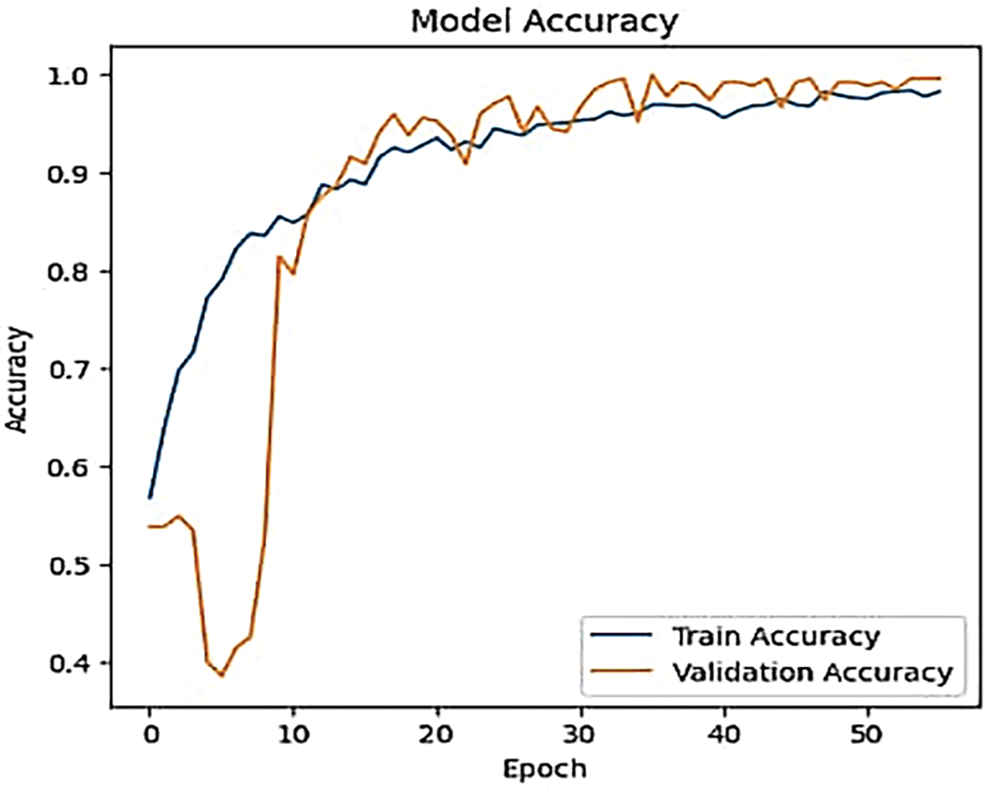
Figure 6: The effect of different epoch count on ResNet-50 accuracy

Figure 7: The effect of different epoch counts on the ResNet-50 loss rate
In general, in the initial epochs, the model learns to identify basic patterns and features. Accuracy typically improves rapidly as the model starts to understand the data. Furthermore, the model’s weights are randomly initialized, and it hasn’t yet learned the patterns in the data. Consequently, the loss rate is typically high as the model makes large prediction errors. In the middle of epochs: As training progresses, the model refines its understanding and starts to capture more complex patterns. The rate of improvement in accuracy may slow down, but continued training allows the model to adjust and optimize its weights further. The loss rate generally continues to decrease, albeit at a slower rate. The model refines its weights and improves its predictions, resulting in reduced errors and lower loss. In the later epochs, the model should ideally converge to a stable state where both training and validation metrics stabilize. Too many epochs can lead to diminishing returns, where further improvements in accuracy become minimal. The loss rate typically stabilizes at a low value. The model should be approaching an optimal state where it makes minimal errors in the training data.
Initially, as the number of epochs increases, the model improves its accuracy by effectively learning and generalizing from the training data. The accuracy increases steadily as the model fine-tunes its weights and captures patterns in the data. After a certain number of epochs, the accuracy curve may plateau. This plateau indicates that the model has reached a point where further training yields minimal improvements in accuracy. The model’s learning rate decreases, and it starts to achieve optimal performance. The most important reason for fluctuations in the accuracy curve before reaching the plateau phase is inadequate learning rate adjustments. During the initial phases of training, the learning rate plays a critical role in determining how effectively the model learns from the data. If the learning rate is too high, it can cause the model to overshoot optimal weight values, leading to oscillations in accuracy as it alternates between different solutions. Conversely, if the learning rate is too low, the model may progress too slowly, resulting in slower and less consistent accuracy improvements. Properly tuning the learning rate and potentially using learning rate schedules or adaptive learning rate methods can help stabilize the accuracy curve and reduce fluctuations before reaching the plateau phase. The same mechanism can explain the fluctuations in the loss curve when the number of epoch increases.
4.6 YOLOv8 Tumor Localization Accuracy Evaluation
To verify the YOLOv8 model’s performance in tumor localization, an experiment was designed to assess its ability to accurately detect and localize lung tumors by comparing predicted bounding boxes with ground truth annotations in a labeled medical image dataset. Using the Comprehensive CT and PET/CT Dataset (Lung-PET-CT-Dx), which contains well-annotated tumor regions, the images will be preprocessed to meet YOLOv8’s input specifications (e.g., resized to 640 × 640 pixels and normalized). The YOLOv8 model will be evaluated using the Intersection over Union (IoU) as the key metric that measures the overlap between the predicted bounding box and the ground truth. An IoU threshold of 0.5 will determine valid detections, with values closer to 1 indicating higher precision. The model will be trained using a 70-15-15 split of training, validation, and testing sets. After training, the IoU for each predicted tumor will be calculated; the results are summarized in Table 5.

The results presented in Table 5 indicate that the YOLOv8 model performs well in detecting and localizing lung tumors, as demonstrated by the high IoU scores, with an average of 0.85. For most images, the model accurately predicted the correct number of tumors, and the IoU scores ranged from 0.82 to 0.87, reflecting precise alignment between the predicted and ground truth bounding boxes. The slight deviation in Image 4, with an IoU of 0.82, could be due to minor inaccuracies in bounding box placement or overlap, potentially caused by factors such as irregular tumor shapes, size variations, or borderline cases where the tumor’s boundary is not clearly defined. Overall, the high average IoU score of 0.85 confirms the model’s effectiveness in tumor detection and localization, validating the robustness of YOLOv8 for this task.
4.7 Impact of DCGAN-Based Synthetic Data Augmentation on CNN Classification and YOLOv8 Detection Performance
To determine whether incorporating synthetic images generated by a DCGAN improves the classification accuracy of a CNN and the detection performance of YOLOv8, an experiment was conducted to compare the results of the models trained with and without DCGAN-based synthetic data augmentation. We utilize the IQ-OTHNCCD Lung Cancer Dataset, which contains CT images categorized into normal, benign, and malignant classes and use DCGAN to generate synthetic images for minority classes (e.g., benign tumors), thus balancing the dataset. Table 6 compares the classification accuracy of ResNet-50 with and without data augmentation using DCGAN. The table also includes different augmentation data sizes to analyze how the size of augmented data impacts the model’s performance. The table shows the impact of data augmentation using DCGAN on the classification accuracy of ResNet-50. Without any augmentation, ResNet-50 achieves an accuracy of 85.23%. However, as synthetic images are added in increments (starting with 10% and increasing to 50% of the original dataset), the classification accuracy improves significantly. With 100 DCGAN-generated images (10% augmentation), accuracy rises to 88.45%, and with 500 synthetic images (50% augmentation), the accuracy reaches 95.65%. This demonstrates that increasing the number of synthetic images generated by DCGAN leads to a consistent and substantial improvement in the model’s performance, highlighting the effectiveness of data augmentation for enhancing classification accuracy.

By using DCGAN to generate synthetic images, data augmentation helps increase the diversity of the training data. Each DCGAN-generated image provides new variations that help the model learn broader patterns rather than memorizing the limited real images. This helps in reducing overfitting. This improvement suggests that the model is learning more generalizable features with the help of augmented data, reducing overfitting. In general, there is a trade-off between the size of augmented data and its impact on accuracy. While data augmentation can significantly improve model performance by providing more diverse training examples and reducing overfitting, there are diminishing returns and even potential reductions in accuracy if augmentation is overused. Beyond a certain point, however, adding too many synthetic images can lead to saturation—the model already learns most of the important features from the data, and adding more synthetic images may not provide new, meaningful information. This can lead to a plateau or diminishing returns in accuracy improvements. There is an optimal balance in augmentation size where the model achieves maximum accuracy, as seen when augmenting the dataset by 50% in our results. Beyond this, adding more synthetic images does not necessarily improve accuracy and can even reduce it if the model starts to overfit to the synthetic data [43,44].
The dual-model framework, combining a CNN for categorization and YOLOv8 for real-time tumor detection, effectively reduces overfitting, a common issue in clinical settings with limited, imbalanced data. Medical imaging often struggles with data scarcity due to privacy concerns and the rarity of certain conditions. To address this, DCGAN-based data augmentation generates synthetic images that introduce realistic variability, expanding the dataset and improving model generalization. This allows both models to learn from a broader range of conditions and tumor shapes, preventing overfitting. YOLOv8 improves its ability to detect tumors at different scales and positions, while CNN enhances its capacity to distinguish between healthy and cancerous tissues, ensuring robust performance across diverse clinical cases.
ResNet addresses overfitting through its structural design and regularization techniques, particularly residual learning and L2 regularization. Residual connections between layers enable deeper networks to avoid the vanishing gradient problem, ensuring smoother gradient flow and promoting the learning of meaningful, generalizable features rather than memorizing noise. L2 regularization, or weight decay, further reduces overfitting by penalizing large weights, which can lead to overly complex models that fit noise in the training data. By adding a penalty term proportional to the square of the weights, L2 regularization encourages smoother, simpler decision boundaries and forces the network to focus on learning general patterns. This approach, combined with ResNet’s efficient residual connections, ensures the model maintains high performance without overfitting, even in deep architectures [1,4,23,24].
4.8 Comparative Analysis of Methodologies for Lung Cancer Classification across Datasets
Table 7 presents a comparison of different methodologies for lung cancer classification across various datasets and approaches. It includes well-established deep learning models like traditional CNN without a residual block, 3D-CNN, and AlexNet, applied to different types of medical imaging datasets along with the proposed methodology that employs a CNN model with a residual block. The method by Toğaçar et al. [6] achieved the highest accuracy in the table, with a remarkable 96.74% using a CNN applied to tomography data. Tomography images, which offer detailed cross-sectional scans, provide a wealth of information that can be effectively leveraged by deep learning models. The high accuracy is likely due to the quality and detailed structure of the tomography dataset, making it easier for CNN to capture important diagnostic features.

In contrast, Zhao et al. [9] employed a CNN with multi-scale feature fusion for the detection of pulmonary nodules, reaching an accuracy of 93.92%. Multi-scale feature fusion enhances feature extraction by analyzing information at various resolutions, which is particularly useful for identifying nodules. However, despite these enhancements, the model’s accuracy is slightly lower than other approaches, possibly due to the inherent challenges of pulmonary nodule detection or the specific limitations of the dataset. Alakwaa et al. [7] adopted a 3D CNN approach, achieving 95.96% accuracy on the DSB dataset. The use of 3D CNNs allows the model to take advantage of spatial information from 3D CT scans, which provides a more comprehensive view of the lung structures. The DSB dataset is a well-known benchmark for lung cancer detection, and the application of 3D CNNs enabled Alakwaa et al. [7] to achieve high performance by effectively utilizing this spatial context.
Mehmood et al. [8] applied the classical AlexNet architecture to histopathology images, obtaining an accuracy of 89.23%. Although AlexNet is a foundational CNN model, it is relatively shallow compared to more modern architectures. The lower accuracy in this case might be due to the complexity and variability present in histopathology images, making it more challenging for the network to classify the data accurately. AL-Huseiny et al. [45] employed GoogLeNet, utilizing inception modules, which enhance computational efficiency and performance. This method achieved an accuracy of 94.38% on the IQ-OTHNCCD Lung Cancer Dataset, which is specifically designed for lung cancer detection. The success of GoogLeNet in this case highlights the strength of inception-based architectures for handling medical image data.
The proposed methodology (2024) outperforms many of the other methods with an accuracy of 97.82%, utilizing a CNN model with residual blocks. Residual blocks, which are a key feature of ResNet architectures, help address the vanishing gradient problem, allowing for the training of deeper networks. This capability, combined with the structured nature of the IQ-OTHNCCD dataset, enables the model to extract more meaningful and deeper features, leading to a higher classification performance compared to simpler CNN models. The proposed methodology surpasses other methods by achieving a high accuracy of 97.82%, leveraging a CNN model with residual blocks, which are designed to address the vanishing gradient problem and enable the training of deeper, more efficient networks. This deeper feature extraction capability allows the model to capture more complex patterns in the IQ-OTHNCCD Lung Cancer Dataset, outperforming traditional CNNs and architectures like AlexNet, GoogLeNet, and even 3D CNNs. The superior performance is due to the model’s ability to balance depth and complexity, extracting more nuanced diagnostic features from the dataset while maintaining high accuracy.
Fig. 8 presents a graphical representation comparing the accuracy of several convolutional neural network (CNN) architectures, confirming the performance results across different models. The chart includes various architectures such as standard CNN, CNN with multi-scale feature fusion, 3D CNN, AlexNet, GoogLeNet, and the proposed CNN model with residual blocks. The proposed model achieves one of the highest accuracy rates, alongside the standard CNN, demonstrating the effectiveness of incorporating residual blocks. Other models, such as AlexNet and GoogLeNet, show lower performance, with AlexNet achieving the lowest accuracy. This figure visually reinforces the conclusion that the proposed model with residual blocks outperforms many of the other architectures, particularly in terms of accuracy.

Figure 8: The comparison of different CNN architectures (e.g., CNN, 3D CNN, AlexNet, GoogLeNet, and CNN with residual block) based on their accuracy performance
Residual blocks play a critical role in enabling deeper CNNs by introducing skip connections, which allow the network to learn residuals (the difference between input and output) rather than directly mapping inputs to outputs. This design makes it easier to train deep networks by improving gradient flow during backpropagation, preventing gradients from becoming too small as they pass through many layers. By allowing information to bypass certain layers, residual blocks also address the degradation problem, where deeper networks perform worse than shallower ones, by facilitating identity learning when extra layers are unnecessary. The skip connections promote feature reuse, enabling the network to focus on new information while leveraging earlier learned features, making learning more efficient. Additionally, this architecture accelerates convergence during training, leading to faster optimization and improved accuracy [14,35,43].
The results in Fig. 8 underscore the critical role of the residual block in enhancing the performance of the proposed model, which significantly outperforms the method described in Reference [6], where a CNN without residual blocks was used. Specifically, the proposed model achieves approximately a 1.5% improvement in accuracy across different datasets. This improvement highlights the effectiveness of residual blocks in optimizing learning and generalization, particularly with more complex datasets. While the comparative model in Reference [6] was tested on a relatively simple dataset (Tomography dataset), the proposed model demonstrates its superiority using the more challenging and diverse IQ-OTHNCCD Lung Cancer Dataset. The complexity of the latter dataset, combined with the advantages of residual learning, showcases the robustness of the suggested approach in handling intricate medical data and yielding higher accuracy.
Furthermore, to compare the suggested model in which CNN learns hierarchical features from images, and YOLOv8 captures multi-scale features for real-time tumor detection with state-of-the-art-dual-models presented in Reference [46] in which multiple CNNs use various layers, kernels, and pooling techniques to learn diverse features in terms of lung cancer detection performance. The results presented in Table 8 highlight the comparative strengths of the proposed model and the Ensemble 2D CNN approach for lung cancer detection. The proposed model, which combines CNN for classification with YOLOv8 for real-time tumor localization, outperforms the Ensemble 2D CNN approach with a classification accuracy of 97.67%, compared to 95% for the latter. Additionally, the proposed model achieves an impressive Intersection over Union (IoU) score of 0.85 for precise tumor localization, a feature absents in the Ensemble approach. The use of DCGAN-generated synthetic images also boosts the performance of the proposed model by 10%, further enhancing its classification and detection capabilities. Moreover, the proposed model supports real-time tumor detection, making it more suitable for clinical applications, while the Ensemble 2D CNN approach focuses solely on classification without real-time processing.

By integrating the YOLOv8 model for precise tumor detection and localization alongside the classification capabilities of the CNN model, we have developed a robust and comprehensive approach to lung cancer analysis. YOLOv8’s real-time detection allows the system to accurately identify and pinpoint tumor regions within images, while the CNN, enhanced with residual blocks, excels at classifying these regions with high accuracy. This combination outperforms comparative methods by addressing both detection and classification in a unified framework, offering superior diagnostic insights. The advanced feature extraction of the CNN, combined with YOLOv8’s spatial accuracy, provides a more holistic and precise solution compared to standalone models, leading to improved accuracy and efficiency in lung cancer detection and classification.
4.9 Computational Cost and Processing Time
The combination of a ResNet for categorization and YOLOv8 for real-time tumor detection in lung cancer introduces a significant computational cost, particularly in terms of O-notation. For the ResNet the complexity depends on the number of layers, neurons, and filter sizes, typically expressed as
Below is a comparative table summarizing the estimated processing times for the proposed model (CNN + YOLOv8) and the Ensemble 2D CNN approach [46], based on the hardware configuration. Table 9 confirms that the proposed model’s combination of CNN and YOLOv8 adds computational complexity due to the high-dimensional feature extraction by the CNN and the multi-scale tumor localization performed by YOLOv8. When executed on a CPU, which is less optimized for deep learning tasks, the processing time increases significantly compared to GPU-based systems, as YOLOv8, despite being designed for real-time detection, experiences considerable slowdowns on a CPU. In contrast, the Ensemble 2D CNN approach, while utilizing multiple CNNs, avoids the additional computational demands of real-time object detection and localization. By focusing solely on classification without the YOLOv8 component, this method results in a lower overall computational cost and faster per-image processing times.
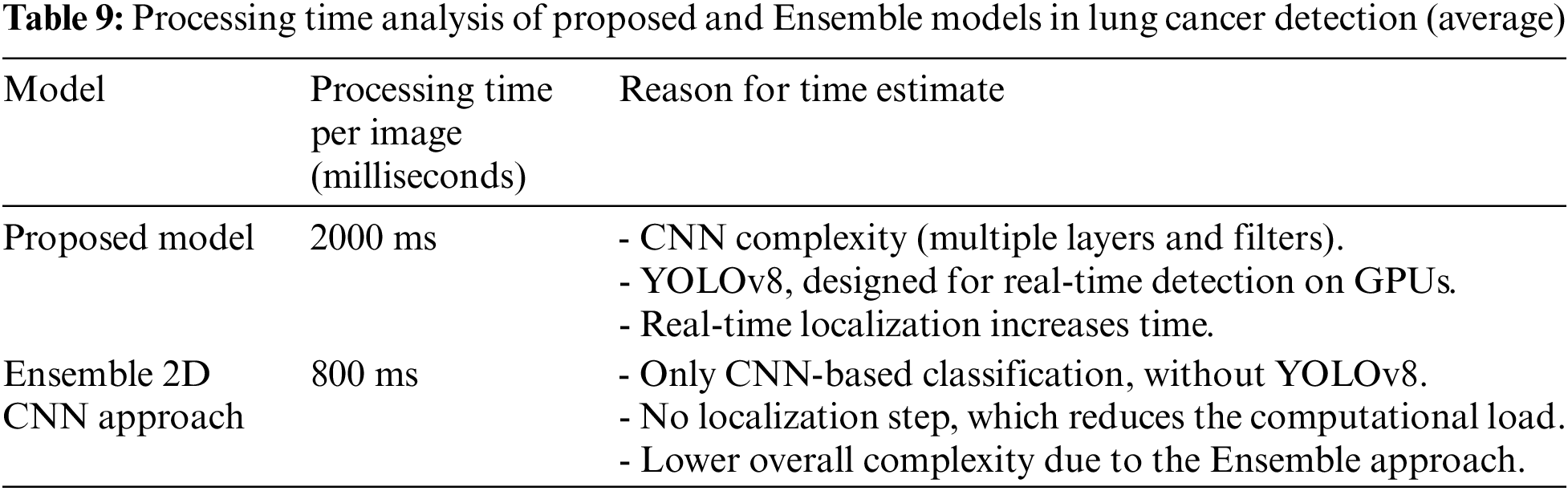
GPU-based systems can significantly reduce processing time for lung cancer detection by leveraging parallel processing capabilities, allowing thousands of calculations to occur simultaneously, which is particularly beneficial for deep learning tasks like convolutions in CNNs. They are optimized for deep learning frameworks, utilize high memory bandwidth for faster data transfer, and efficiently perform matrix operations, crucial for handling large image datasets. Additionally, GPUs enable real-time detection through high frame rates and can process multiple images at once using batch processing, further decreasing overall processing time and facilitating quicker diagnoses.
While the proposed dual-model approach combining CNNs and YOLOv8 offers notable improvements in lung cancer detection, several limitations must be considered. The reliance on synthetic data augmentation using DCGAN, though beneficial for enhancing training data, may not fully capture the complexity of real-world medical images, potentially leading to overfitting or biased performance on unseen clinical datasets. Additionally, the computational demands of the CNN-YOLOv8 architecture, particularly for real-time tumor localization, may hinder its application in resource-limited clinical settings. The model, despite achieving a high IoU score, may still struggle with edge cases such as irregularly shaped tumors or those near complex anatomical structures. Moreover, the lack of testing on a large, diverse patient dataset limits its generalizability, underscoring the need for further validation on broader and more heterogeneous populations.
4.11 Discussion: Enhancing Model Interpretability for Clinical Decision-Making
To enhance the interpretability of a dual-model approach integrating complex deep learning algorithms for clinical use, several strategies can be employed to make predictions transparent and meaningful for end-users. Shapley Additive Explanations (SHAP) can be used to interpret lung cancer detection in a dual-model deep learning approach by quantifying the contribution of each input feature (such as pixel values from CT or PET scans) to the final model prediction. In a dual-model setup, where one model may handle tumor categorization (e.g., benign vs. malignant) and the other handles tumor localization or identification (e.g., using CNNs and YOLOv8), SHAP helps by breaking down the prediction of each model into contributions from individual features. For example, SHAP can show how specific pixel regions in a CT scan, influenced by the first model, and corresponding feature activations from the second model, affect the likelihood of the tumor being malignant. By assigning Shapley values to features, SHAP provides a global and local explanation—global for understanding overall feature importance and local for individual case analysis—offering clinicians a clear understanding of how the dual-model arrived at its decision. This transparency helps end-users interpret which image regions or features are driving the model’s prediction, building trust in the AI system for clinical use [47].
Furthermore, Grad-CAM (Gradient-weighted Class Activation Mapping) is particularly useful for CNN-based models, as it highlights specific areas of an image that are most influential in the model’s predictions. For lung cancer detection, Grad-CAM can generate heatmaps over CT images, visually indicating which regions of the tumor or surrounding tissue led to the classification, thereby increasing clinicians’ trust by aligning the model’s focus with their medical expertise. By applying Grad-CAM to the CNN, heatmaps can be generated to visually highlight the specific regions in CT or PET scans that influence the model’s decision to classify a tumor as benign or malignant. Similarly, Grad-CAM can be applied to YOLOv8 to show how the model identifies and pinpoints tumor regions during real-time detection. This dual application of Grad-CAM provides interpretable visual explanations, allowing clinicians to see how both the categorization and localization processes align with medical understanding, thereby enhancing trust and transparency in the model’s predictions [48].
Lung cancer remains one of the leading causes of cancer-related mortality worldwide, underscoring the importance of early and accurate detection to improve patient outcomes. This research has demonstrated that combining two powerful deep learning models—a CNN for categorization and the YOLOv8 architecture for real-time tumor identification and localization—can significantly enhance the detection and classification of lung cancer. CNN effectively extracted hierarchical features from medical images, achieving an accuracy of 97.67% in distinguishing between healthy and cancerous lung tissues. Meanwhile, the YOLOv8 model’s multi-scale feature extraction allowed for precise tumor localization, achieving an IoU score of 0.85, making it particularly adept at detecting small or irregularly shaped tumors. One of the key advantages of this dual-model approach is its robustness in handling the variability present in medical imaging, including differences in imaging conditions, patient anatomy, and tumor characteristics. Additionally, the integration of synthetic data generated by DCGAN helped address the issue of limited training data, resulting in a 10% improvement in both classification and detection performance. Future research could focus on refining the models by incorporating more advanced data augmentation techniques to further enhance their generalization capabilities across different patient populations and imaging modalities. Another area of exploration could involve integrating 3D imaging data for a more comprehensive analysis of tumor volume and progression. Additionally, investigating the potential of explainable AI (XAI) methods would help to provide clearer insights into the decision-making process of these models, increasing their trustworthiness for clinical use.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Saad Mohamed Darwish, Sumaia Mohamed Elhassan; data collection: Sumaia Mohamed Elhassan; analysis and interpretation of results: Saleh Mesbah Elkaffas, Sumaia Mohamed Elhassan; draft manuscript preparation: Saad Mohamed Darwish, Sumaia Mohamed Elhassan. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data is available on both CT and PET/CT, and IQ-OTH/NCCD Lung Cancer Datasets, a public benchmark datasets available at: https://www.cancerimagingarchive.net/collection/lung-pet-ct-dx/ (accessed on 14 November 2024) and https://www.kaggle.com/datasets/hamdallak/the-iqothnccd-lung-cancer-dataset (accessed on 14 November 2024). The authors confirm others would be able to access these data in the same manner as the authors and that the authors did not have any special access privileges that others would not have.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Thanoon MA, Zulkifley MA, Mohd Zainuri MA, Abdani SR. A review of deep learning techniques for lung cancer screening and diagnosis based on CT images. Diagnostics. 2023;13(16):2617. doi:10.3390/diagnostics13162617. [Google Scholar] [PubMed] [CrossRef]
2. Zhang C, Sun X, Dang K, Li K, Guo XW, Chang J, et al. Toward an expert level of lung cancer detection and classification using a deep convolutional neural network. Oncologist. 2019;24(9):1159–65. doi:10.1634/theoncologist.2018-0908. [Google Scholar] [PubMed] [CrossRef]
3. Dlamini S, Chen Y-H, Kuo C-FJ. Complete fully automatic detection, segmentation and 3D reconstruction of tumor volume for non-small cell lung cancer using YOLOv4 and region-based active contour model. Expert Syst Appl. 2023;212:118661. doi:10.1016/j.eswa.2022.118661. [Google Scholar] [CrossRef]
4. Kumar Y, Koul A, Singla R, Ijaz MF. Artificial intelligence in disease diagnosis: a systematic literature review, synthesizing framework and future research agenda. J Ambient Intell Humaniz Comput. 2023;14(7):8459–86. doi:10.1007/s12652-021-03612-z. [Google Scholar] [PubMed] [CrossRef]
5. Cui F, Li Y, Luo H, Zhang C, Du H. SF2T: Leveraging swin transformer and two-stream networks for lung nodule detection. Biomed Signal Process Control. 2024;95:106389. doi:10.1016/j.bspc.2024.106389. [Google Scholar] [CrossRef]
6. Toğaçar M, Ergen B, Cömert Z. Detection of lung cancer on chest CT images using minimum redundancy maximum relevance feature selection method with convolutional neural networks. Biocybern Biomed Eng. 2020;40(1):23–39. doi:10.1016/j.bbe.2019.11.004. [Google Scholar] [CrossRef]
7. Alakwaa W, Nassef M, Badr A. Lung cancer detection and classification with 3D convolutional neural network (3D-CNN). Int J Adv Comput Sci Appl. 2017;8(8):409–17. doi:10.14569/IJACSA.2017.080853. [Google Scholar] [CrossRef]
8. Mehmood S, Ghazal TM, Khan MA, Zubair M, Naseem MT, Faiz T, et al. Malignancy detection in lung and colon histopathology images using transfer learning with class selective image processing. IEEE Access. 2022;10:25657–68. doi:10.1109/ACCESS.2022.3150924. [Google Scholar] [CrossRef]
9. Zhao J, Zhang C, Li D, Niu J. Combining multi-scale feature fusion with multi-attribute grading, a CNN model for benign and malignant classification of pulmonary nodules. J Digit Imaging. 2020;33:869–78. doi:10.1007/s10278-020-00333-1. [Google Scholar] [PubMed] [CrossRef]
10. Khanmohammadi A, Aghaie A, Vahedi E, Qazvini A, Ghanei M, Afkhami A, et al. Electrochemical biosensors for the detection of lung cancer biomarkers: a review. Talanta. 2020;206:120251. doi:10.1016/j.talanta.2019.120251. [Google Scholar] [PubMed] [CrossRef]
11. Salama WM, Shokry A, Aly MH. A generalized framework for lung cancer classification based on deep generative models. Multimed Tools Appl. 2022;81(23):32705–22. doi:10.1007/s11042-022-13005-9. [Google Scholar] [CrossRef]
12. Naseer I, Masood T, Akram S, Jaffar A, Rashid M, Iqbal MA. Lung cancer detection using modified AlexNet architecture and support vector machine. Comput Mater Contin. 2023;74(1):2039–54. doi:10.32604/cmc.2023.032927. [Google Scholar] [CrossRef]
13. Liu S, Yao W. Prediction of lung cancer using gene expression and deep learning with KL divergence gene selection. BMC Bioinform. 2022;23(1):175. doi:10.1186/s12859-022-04689-9. [Google Scholar] [PubMed] [CrossRef]
14. Reddy S, Bhuvaneshwari V, Tikariha AK, Amballa YS, Raj BSS. Automatic pulmonary nodule detection in CT scans using Xception, ResNet50 and advanced convolutional neural networks models. Int J Eng Res Technology. 2022;9(4):3226–37. doi:10.1109/TMI.2019.2935553. [Google Scholar] [CrossRef]
15. Zhan X, Long H, Gou F, Duan X, Kong G, Wu J. A convolutional neural network-based intelligent medical system with sensors for assistive diagnosis and decision-making in non-small cell lung cancer. Sensors. 2021;21(23):7996. doi:10.3390/s21237996. [Google Scholar] [PubMed] [CrossRef]
16. Nageswaran S, Arunkumar G, Bisht AK, Mewada S, Kumar JS, Jawarneh M, et al. Lung cancer classification and prediction using machine learning and image processing. BioMed Res Int. 2022;2022(1):1755460. doi:10.1155/2022/1755460. [Google Scholar] [PubMed] [CrossRef]
17. Zhang G, Lin L, Wang J. Lung nodule classification in CT images using 3D DenseNet. J Phy: Conf Ser. 2021;1827:012155. doi:10.1088/1742-6596/1827/1/012155. [Google Scholar] [CrossRef]
18. Qiao T, Shao H, Xie S, Shi R. Unsupervised generative fake image detector. IEEE Trans Circuits Syst Video Technology. 2024;34(9):6900–15. [Google Scholar]
19. Swathi K, Kodukula S. XGBoost classifier with Hyperband optimization for cancer prediction based on gene selection by using machine learning techniques. Revue d’Intelligence Artificielle. 2022;36(5):665–80. [Google Scholar]
20. Bhalerao RY, Jani HP, Gaitonde RK, Raut V. A novel approach for detection of lung cancer using digital image processing and convolution neural networks. In: 5th International Conference on Advanced Computing & Communication Systems (ICACCS), 2019 Mar 15; Coimbatore, India; p. 577–83. [Google Scholar]
21. Nadkarni NS, Borkar S. Detection of lung cancer in CT images using image processing. In: 3rd International Conference on Trends in Electronics and Informatics (ICOEI), 2019 Apr 23; Tirunelveli, India; p. 863–6. [Google Scholar]
22. Sori WJ, Feng J, Godana AW, Liu S, Gelmecha DJ. DFD-Net: lung cancer detection from denoised CT scan image using deep learning. Front Comput Science. 2021;15:1–13. doi:10.1007/s11704-020-9050-z. [Google Scholar] [CrossRef]
23. Forte GC, Altmayer S, Silva RF, Stefani MT, Libermann LL, Cavion CC, et al. Deep learning algorithms for diagnosis of lung cancer: a systematic review and meta-analysis. Cancers. 2022;14(16):3856. doi:10.3390/cancers14163856. [Google Scholar] [PubMed] [CrossRef]
24. Han K, Sheng VS, Song Y, Liu Y, Qiu C, Ma S, et al. Deep semi-supervised learning for medical image segmentation: a review. Expert Syst Applications. 2024;245:123052. doi:10.1016/j.eswa.2023.123052. [Google Scholar] [CrossRef]
25. Rathod B, Seema M, Ragha L. DLCTLungDetectNet: deep learning for lung tumor detection in CT scans. Int J Comput Digit Systems. 2024 Jan 22;15(1):1–9. doi:10.52783/jes.1771. [Google Scholar] [CrossRef]
26. Narotamo H, Dias M, Santos R, Carreiro AV, Gamboa H, Silveira M. Deep learning for ECG classification: a comparative study of 1D and 2D representations and multimodal fusion approaches. Biomed Signal Proces Control. 2024 Jul 1;93:106141. doi:10.1016/j.bspc.2024.106141. [Google Scholar] [CrossRef]
27. Mirza A, Shaikh B. A deep learning based dual model developed for automated detection of glioma-brain tumor. In: Proceedings of the 4th International Conference on Advances in Science & Technology (ICAST2021), 2021 May 7; Mumbai, India; p. 1–6. [Google Scholar]
28. Kiran A, Ramesh JV, Rahat IS, Khan MA, Hossain A, Uddin R. Advancing breast ultrasound diagnostics through hybrid deep learning models. Comput Biol Medicine. 2024 Sep 1;180:108962. doi:10.1016/j.compbiomed.2024.108962. [Google Scholar] [PubMed] [CrossRef]
29. Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomic study. PLoS Med. 2018;15(11):e1002711. doi:10.1371/journal.pmed.1002711. [Google Scholar] [PubMed] [CrossRef]
30. Gatidis S, Hepp T, Früh M, La Fougère C, Nikolaou K, Pfannenberg C, et al. A whole-body FDG-PET/CT dataset with manually annotated tumor lesions. Sci Data. 2022 Oct 4;9(1):601. doi:10.1038/s41597-022-01718-3. [Google Scholar] [PubMed] [CrossRef]
31. Reser C, Reich C. DCGAN-based data augmentation for enhanced performance of convolution neural networks. In: Data Analytics 2020: The Ninth International Conference on Data Analytics, 2020 Oct 25–29; Nice, France; p. 47–53. [Google Scholar]
32. Bali M, Mahara T. Comparison of affine and DCGAN-based data augmentation techniques for chest X-ray classification. Procedia Comput Sci. 2023 Jan 1;218(11):283–90. doi:10.1016/j.procs.2023.01.010. [Google Scholar] [CrossRef]
33. Wang S, Dong L, Wang X, Wang X. Classification of pathological types of lung cancer from CT images by deep residual neural networks with transfer learning strategy. Open Med. 2020 Mar 8;15(1):190–7. doi:10.1515/med-2020-0028. [Google Scholar] [PubMed] [CrossRef]
34. Wu P, Sun X, Zhao Z, Wang H, Pan S, Schuller B. Classification of lung nodules based on deep residual networks and migration learning. Comput Intell Neurosci. 2020;2020(1):8975078. doi:10.1155/2020/8975078. [Google Scholar] [PubMed] [CrossRef]
35. Khanna A, Londhe ND, Gupta S, Semwal A. A deep Residual U-Net convolutional neural network for automated lung segmentation in computed tomography images. Biocybern Biomed Eng. 2020 Jul 1;40(3):1314–27. doi:10.1016/j.bbe.2020.07.007. [Google Scholar] [CrossRef]
36. Yuvaneswaren RS, Prakash ST, Sudha K. A YOLOv8-based model for precise corrosion segmentation in industrial imagery. In: 3rd International Conference on Artificial Intelligence for Internet of Things (AIIoT), 2024 May 3; Tamil Nadu, India; p. 1–6. [Google Scholar]
37. Tasnim F, Islam MT, Maisha AT, Sultana I, Akter T, Islam MT. Comparison of brain tumor detection techniques by using different machine learning YOLO algorithms. In: Congress on Intelligent Systems, 2023 Sep 4; Singapore: Springer Nature Singapore; p. 51–65. [Google Scholar]
38. Ragab M, Abdulkader S, Muneer A, Alqushaibi A, Sumiea EH, Qureshi R, et al. A comprehensive systematic review of YOLO for medical object detection (2018 to 2023). IEEE Access. 2024 Apr 10;12:57815–36. [Google Scholar]
39. Şaman Cİ, Çelikbaş ŞE. YOLOv8-based lung nodule detection: a novel hybrid deep learning model proposal. Int Res J Eng Technology. 2023;10(8):230–7. [Google Scholar]
40. Mammeri S, Amroune M, Haouam MY, Bendib I, Corrêa Silva A. Early detection and diagnosis of lung cancer using YOLOv7, and transfer learning. Multimed Tools Applications. 2024 Mar;83(10):30965–80. doi:10.1007/s11042-023-16864-y. [Google Scholar] [CrossRef]
41. Huang W, Chen Y, Zhu X. YOLOv8: A new method for object detection of kidney stone images. In: Proceedings of the 2024 6th International Conference on Intelligent Medicine and Image Processing, 2024 Apr 26; Bali, Indonesia; p. 36–40. [Google Scholar]
42. Saranya G, Sujatha C. A novel framework leveraging adam optimization techniques, coupled with mask R-CNN deep learning mechanisms for cervical cancer detection system. In: 3rd International Conference on Applied Artificial Intelligence and Computing (ICAAIC), 2024 Jun 5; Tamil Nadu, India; p. 233–45. Available from: IEEE. [Google Scholar]
43. Du A, Zhou Q, Dai Y. Methodology for evaluating the generalization of ResNet. Appl Sciences. 2024 May 6;14(9):3951. doi:10.3390/app14093951. [Google Scholar] [CrossRef]
44. Beltran-Royo C, Llopis-Ibor L, Pantrigo JJ, Ramírez I. DC neural networks avoid overfitting in one-dimensional nonlinear regression. Knowl-Based Systems. 2024 Jan 11;283:111154. doi:10.1016/j.knosys.2023.111154. [Google Scholar] [CrossRef]
45. Al-Huseiny MS, Sajit AS. Transfer learning with GoogLeNet for detection of lung cancer. Indones J Electr Eng Comput Sci. 2021;22(2):1078–86. doi:10.11591/ijeecs.v22.i2.pp1078-1086. [Google Scholar] [CrossRef]
46. Shah AA, Malik HA, Muhammad A, Alourani A, Butt ZA. Deep learning ensemble 2D CNN approach towards the detection of lung cancer. Sci Reports. 2023 Feb 20;13(1):2987. doi:10.1038/s41598-023-29656-z. [Google Scholar] [PubMed] [CrossRef]
47. Nohara Y, Matsumoto K, Soejima H, Nakashima N. Explanation of machine learning models using Shapley additive explanation and application for real data in hospital. Comput Meth Prog Bio. 2022 Feb 1;214:106584. doi:10.1016/j.cmpb.2021.106584. [Google Scholar] [PubMed] [CrossRef]
48. Qiu Z, Rivaz H, Xiao Y. Is visual explanation with Grad-CAM more reliable for deeper neural networks? A case study with automatic pneumothorax diagnosis. In: International Workshop on Machine Learning in Medical Imaging, 2023 Oct 8; Vancouver, BC, Canada; p. 224–33. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools