 Open Access
Open Access
ARTICLE
Congruent Feature Selection Method to Improve the Efficacy of Machine Learning-Based Classification in Medical Image Processing
1 Department of Computer Engineering, Aligarh Muslim University, Aligarh, 202002, India
2 College of Computer Science, King Khalid University, Abha, 61413, Saudi Arabia
3 School of Computing, Gachon University, Seongnam, 13120, Republic of Korea
4 Department of Computer Science and Information Systems, College of Applied Sciences, AlMaarefa University, Riyadh, 13713, Saudi Arabia
5 Department of Computer Engineering and Information, College of Engineering in Wadi Alddawasir, Prince Sattam bin Abdulaziz University, Al-Kharj, 16273, Saudi Arabia
* Corresponding Author: Hong Min. Email:
Computer Modeling in Engineering & Sciences 2025, 142(1), 357-384. https://doi.org/10.32604/cmes.2024.057889
Received 30 August 2024; Accepted 12 November 2024; Issue published 17 December 2024
Abstract
Machine learning (ML) is increasingly applied for medical image processing with appropriate learning paradigms. These applications include analyzing images of various organs, such as the brain, lung, eye, etc., to identify specific flaws/diseases for diagnosis. The primary concern of ML applications is the precise selection of flexible image features for pattern detection and region classification. Most of the extracted image features are irrelevant and lead to an increase in computation time. Therefore, this article uses an analytical learning paradigm to design a Congruent Feature Selection Method to select the most relevant image features. This process trains the learning paradigm using similarity and correlation-based features over different textural intensities and pixel distributions. The similarity between the pixels over the various distribution patterns with high indexes is recommended for disease diagnosis. Later, the correlation based on intensity and distribution is analyzed to improve the feature selection congruency. Therefore, the more congruent pixels are sorted in the descending order of the selection, which identifies better regions than the distribution. Now, the learning paradigm is trained using intensity and region-based similarity to maximize the chances of selection. Therefore, the probability of feature selection, regardless of the textures and medical image patterns, is improved. This process enhances the performance of ML applications for different medical image processing. The proposed method improves the accuracy, precision, and training rate by 13.19%, 10.69%, and 11.06%, respectively, compared to other models for the selected dataset. The mean error and selection time is also reduced by 12.56% and 13.56%, respectively, compared to the same models and dataset.Keywords
Machine learning (ML) is a subset of artificial intelligence that enables machines to learn from data without being explicitly programmed, making decisions based on prior learning. Computer vision (CV) is a specific domain of ML that teaches machines to interpret and understand visual data from the world around them. It involves using algorithms and mathematical models to analyze and interpret images or videos and then make decisions or take actions based on that analysis [1]. CV aims to enable computers to see and interpret the visual world as humans do and to use that understanding to perform a wide range of tasks, from object recognition and tracking to autonomous navigation and decision-making. In medical imaging, ML algorithms extract relevant features and characteristics from images to support medical diagnosis and treatment processes [2,3]. These algorithms help reduce errors in medical image processing, thereby improving the accuracy of the diagnosis [4,5].
Medical images are complex and challenging to interpret, and even experienced radiologists can make errors or overlook important details. ML algorithms are used to automate various tasks in medical image processing, such as image segmentation and feature extraction, helping to reduce human error and improve analysis accuracy [6]. These algorithms are trained to recognize and segment-specific structures or patterns within medical images, such as tumours, blood vessels, or regions of inflammation. They then analyze these structures to extract quantitative features—such as size, shape, texture, and intensity used to support medical diagnosis and treatment planning [7,8]. Medical images are typically captured using specialized imaging modalities such as X-rays, computed tomography (CT), magnetic resonance imaging, ultrasound, and positron emission tomography, which produce detailed images of specific organs or structures within the body [9]. Applying ML algorithms reduces the time and effort required for image analysis [10]. These algorithms enhance accuracy, efficiency, objectivity, speed, and treatment planning [11].
Feature extraction is a process in ML that involves identifying and extracting important features or patterns from an image relevant to a specific task, application, or object [12]. These features may include colour, texture, shape, or other visual characteristics of the image. During feature extraction, a region of interest (ROI) is typically identified and isolated from the rest of the image [13]. This ROI contains the specific object or pattern of interest, such as a tumour or blood vessel in a medical image [14]. Once the ROI is identified, feature extraction algorithms analyze its visual characteristics to extract relevant features or patterns [15]. Feature extraction techniques can be applied to high-resolution and low-resolution images, but their effectiveness depends on the image’s quality and complexity [16]. Generative algorithms are also used for image super-resolution, where low-resolution medical images are upscaled to higher resolution using a generative model. These algorithms have been used to generate synthetic medical images for training deep learning (DL) models, augmenting small datasets and improving model performance. In generative algorithms, feature extraction techniques are applied to extract relevant and informative features from images, which are then used to generate new data. Feature extraction reduces latency in classification, segmentation, and identification processes, enhancing detection accuracy and efficiency [17].
Feature selection is a pre-processing technique that involves selecting a subset of features from a larger set of available features in an image or dataset [18]. Feature selection aims to identify the most important and informative features related to specific tasks, such as image classification or object detection. This process helps reduce the data’s dimensionality, improving the analysis’s efficiency and accuracy [19]. Several feature selection methods exist, including statistical tests, correlation analysis, and ML algorithms [20]. These methods evaluate the importance of different features based on their relevance to the task at hand, their correlation with other features, and their ability to enhance the performance of an ML model [21]. Therefore, feature selection is an important technique in medical image processing, as it reduces the number of input variables and enhances the effectiveness and performance of the systems [22]. This process demonstrates that precise feature selection in CV applications is a critical task that directly impacts pattern detection and region classification. There is now a need for a more accurate and flexible feature selection method that can identify relevant patterns across different medical image datasets. Therefore, a Congruent Feature Selection Method (CFSM) using an analytical learning paradigm is proposed to enhance feature selection for CV applications. The CFSM algorithm employs a learning paradigm that computes correlation and similarity-related features to identify pixel distributions and intensities. These distribution patterns are then used to detect diseases with maximum recognition accuracy.
The following objectives are established for the implementation of the proposed method’s contribution and novelty:
1. Designing the Congruent Feature Selection Method (CFSM) for maximizing the accuracy of processing medical images, regardless of intensity and pixel distributions.
2. Developing a pattern distribution and classification process for congruency verification, enhancing feature selection precision.
3. Validating the performance of the proposed method through experimental analysis using various medical images and regions.
The research paper is structured into six main sections. Section 1 provides a brief overview of the research problem and sets the stage for the rest of the paper. Section 2 reviews existing literature and research related to the problem. Section 3 presents the proposed CFSM to address these issues. Section 4 outlines the experimental analysis conducted to evaluate the proposed method, while Section 5 compares the study’s findings with those of other state-of-the-art models. Finally, Section 6 summarizes the conclusions of the study.
2.1 Feature Selection in Medical Image Classification
Sun et al. [23] developed an Adaptive Feature Selection-guided Deep Forest model to detect and classify COVID-19 infections. The model extracted location-specific features from CT images and employed a deep forest approach to determine a high-level representation. The proposed feature selection method reduced feature redundancy, which was adaptively integrated into the model. However, this method consumed significant computation time when analyzing large real-time datasets. Similarly, in [24], authors proposed an IoT-based optimization-driven deep belief networks model that leverages the Mayfly optimization algorithm for feature selection, aiming to achieve precise and reliable classifications that assist clinicians in early disease detection and reduce diagnostic errors. This propsed model incorporates advanced feature extraction and selection methods to refine classification outcomes. A novel approach for Multi-Objective Feature Selection using a Genetic Algorithm and a 3-Dimensional Compass was introduced for binary medical classification [25]. The proposed method outperformed other competitive Multi-Objective Feature Selection methods. These methods must identify the essential medical data features and patterns from the images. A genetic model was employed to reduce latency in the classification process; however, it incurred high computational costs.
2.2 Advances in Segmentation and Disease Detection
Yue et al. [26] designed an automated polyp segmentation method called LFSRNet to segment polyps in colonoscopy images accurately. The proposed model architecture includes a lesion-aware feature refinement module to perform the feature selection process. This module detects the characteristics and variables essential for disease detection and prediction, decoding the data hidden within the images. The method demonstrated better generalization than competing approaches when tested on other datasets. A novel feature selection method utilizing a wrapper technique and binary bat algorithm was also developed for classifying diabetic retinopathy [27]. The binary bat algorithm selected feasible information and features extracted by the EfficientNet and DenseNet models. An equilibrium optimizer was employed to classify the specific types of features based on certain conditions and functions. This study demonstrated that DL techniques and wrapper methods could provide an accurate and cost-effective approach to diagnosing diabetic retinopathy. In [28], the authors proposed an intelligent approach to diagnosing colorectal cancer using a group teaching optimization algorithm for feature selection and a multilayer artificial neural network to classify images. The experiments demonstrated that the proposed method outperformed classification methods such as 3-layer convolutional neural network (CNN), Random Forest, and CNN DropBlock.
2.3 Deep Learning in Medical Image Fusion and Classification
Artificial intelligence and the Internet of Things in medical imaging for early disease detection and patient monitoring have revealed promising solutions. Therefore, Khan et al. [29] developed a new feature selection method for computer-aided gastrointestinal disease analysis using DL feature fusion and selection. The proposed system utilized a VGG16 architecture to extract crucial features, fused using an array-based technique. The best individual was selected using particle swarm optimization with a mean value-based fitness function. While combining the radiomics framework with DL networks in medical image classification has shown promise, it can be hindered by challenges such as overfitting and ineffective feature selection. To address these challenges, a new approach called Deep Semantic Segmentation Feature-based Radiomics (DSFR) has been introduced [30]. The DSFR framework includes feature extraction and selection components that utilize a novel feature similarity adaptation algorithm; however, the system struggles with scalability and reliability. To address these limitations, a novel deep multi-cascade fusion method with a classifier-based feature extraction approach for multi-modal medical images was proposed in [31]. Multi-modal images were utilized to extract relevant features, and a Gaussian high-pass filtering technique was implemented to identify these features and their corresponding maps from the images.
2.4 Novel Architectures for Improved Medical Image Analysis
Li et al. [32] proposed a novel framework called the dual-branch feature-enhanced network (DFENet) for multi-modal medical image fusion. The convolutional neural network algorithm was implemented to detect key patterns and features from the input medical images. This method was based on a decoder-encoder network, which trained on datasets collected from low-resolution images. The DFENet method maximized the accuracy of image classification and identification processes, enhancing diagnostic efficiency. Additionally, a hybrid domain feature learning model for medical image classification was suggested, combining global features in the frequency domain with local features in the spatial domain [33]. This proposed module utilized a windowed fast Fourier convolution pyramid. The authors integrated ResNet, FPN, and an attention mechanism to build the module and employed a genetic algorithm for automatic optimization.
In [34], The authors proposed three variants of an efficient Feature Selection Ensemble Learning approach for medical image classification systems. This method accurately estimated the content and patterns of medical images, reducing energy consumption during computation. Additionally, spatial and temporal features were detected within the images. Peng et al. [35] proposed a new method called MShNet for medical image segmentation, which combined multi-scale features with an h-network architecture. The network framework included an encoder, two decoders, and an enhanced down-sampling module integrated into the encoder. A fusion convolutional pyramid pooling module was also designed for multi-scale feature fusion. In [36], The authors introduced a similar segmentation model called the encoder-decoder structured 2D model (DGFAU-Net), which addresses the challenges of variable lesion areas and complex shapes in medical images by leveraging DenseNet and AtrousCNN networks in the encoder. The decoder comprises two modules: GFAU and PPASE. A hybrid algorithm for multi-modal medical image fusion was also introduced to combine pixel-level and feature-level information [37]. The proposed method utilized a Discrete Wavelet Transform for pixel-level fusion and employed a curvelet transform combined with Principal Component Analysis for high-frequency coefficient fusion. Feature-level fusion was achieved by extracting various features from both coarse and detailed sub-bands.
2.5 Energy-Efficient and Scalable Medical Image Analysis Techniques
Furthermore, a pyramidal feature multi-distillation network was proposed for super-resolution reconstruction in medical imaging systems [38]. The introduced network is commonly utilized in intelligent healthcare systems. Combining pyramidal construction with residual blocks provides valuable information for the classification process. This network maximizes the quality and feasibility of the diagnostic process while enhancing the performance and robustness of various healthcare systems. In [39], the authors presented an energy-efficient framework called ELMAGIC for medical image analysis. This study aimed to reduce computational resource usage while maintaining high performance by leveraging the Forward-Forward Algorithm, along with knowledge distillation and iterative magnitude pruning. The ELMAGIC model demonstrated effective classification and image generation, achieving an F1 score of 87% when tested on the ODIR5K medical image dataset. This approach promotes sustainable medical data analysis with optimized, efficient neural networks.
In [40], the integration of colour deconvolution, self-attention mechanisms, and fusion techniques is analyzed to improve histopathological image recognition. DecT blends multi-modal images through residual connections, enhancing the model’s ability to capture features and compensating for data loss during conversion. The results achieved an accuracy of 93% and an F1 score of 93.8%, although these metrics may vary across different datasets. However, varying datasets and staining sensitivity could hinder generalization. The authors from [41] investigated the effectiveness of traditional ML and DL methods for image classification. Initially, they implemented a Support Vector Machine on a small dataset, achieving an accuracy of 93%; however, they noted limitations due to the dataset’s size. After expanding the dataset through data augmentation, Support Vector Machine accuracy decreased to 82%. They then applied CNNs, achieving a higher accuracy of 93.57%, demonstrating CNN’s superior capability in feature learning and classification accuracy compared to traditional methods. This study underscores the scalability and accuracy benefits of deep learning in image classification tasks.
In [42], the authors developed an advanced artificial intelligence model called RDAG U-Net for detecting SARS-CoV-2 pneumonia lesions in CT scans. This model extends the traditional U-Net by integrating Residual Blocks, Dense Blocks, and Attention Gates to improve accuracy and reduce computational time. Trained on an augmented dataset of 10,560 CT images, RDAG U-Net achieved a lesion recognition accuracy of 93.29% and reduced computational time by 45% compared to other models. It outperformed Attention U-Net and Attention Res U-Net, especially in handling diverse lesion types, and provides both 2D and 3D lesion visualizations, enhancing clinical interpretability for COVID-19 diagnosis. Furthermore, a CNN model was employed to detect brain tumors using magnetic resonance imaging [43]. The authors enhanced classification accuracy by optimizing CNN architectures through hyperparameter tuning and advanced preprocessing techniques. The best-performing model achieved 97.5% accuracy, 99.2% sensitivity, and 97.5% precision, demonstrating robustness across varied datasets. Compared to similar studies, this model balanced high diagnostic precision with computational efficiency, making it suitable for clinical applications. The model’s reliable detection rates highlight its potential to support timely, accurate diagnoses and contribute to improved outcomes in brain tumor treatment.
Kanya Kumari et al. [44] suggested the Weighted Adaptive Binary Teaching Learning Based Optimization (WA-BTLBO) for feature selection to classify mammogram medical images in breast cancer detection. Several classifiers train and evaluate the chosen features, including XGBoost, K-Nearest Neighbor, Random Forest, Artificial Neural Networks, and Support Vector Machines. The research uses medical pictures from mammograms that are accessible to the public via the Mammographic Image Analysis Society. The findings demonstrate that WA-BTLBO with XGBoost classifier is better than other feature selection strategies, such as Binary TLBO (BTLBO) and Particle Swarm Optimization, to categorize MIAS mammography pictures as normal or abnormal. To extend patients’ lives, this study aids physiologists and radiologists in detecting breast cancer in women. Kutluer et al. [45] proposed the ResNet-50, GoogLeNet, InceptionV3, and MobilNetV2 to classify breast tumours. The suggested feature selection deep learning approaches outperformed the majority of the findings in the literature with classification efficiencies of 98.89% on the local binary class dataset and 92.17% on the BACH dataset. The results on both datasets show that the suggested approaches are quite effective in detecting and classifying the type of malignant tissue.
Jafari et al. [46] recommended the CNN-Based Approach with Feature Selection (CNN-FS) for Breast Cancer Detection in Mammography Images. The results show that the NN-based classifier accomplishes a remarkable 92% accuracy on the RSNA dataset. The enhanced performance is attributable, in part, to the newly introduced dataset, which comprises two perspectives and other variables like age. In particular, the author showed that the suggested algorithm was better in terms of sensitivity and accuracy when compared to state-of-the-art approaches. The author reaches an accuracy of 96% for the DDSM dataset and as high as 94.5 per cent for the MIAS dataset. The technology outperforms previous algorithms and correctly diagnoses breast lesions, as shown by these data. Shetty et al. [47] presented the Content-Based Medical Image Retrieval (CBMIR). Aiming to improve CBMIR’s accuracy and efficiency, this strategy leverages deep learning and sophisticated optimization techniques. Train and test are the two main components of the suggested model. All pre-processing, feature extraction and best feature selection happen during training. The database pictures undergo pre-processing employing a Gaussian filter, contrast-limited adaptive Histogram Equalization, and Gaussian smoothing. Following this, the VGG19 and the Inception V3 CNN models extract the deep features from the database photos. Combine the retrieved characteristics and choose the best ones. The author uses the brand-new Coyote-Moth Optimization Algorithm to make these choices. This model is an intellectual synthesis of the well-known Moth-flame optimization and the coyote optimization algorithm, which eliminate lesions and outperform previous methods.
The literature analysis concludes that medical image analysis necessitates precise feature selection to prevent errors and enhance diagnostic accuracy. Consequently, an input image with varying textures and regions requires thorough and consistent analysis for effective disease identification and treatment planning. The methods discussed primarily focus on single images and limited feature analyses to improve congruency. They analyze distribution based on pixel concentration to mitigate errors related to disease diagnosis and intensity congruency. In contrast, the proposed method comprehensively identifies features, pixel representations, and intensities.
3 Proposed Congruent Feature Selection Method
Machine learning (ML) is frequently used in medical image processing, facilitating effective learning and analysis for diagnosing various conditions, including those related to the brain, lungs, and eyes. A crucial aspect of successful ML implementation is accurately selecting resilient image features for effective exemplar revelation and region categorization. This article employs an analytical learning paradigm to design a CFSM. Feature selection reduces the number of input variables in the image processing pipeline when developing forecasting models. This reduction helps minimize the analytical cost of sculpting and, in some cases, enhances model performance. Texture analysis involves characterizing regions in an image based on their texture content, aiming to quantify perceptual qualities described by terms such as rough, smooth, silky, or bumpy as a function of spatial variations in pixel intensities. Region detection is the process of grouping and labelling all pixels corresponding to an object, denoting that they belong to a specific region. Pixels are assigned to regions using evaluations that distinguish them from the rest of the image. ML is a field dedicated to understanding and developing methods that leverage data to enhance performance on specific tasks. It is considered a part of artificial intelligence, particularly in selecting images with high pixel distribution. The process flow of the proposed method is illustrated in Fig. 1.
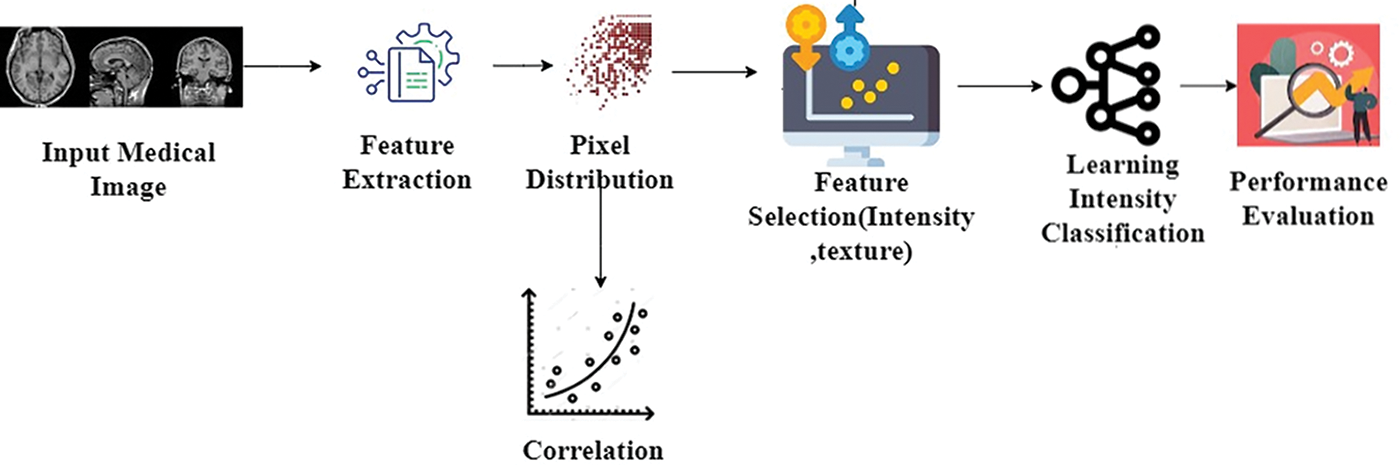
Figure 1: Proposed methods’ process flow
The CFSM process begins with input medical images, from which relevant features like intensity and texture are extracted. These features undergo pixel distribution analysis to understand how various image characteristics are spread out, followed by correlation analysis to identify relationships between the features. Based on this, key features are selected, reducing data complexity while preserving essential information. These selected features are then fed into a learning model for intensity classification, and the method’s effectiveness is assessed through performance evaluation metrics to ensure accurate and efficient classification results.
The analysis begins with observing a medical image, leading to feature extraction and texture analysis. These techniques identify and extract picture data features and textural qualities. After extracting features, the learning system undergoes training using pixel resemblance and matching-based features across textural values and pixel distributions. This training phase helps the model to identify picture data patterns and linkages for analysis and decision-making. After learning, features and textures are used to define the medical image’s intensity and texture-based pixel distribution. This step delineates regions of the image and correlations by examining the spatial variation in pixel brightness. Correlation in the medical processing of images helps understand and analyze fresh medical images using database information and trends. Medical images can be analyzed using the suggested approach to extract features, which include shapes, textures, and patterns. These features are vital for diagnosis and classification. Typically, the stored images are pre-processed or stored as an intermediate step in the pipeline. These images may have undergone noise reduction, augmentation, or feature extraction and are preserved for further analysis or classification. Later model phases or validations might refer to these images for assistance.
The correlation method compares the incoming image’s pixel distribution to database pictures. This comparison helps diagnose diseases and detect anomalies by identifying images with similar features and pixel distributions. The medical image is first analyzed to extract its features and textures, then used to determine pixel distribution based on intensity and texture. This procedure trains the learning exemplar using pixel similarity and matching-based characteristics over different textural intensities and pixel distributions. Medical images are initially examined to extract features and textures, which determine pixel distribution depending on intensity and texture. This stage trains the learning example utilizing pixel resemblance and matching-based characteristics across textural intensities and pixel dispersion. The collected features and textures determine the medical image’s pixel dispersion following intensity and texture. The spatial variation in pixel brightness is analyzed to define the image regions. The correlations between the pixels over the various distribution exemplars with high directories are suggested for disease examination. Later, the matching, depending on the intensity and distribution, is analyzed to improve the congruency in feature selection. Therefore, the more congruent pixels are sorted in the descending order of the selection, which identifies better regions than the distribution. Now, the learning paradigm is trained using ferocity and region-based correlation to increase the chances of selection. The feasibility of feature selection was unconcerned with the textures, and the medical image exemplar was enhanced. Therefore, this improves the consummation of ML applications for different medical image processing. The medical image is used to determine its features and has the method to extract the needed characteristics and textured features from the acquired medical images. This output will be used in the learning paradigm to estimate similar images with high pixel distribution and the same features. The feature extraction procedure estimates the number of pixels in the image. It is also used to show off the different methods to determine the features of the acquired medical images. The image is used in feature extraction to find the given images’ pixel distribution. The methodologies that can be used to examine the given image’s characteristics and enhance the pixel dispense are forecasted in this process. The learning paradigm will use this output for future processes to determine the high-pixel dispensed images. The foremost process involves extracting the features from the medical image for further procedures using the learning technique. The process of extracting the features from the acquired medical image is explained by the following Equation given below:
where
The function

Figure 2: Feature extraction and pixel distribution in medical image analysis
A number of essential components and procedures are involved in the distribution extraction process. Starting from the beginning, the input consists of acquired medical images, which are represented by
A learning paradigm is utilized throughout these operations to improve the precision and effectiveness of feature extraction and correlation. This deep analytical learning method helps refine the correlation procedure by iteratively increasing the degree of matching of pixel dispersion between the photos that have been acquired and the images that have been stored. This distribution of the pixels in an image will be useful in upcoming processes to determine the intensity of an image (Fig. 2). The deep analytical process is used in the pixel distribution to detect similar images with precise intensity. The deep analytical learning paradigm is processed based on the output of the feature extraction procedure from the acquired medical image. With the help of the learning technique, the determined images are correlated, and then further procedures take place with its output. The process of determining the pixel distribution of the image from the output of the feature extraction process is explained by the following equations given below:
where
Medical images are involved with the Gabor kernel at numerous scales and orientations to apply the Gabor filter. This method captures image texture patterns. They are great for capturing texture details at multiple scales and orientations. 2D Gabor filter equation:
where
Now, the correlation process is implied by the stored images. Similar images have the same features, and the pixel distribution is figured out. Based on the intensity of an image, similar images are identified. Local binary pattern (LBP) compares the intensity of a core pixel to its neighbours to record local texture variations in photos. The study uses LBP to extract texture features from medical images for analysis. The computationally efficient and noise-resistant LBP can analyze complicated medical images with complex textures and variations.
where
Its simplicity and efficacy make it a popular texture feature extraction method. By using LBP for texture-extracted features, medical image processing can improve texture analysis, feature representation, and diagnostic image interpretation. The determined images are checked with the already stored images, which have gone through the image processing technique with that given input. The acquired images will be checked with the stored images to determine the correlated images with a high pixel distribution. This process is done using the DL algorithm, which extracts the correlated images based on the intensity of the images. The stored images have been processed through image processing to acquire the high-featured images with precise intensity. The image, obtained from the feature extraction process, is checked with the stored images to estimate an image’s matching features and textures.
where

Figure 3: Correlation-based on distributions
The learning approach is trained using similarity and correlation-based features over different textural intensities and pixel distributions. The similarity between the pixels over the various distribution exemplars with high pixel distribution is recommended for disease recovery. After that, the correlation based on intensity and pixel distribution is identified to enhance consistency in feature selection (Fig. 3). The process of correlation implied by the stored images based on the results of the pixel distribution procedure is explained by the following equations given below:
The congruency enhancements in the features are done before the feature selection process, where the intensity and textures of an image are used to determine similar features. Then, the feature’s consistency will be improved to enhance the feature selection in the procedure. This process is also done using the DL technique based on an image’s intensity and texture. Then, the blunders will be eliminated according to the feature selection in an image processing process. The procedure of enhancing the congruency of the feature based on the pixel distribution and the correlation process is explained by the following equations given below:
where

Figure 4: Learning for intensity classification for
The intensity of an image is determined by summing all of the pixels distributed in the image. This is used to measure the intensities of the images acquired for the feature selection process. Based on the similar features that match the stored images, the intensity and the textures are identified for the feature selection process. From this, the congruency can also be enhanced without any blunders. This process is used to identify images’ precise intensity and textures with the help of the deep analytical technique in the ML procedures (Fig. 4). The features that are obtained from the medical images for further processing are used in the procedure of identifying the pixel distribution and correlation process and also in enhancing the congruency of the features. With the help of these processes’ outputs, an image’s intensity and texture are determined. Then, the outcome of the identified intensity, textures, and highly similar features are estimated, with an increased pixel distribution rate and the textures without blurring. The process of estimating the intensity and texture of the image from the outcome of the previous process to determine similar features is explained by the following equations given below:
where
Figure 5: Feature selection process
The experimental analysis uses the image data in [48]; 100 TIF images of different CT scan outputs are provided. The images are analyzed using MATLAB experiments by extracting 17 textural features and identifying 12 regions in each image. For the proposed method’s efficiency, four varied CT images are used for feature extraction and precise selection condition verification. The collected images are developed using the MATLAB tool, which consists of several built-in functions that help to identify the images effectively. This study uses the SSE2 instruction set processors, 8 GB RAM for processing large volumes of data, 1 GB VRAM graphics card, 2.5 GB disk space, Windows operating systems, and other additional hardware settings to implement the process. The created system uses the 10-fold cross-validation process to evaluate the system’s efficiency. The experimental setup uses 10-fold cross-validation for training sets. The 100 TIF images are partitioned into 10 sets, with each subset acting as a set for validation once and a training set nine times. Each of the 10 training sets has 90 images. This partitioning maximizes training data while evaluating the suggested method’s effectiveness across dataset subsets. Table 1 shows the type of image and its extractable features.
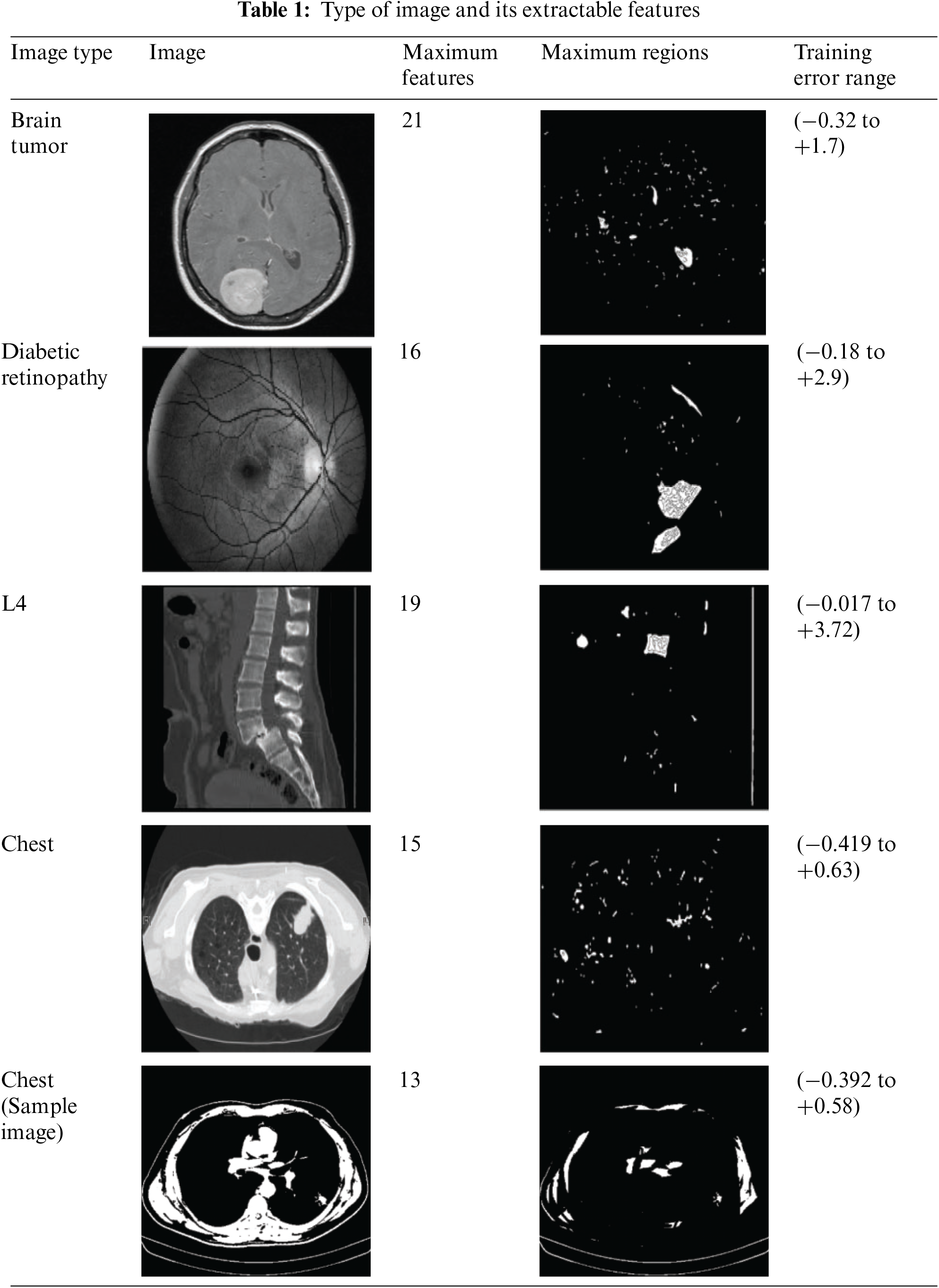
The above table is a representation of the features extractable and their corresponding
The Maximum Features
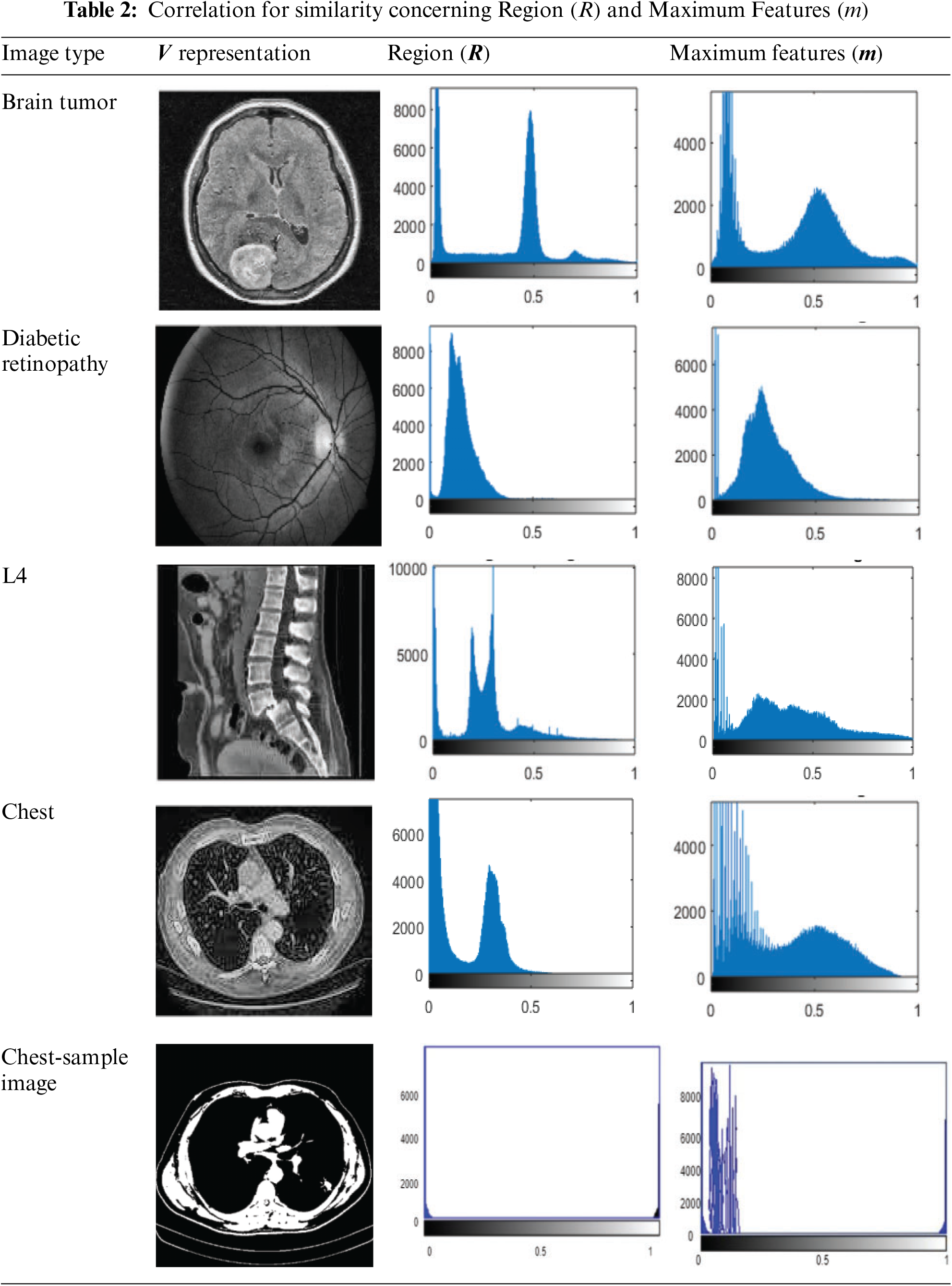
Different types of images or categories are represented by the values located on the horizontal axis (x-axis) of the graph presented in Table 2. To be more specific, the numbers 0 and 0.5 are likely to correspond to several categories of medical imaging, such as diabetic retinopathy, brain tumours, and other similar conditions. There is a high probability that every value on the axis that is horizontal relates to a certain category or type of image that is being reviewed to determine its
In Table 3, the horizontal axis represented in the graph describes the regions
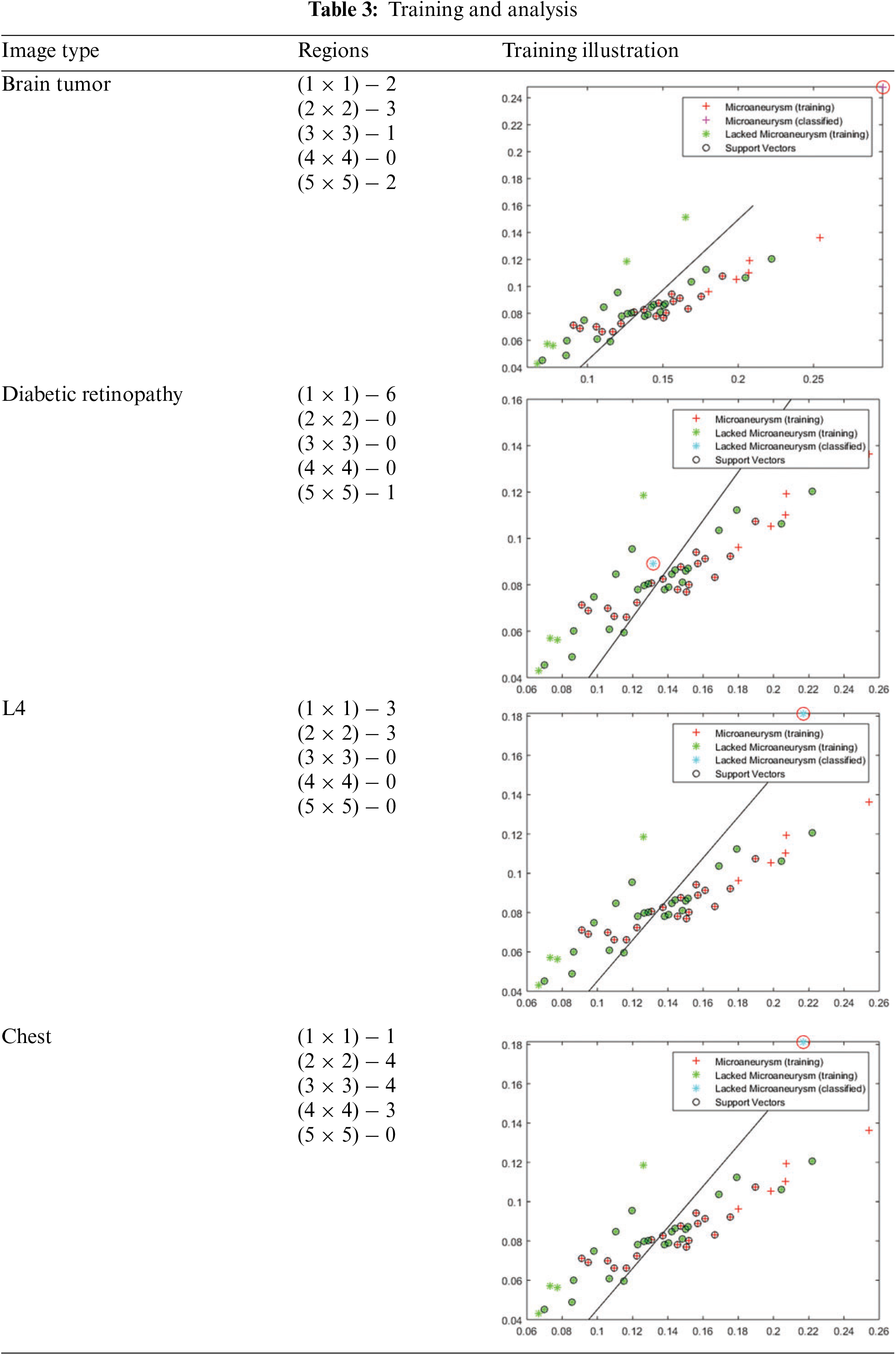
The training error is the difference between the expected and actual values in the training set; it measures how well an ML model matches the data used for training. It is usually expressed as the misclassification rate for classification problems, where the percentage of inaccurate predictions is relative to the total training samples. Measures that quantify the difference between anticipated and actual values in regression tasks include mean squared and absolute errors. While reducing training error is critical, an overfitting model, that is, doing well on the training data but failing to generalize to new data, occurs when the training error is very low. The training and analysis for the above representations are presented in Table 3.
The variations (causing the error) and the actual distribution are analyzed for the varying regions using MATLAB experiments. The variation in regions is observed using multiple factors that impact the accuracy and precision selection. The selected regions and the long-lasting features are used for handling congruency and maximizing
In this section, the statistical analysis is discussed using a comparative study. Different from the above section, metrics such as selection accuracy, selection precision, mean error, selection time, and training rate are considered in this analysis. These metrics evaluate the system’s efficiency, which helps identify how effectively the proposed approach selects the features from the images. The maximum number of features extracted is 16, and the regions considered are 11. The existing CNN-FS [46], WA-BTLBO [44], and CBMIR [47] methods are considered alongside the proposed CFSM. The various methods [44,46,47] are selected for comparison because they effectively analyze the image with minimum computation complexity and attain scalability.
The selection accuracy is productive with the help of the deep analytical algorithm based on the image’s intensity and texture. The features are extracted from the acquired medical images to determine the pixel distribution and the correlation implied with the stored images. Based on the image’s texture and intensity, similar features are identified in the selection process. The image should have a high pixel distribution and the same features as the stored images. Then, it will be selected in the process for the upcoming procedures, and the congruencies of the feature will also be enhanced in this procedure. The pixel distribution process will help maximize the similar features of the acquired medical images. It will be sent to the correlation process to estimate the matching images based on the stored images that have already gone through the image processing. Then, from the outcomes of these processes, which are done using the learning paradigm, the selection process will take place, and the enhancements of the features are also done. When compared to the other approaches that were reviewed previously, like CNN-FS [46], WA-BTLBO [44], and CBMIR [47], the proposed method, CFSM, has a significantly greater selection accuracy, as shown in Fig. 6. It has been stated that the accuracy of CFSM is 0.9389, which is a significant improvement above the accuracy of the other approaches.
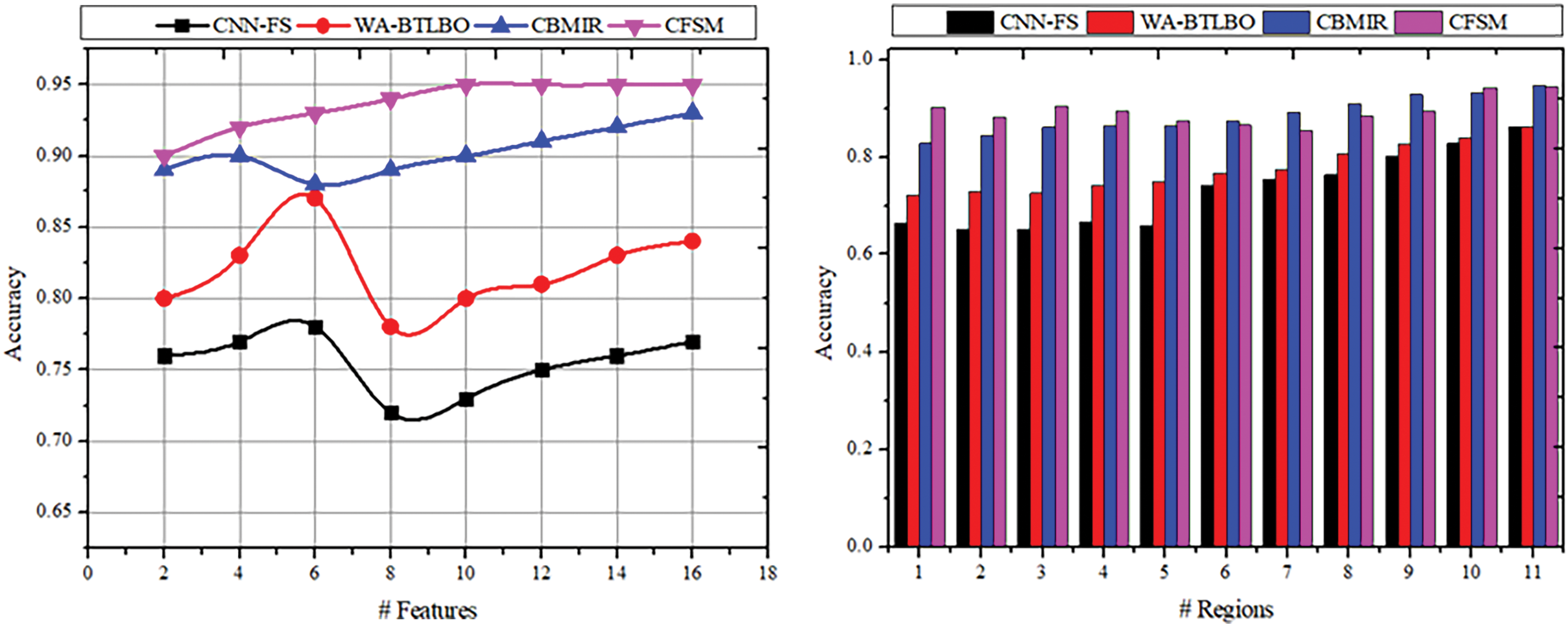
Figure 6: Selection accuracy comparisons
The precision of the selection of the features is done efficiently with the output of the image’s pixel count and its intensity. The textures and features will be extracted from the medical images acquired for the procedures. It also has methods to extract the textures and features of an image. The correlation-based and region-based images will be useful in the selection process where highly similar features are identified. The images with precise intensity and congruency will also be selected based on the pixel distribution. The precision of the selected features will be high, according to the outcome of the previous procedures. The results of the pixel and correlation processes enhance the features’ congruency; thus, they will be used to determine the images’ intensity and textures. This way, precision is achieved in the acquired medical image selection, which has a high pixel distribution and precise intensity. Along the same lines as the accuracy, the CFSM approach exceeds the other methods that are taken for comparison, as shown in Fig. 7, and it can reach a precision of 0.9574, which indicates a significant improvement in the precision of feature selection compared to other methods.
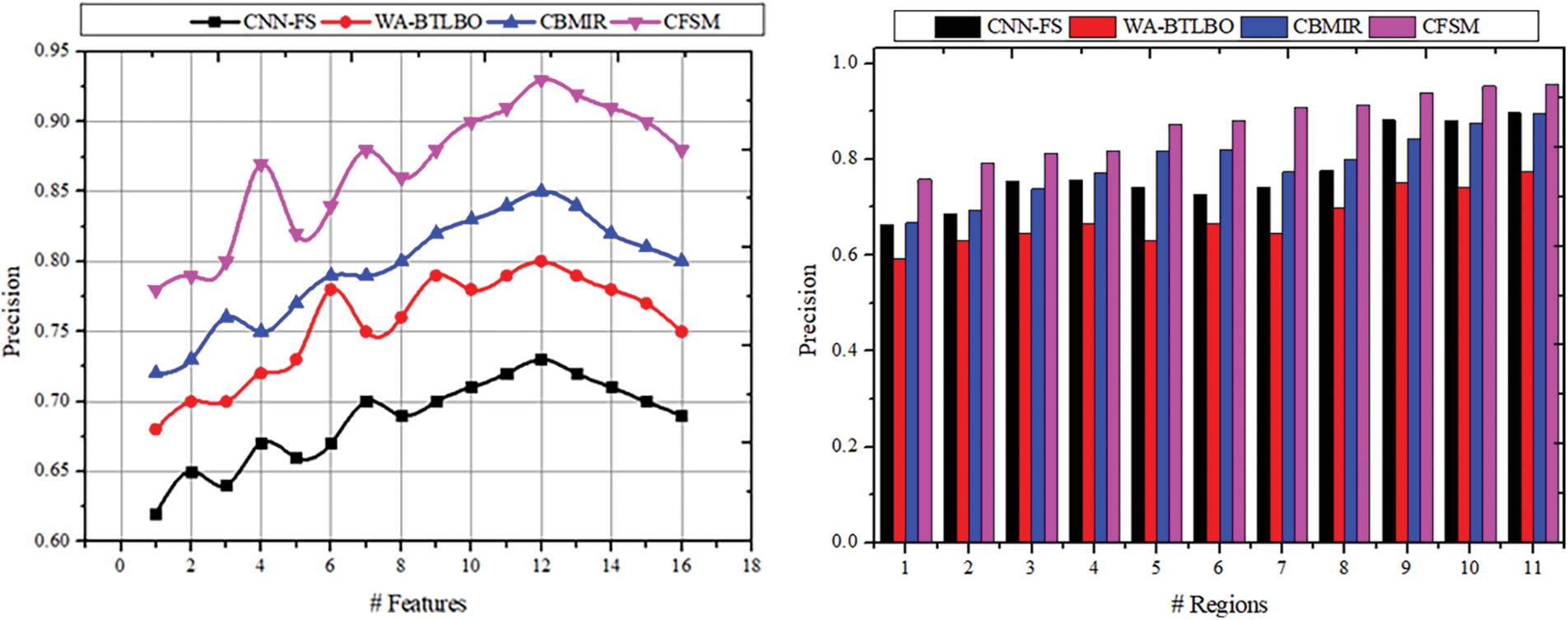
Figure 7: Selection precision comparisons
The errors in the process are reduced by using the learning paradigm and the congruency of the features. The errors are reduced in this process while highly similar features are selected. The precision of the intensity and the textures are enhanced with the deep analytical technique based on the intensity and pixel distribution outcomes. The pixel shows the representation of an original image with the perfect intensity and textures of an image. Thus, this will be helpful in the selection process. From the outputs of the correlation process, the intensity and the textures are identified to make from the errorless features. By increasing the congruency of the feature, the errors and blunders can be reduced. Thus, in the outputs of the DL method procedures, the precise features are selected based on the high pixel distribution and the textures of an image. The correlation process takes place based on intensity, which will help enhance the congruency of the features. Accordingly, the same or similar features are selected in the selection process. A result of 0.0658 indicates that the CFSM technique has a significantly lower mean error than other methods, as in Fig. 8. In light of this, the method provided appears superior to the other ways to reduce the number of errors that occur during the feature selection process.

Figure 8: Mean error comparisons
The time taken for the selection process based on the output of the intensity and the textures is less according to the outcome of pixel distribution—the correlation and the pixel distribution outcome help in the selection process with precise intensity and textures. The similarity-checking procedure is done depending on the intensity of the medical image. From this output, the selection of the imaging procedure is made based on the intensity and texture of the image. After that, the correlation based on intensity and pixel distribution is identified to enhance consistency in feature selection. Congruency is used to identify the significance of the determined features based on the pixels distributed in the images. The pixel distribution process and the correlation process implied with the stored images are done to enhance the consistency of an image without any inaccuracy in the results. This congruency enhancement in the feature helps find highly similar images with increased pixel distribution and precise intensity. Through these processes, the time consumption for the selection process is reduced. Indicating that the suggested technique is more effective in choosing features from the images, the selection time for the CFSM technique is significantly shorter than that of the other approach observed from existing related works, as depicted in Fig. 9. Applications that can benefit from this include those that require real-time processing or time-sensitive processing.
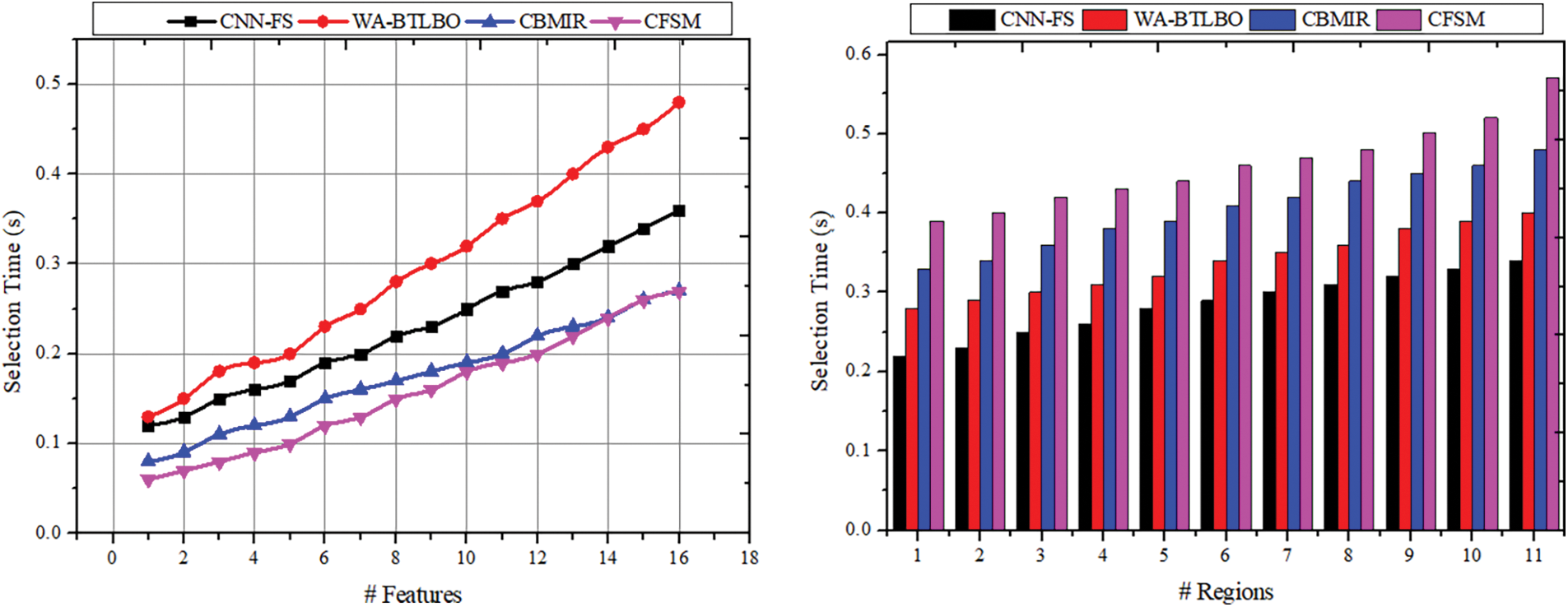
Figure 9: Selection time comparisons
The training for deep analytical learning is efficacious with the help of the correlation outputs and the pixel distribution process. In this process, the dispensation of the pixels in images is determined; thus, the correlation procedure will be done with this output. The pixel in an image is a sample of the original image, whereas more specimens typically produce a more precise impersonation of the original image. The pixel distribution is used to determine the number of pixels provided for the medical image that is acquired for the process. The correlation process shows similar images with the same features and pixel distribution. Later, the correlation based on intensity and distribution is analyzed to improve feature selection congruency. Therefore, the more congruent pixels are sorted in the descending order of the selection, which identifies better regions than the distribution. The DL technique is trained based on intensity and region-based similarity to maximize the chances of feature selection. Tables 4 and 5 present the comparative analysis summary for the features and regions. The illustration in Fig. 10 shows the training rate, including the comparisons of training rates, and demonstrates that the suggested CFSM approach enhances the training rate by approximately 11.06% compared to the existing methods. Based on this, it can be concluded that the CFSM technique is more efficient in learning and adapting to the input data, which ultimately results in improved performance.
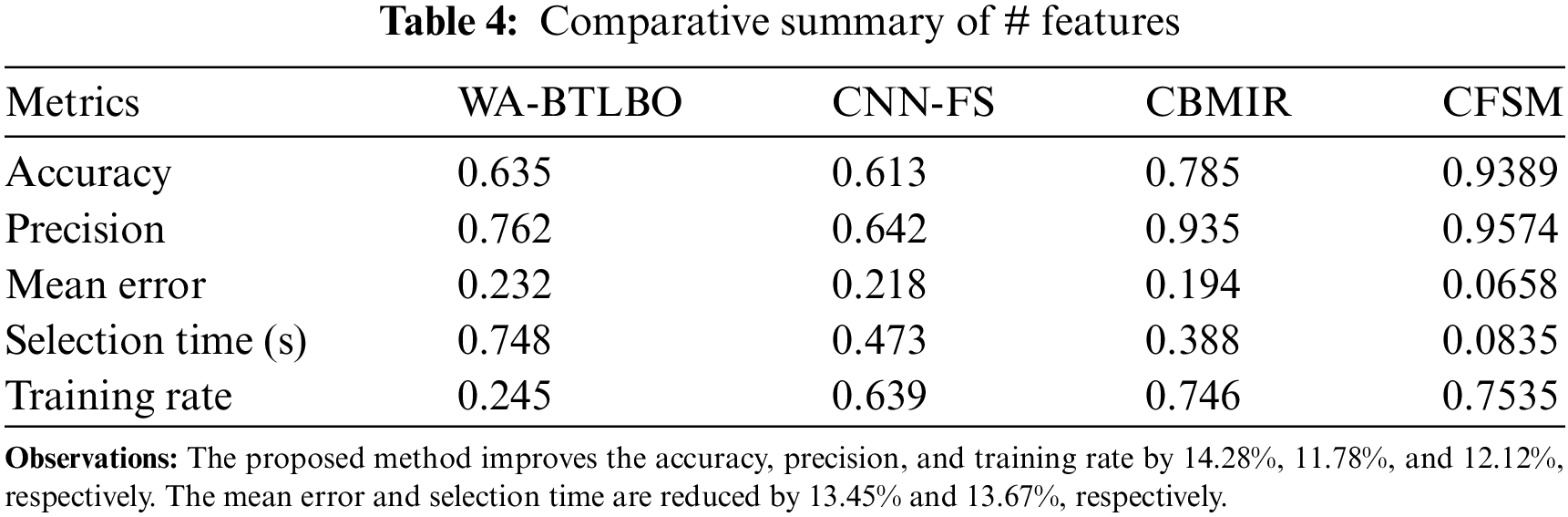
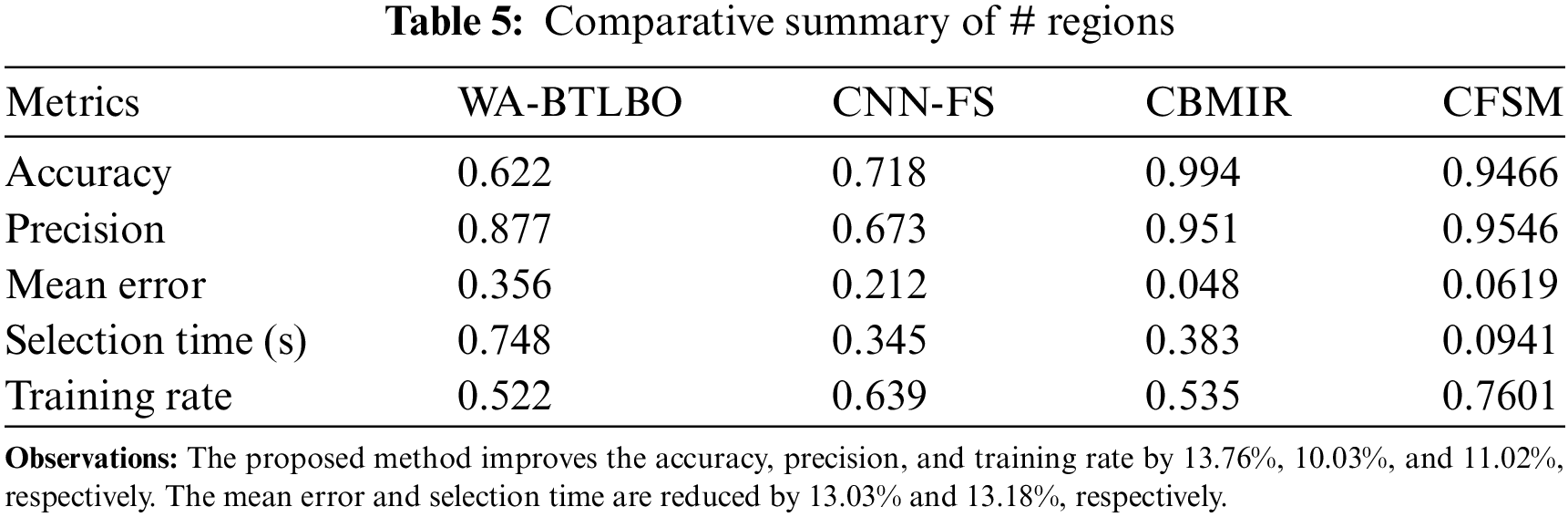
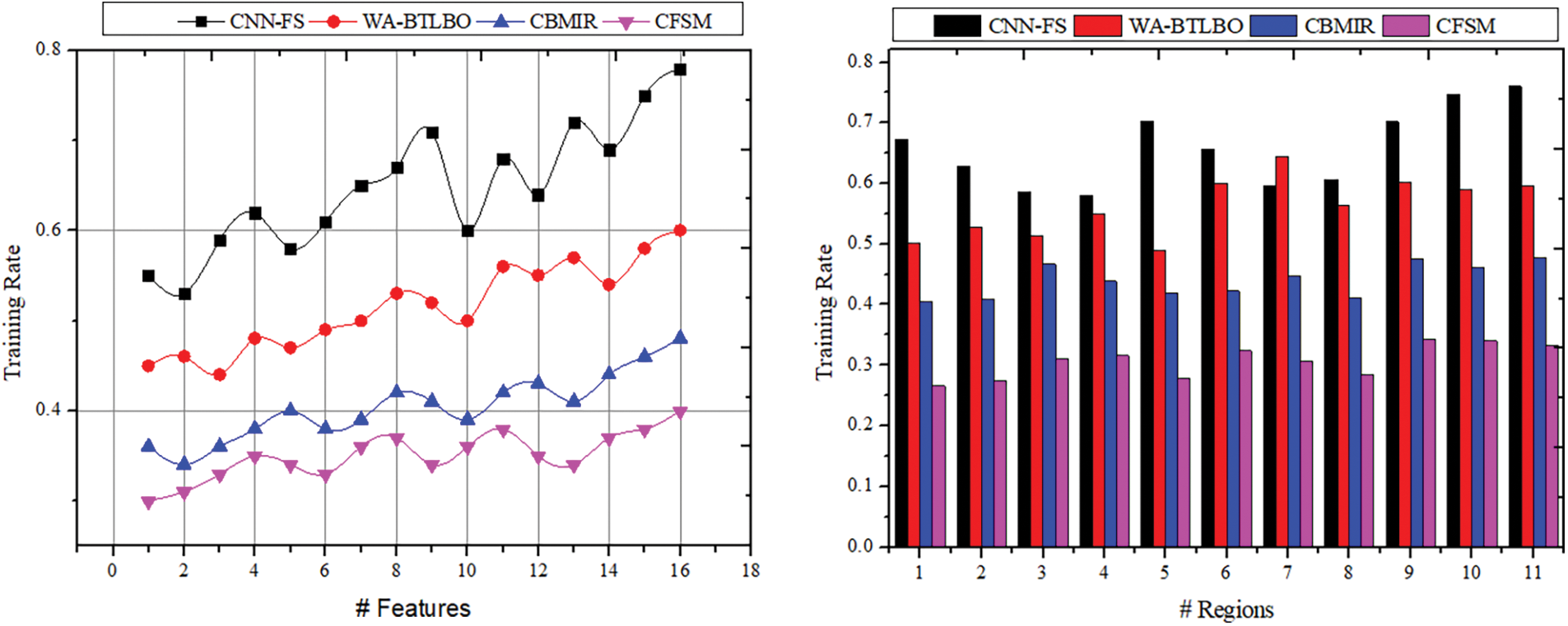
Figure 10: Training rate comparisons
This article introduces the CFSM for improving medical image analysis. The congruency of feature analysis and similarity-based selection is encouraged in this proposed method. The different patterns and regions based on pixel distribution and intensities are used for feature selection. The proposed process relies on analytical learning for recurrent training and similarity for reducing errors. Compared to the consecutive feature identification, the correlation and similarity are performed using the stored and existing image extractions. This process verifies the maximum similarity across different training instances to improve the descending and variational pixel distributions. The congruency in pixel selection and distribution is utilized to maximize selection chances. The learning process is responsible for retaining precision across different new and overlapping features and regions extracted. However, high-dimensional medical images require substantial computational resources for both feature selection and classification, which can lead to increased processing times. For future work, we plan to explore the generality of our method by extending its use to 3D tasks. One particularly exciting direction is applying our feature selection technique in 3D engineering and medical imaging tasks, such as 3D reconstruction.
Acknowledgement: The authors extend their appreciation to the Deanship of Scientifc Research at King Khalid University for funding this work through large group Research Project under grant number RGP2/421/45. This study is supported via funding from Prince Sattam bin Abdulaziz University project number (PSAU/2024/R/1446). This work was supported by the Researchers Supporting Project Number (UM-DSR-IG-2023-07) Almaarefa University, Riyadh, Saudi Arabia.
Funding Statement: This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2021R1F1A1055408).
Author Contributions: Study conception and design: Mohd Anjum, Hong Min; data collection: Yousef Ibrahim Daradkeh; analysis and interpretation of results: Naoufel Kraiem, Ashit Kumar Dutta, Yousef Ibrahim Daradkeh; draft manuscript preparation: Mohd Anjum, Hong Min, Ashit Kumar Dutta. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data used in the study is publicly available at Mader KS. CT Medical Images | Kaggle: https://www.kaggle.com/datasets/kmader/siim-medical-images?select=full_archive.npz (accessed on 22 October 2024).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Tang W, He F, Liu Y, Duan Y. MATR: multimodal medical image fusion via multiscale adaptive transformer. IEEE Trans Image Process. 2022;31:5134–49. [Google Scholar] [PubMed]
2. Cai Y, Chen H, Yang X, Zhou Y, Cheng KT. Dual-distribution discrepancy with self-supervised refinement for anomaly detection in medical images; 2022. doi:10.48550/arXiv.2210.04227. [Google Scholar] [CrossRef]
3. Wei X, Zhang W, Yang B, Wang J, Xia Z. Fragile watermark in medical image based on prime number distribution theory. J Digit Imaging. 2021;34(6):1447–62. doi:10.1007/s10278-021-00524-4. [Google Scholar] [PubMed] [CrossRef]
4. Xu Y, Li Y, Shin BS. Medical image processing with contextual style transfer. Hum-Centric Comput Inf Sci. 2020;10(1):46. doi:10.1186/s13673-020-00251-9. [Google Scholar] [CrossRef]
5. Zeng X, Tong S, Lu Y, Xu L, Huang Z. Adaptive medical image deep color perception algorithm. IEEE Access. 2020;8:56559–71. [Google Scholar]
6. Elmi S, Elmi Z. A robust edge detection technique based on Matching Pursuit algorithm for natural and medical images. Biomed Eng Adv. 2022;4:100052. doi:10.1016/j.bea.2022.100052. [Google Scholar] [CrossRef]
7. Qiao S, Yu Q, Zhao Z, Song L, Tao H, Zhang T, et al. Edge extraction method for medical images based on improved local binary pattern combined with edge-aware filtering. Biomed Signal Process Control. 2022;74(1):103490. doi:10.1016/j.bspc.2022.103490. [Google Scholar] [CrossRef]
8. Aljuaid A, Anwar M. Survey of supervised learning for medical image processing. SN Comput Sci. 2022;3(4):1–22. doi:10.1007/s42979-022-01166-1. [Google Scholar] [PubMed] [CrossRef]
9. Zhang Z, Sejdić E. Radiological images and machine learning: trends, perspectives, and prospects. Comput Biol Med. 2019;108:354–70. [Google Scholar] [PubMed]
10. Feng X, Jiang Y, Yang X, Du M, Li X. Computer vision algorithms and hardware implementations: a survey. Integration. 2019;69:309–20. [Google Scholar]
11. O’Mahony N, Campbell S, Carvalho A, Harapanahalli S, Hernandez GV, Krpalkova L, et al. Deep learning vs. traditional computer vision. In: Advances in computer vision. Springer; 2020. vol. 943, p. 128–44. doi:10.1007/978-3-030-17795-9_10. [Google Scholar] [CrossRef]
12. Alzubaidi MA, Otoom M, Jaradat H. Comprehensive and comparative global and local feature extraction framework for lung cancer detection using CT scan images. IEEE Access. 2021;9:158140–54. doi:10.1109/ACCESS.2021.3129597. [Google Scholar] [CrossRef]
13. Kuwil FH. A new feature extraction approach of medical image based on data distribution skew. Neurosci Inform. 2022;2(3):100097. doi:10.1016/j.neuri.2022.100097. [Google Scholar] [CrossRef]
14. Yu R, Tian Y, Gao J, Liu Z, Wei X, Jiang H, et al. Feature discretization-based deep clustering for thyroid ultrasound image feature extraction. Comput Biol Med. 2022;146(5):105600. doi:10.1016/j.compbiomed.2022.105600. [Google Scholar] [PubMed] [CrossRef]
15. Jalali V, Kaur D. A study of classification and feature extraction techniques for brain tumor detection. Int J Multimed Inf Retr. 2020;9(4):271–90. doi:10.1007/s13735-020-00199-7. [Google Scholar] [CrossRef]
16. Zhou S, Nie D, Adeli E, Yin J, Lian J, Shen D. High-resolution encoder-decoder networks for low-contrast medical image segmentation. IEEE Trans Image Process. 2020;29:461–75. doi:10.1109/TIP.2019.2919937. [Google Scholar] [PubMed] [CrossRef]
17. Hamid MAA, Khan NA. Investigation and classification of MRI brain tumors using feature extraction technique. J Med Biol Eng. 2020;40(2):307–17. doi:10.1007/s40846-020-00510-1. [Google Scholar] [CrossRef]
18. Ershadi MM, Seifi A. Applications of dynamic feature selection and clustering methods to medical diagnosis. Appl Soft Comput. 2022;126(350):109293. doi:10.1016/j.asoc.2022.109293. [Google Scholar] [CrossRef]
19. Hu G, Zhong J, Wang X, Wei G. Multi-strategy assisted chaotic coot-inspired optimization algorithm for medical feature selection: a cervical cancer behavior risk study. Comput Biol Med. 2022;151(1):106239. doi:10.1016/j.compbiomed.2022.106239. [Google Scholar] [PubMed] [CrossRef]
20. Bolón-Canedo V, Remeseiro B. Feature selection in image analysis: a survey. Artif Intell Rev. 2020;53(4):2905–31. doi:10.1007/s10462-019-09750-3. [Google Scholar] [CrossRef]
21. Narin A. Accurate detection of COVID-19 using deep features based on X-Ray images and feature selection methods. Comput Biol Med. 2021;137:104771. doi:10.1016/j.compbiomed.2021.104771. [Google Scholar] [PubMed] [CrossRef]
22. Bidgoli AA, Rahnamayan S, Dehkharghanian T, Riasatian A, Kalra S, Zaveri M, et al. Evolutionary deep feature selection for compact representation of gigapixel images in digital pathology. Artif Intell Med. 2022;132(1):102368. doi:10.1016/j.artmed.2022.102368. [Google Scholar] [PubMed] [CrossRef]
23. Sun L, Mo Z, Yan F, Xia L, Shan F, Ding Z, et al. Adaptive feature selection guided deep forest for COVID-19 classification with chest CT. 2020;2194(c):1–8. [Google Scholar]
24. Balaji P, Revathi BS, Gobinathan P, Shamsudheen S, Vaiyapuri T. Optimal IoT based improved deep learning model for medical image classification. Comput Mater Contin. 2022;73(2):2275–91. doi:10.32604/cmc.2022.028560. [Google Scholar] [CrossRef]
25. Gutowski N, Schang D, Camp O, Abraham P. A novel multi-objective medical feature selection compass method for binary classification. Artif Intell Med. 2022;127(6):102277. doi:10.1016/j.artmed.2022.102277. [Google Scholar] [PubMed] [CrossRef]
26. Yue G, Han W, Li S, Zhou T, Lv J, Wang T. Automated polyp segmentation in colonoscopy images via deep network with lesion-aware feature selection and refinement. Biomed Signal Process Control. 2022;78(3):103846. doi:10.1016/j.bspc.2022.103846. [Google Scholar] [CrossRef]
27. Canayaz M. Classification of diabetic retinopathy with feature selection over deep features using nature-inspired wrapper methods. Appl Soft Comput. 2022;128:109462. doi:10.1016/j.asoc.2022.109462. [Google Scholar] [CrossRef]
28. Eysa AB, kurnaz S. Diagnose colon disease by feature selection based on artificial neural network and group teaching optimization algorithm. Optik. 2022;271(20):170166. doi:10.1016/j.ijleo.2022.170166. [Google Scholar] [CrossRef]
29. Khan MA, Kadry S, Alhaisoni M, Nam Y, Zhang Y, Rajinikanth V, et al. Computer-aided gastrointestinal diseases analysis from wireless capsule endoscopy: a framework of best features selection. IEEE Access. 2020;8:132850–9. [Google Scholar]
30. Huang B, Tian J, Zhang H, Luo Z, Qin J, Huang C, et al. Deep semantic segmentation feature-based radiomics for the classification tasks in medical image analysis. IEEE J Biomed Heal Informatics. 2021;25(7):2655–64. [Google Scholar]
31. Zuo Q, Zhang J, Yang Y. DMC-fusion: deep multi-cascade fusion with classifier-based feature synthesis for medical multi-modal images. IEEE J Biomed Heal Inform. 2021;25(9):3438–49. [Google Scholar] [PubMed]
32. Li W, Zhang Y, Wang G, Huang Y, Li R. DFENet: a dual-branch feature enhanced network integrating transformers and convolutional feature learning for multimodal medical image fusion. Biomed Signal Process Control. 2023;80:104402. doi:10.1016/j.bspc.2022.104402. [Google Scholar] [CrossRef]
33. Han Q, Hou M, Wang H, Wu C, Tian S, Qiu Z, et al. EHDFL: evolutionary hybrid domain feature learning based on windowed fast Fourier convolution pyramid for medical image classification. Comput Biol Med. 2023;152(10):106353. doi:10.1016/j.compbiomed.2022.106353. [Google Scholar] [PubMed] [CrossRef]
34. Khoder A, Dornaika F. Ensemble learning via feature selection and multiple transformed subsets: application to image classification. Appl Soft Comput. 2021;113(9):108006. doi:10.1016/j.asoc.2021.108006. [Google Scholar] [CrossRef]
35. Peng Y, Yu D, Guo Y. MShNet: multi-scale feature combined with h-network for medical image segmentation. Biomed Signal Process Control. 2023;79(P2):104167. doi:10.1016/j.bspc.2022.104167. [Google Scholar] [CrossRef]
36. Peng D, Yu X, Peng W, Lu J. DGFAU-Net: global feature attention upsampling network for medical image segmentation. Neural Comput Appl. 2021;33(18):12023–37. [Google Scholar]
37. Tawfik N, Elnemr HA, Fakhr M, Dessouky MI, Abd El-Samie FE. Hybrid pixel-feature fusion system for multimodal medical images. J Ambient Intell Humaniz Comput. 2021;12(6):6001–18. doi:10.1007/s12652-020-02154-0. [Google Scholar] [CrossRef]
38. Ren S, Guo K, Ma J, Zhu F, Hu B, Zhou H. Realistic medical image super-resolution with pyramidal feature multi-distillation networks for intelligent healthcare systems. Neural Comput Appl. 2021;35(31):22781–96. doi:10.1007/s00521-021-06287-x. [Google Scholar] [PubMed] [CrossRef]
39. Barua S, Rahman M, Saad MU, Islam R, Sadek MJ. ELMAGIC: energy-efficient lean model for reliable medical image generation and classification using forward forward algorithm. In: 2024 IEEE 3rd International Conference on Computing and Machine Intelligence (ICMI), 2024; Mt Pleasant, MI, USA; p. 1–5. doi:10.1109/ICMI60790.2024.10585776. [Google Scholar] [CrossRef]
40. He Z, Lin M, Xu Z, Yao Z, Chen H, Alhudhaif A, et al. Deconv-transformer (DecTa histopathological image classification model for breast cancer based on color deconvolution and transformer architecture. Inf Sci. 2022;608:1093–112. doi:10.1016/j.ins.2022.06.091. [Google Scholar] [CrossRef]
41. Chaganti SY, Nanda I, Pandi KR, Prudhvith TGNRSN, Kumar N. Image classification using SVM and CNN. In: 2020 International Conference on Computer Science, Engineering and Applications (ICCSEA), 2020; Gunupur, India; p. 1–5. doi:10.1109/ICCSEA49143.2020.9132851 [Google Scholar] [CrossRef]
42. Lee CH, Pan CT, Lee MC, Wang CH, Chang CY, Shiue YL. RDAG U-Net: an advanced AI model for efficient and accurate CT scan analysis of SARS-CoV-2 pneumonia lesions. Diagnostics. 2024;14(18):2099. doi:10.3390/diagnostics14182099. [Google Scholar] [PubMed] [CrossRef]
43. Martínez-Del-Río-Ortega R, Civit-Masot J, Luna-Perejón F, Domínguez-Morales M. Brain tumor detection using magnetic resonance imaging and convolutional neural networks. Big Data Cognitive Comput. 2024;8(9):123. doi:10.3390/bdcc8090123. [Google Scholar] [CrossRef]
44. Kanya Kumari L, Naga Jagadesh B. An adaptive teaching learning based optimization technique for feature selection to classify mammogram medical images in breast cancer detection. Int J Syst Assur Eng Manag. 2024;15(1):35–48. doi:10.1007/s13198-021-01598-7. [Google Scholar] [CrossRef]
45. Kutluer N, Solmaz OA, Yamacli V, Eristi B, Eristi H. Classification of breast tumors by using a novel approach based on deep learning methods and feature selection. Breast Cancer Res Treat. 2023;200(2):183–92. doi:10.1007/s10549-023-06970-8. [Google Scholar] [PubMed] [CrossRef]
46. Jafari Z, Karami E. Breast cancer detection in mammography images: a CNN-based approach with feature selection. Information. 2023;14(7):410. doi:10.3390/info14070410. [Google Scholar] [CrossRef]
47. Shetty R, Bhat VS, Pujari J. Content-based medical image retrieval using deep learning-based features and hybrid meta-heuristic optimization. Biomed Signal Process Control. 2024;92(4):106069. doi:10.1016/j.bspc.2024.106069. [Google Scholar] [CrossRef]
48. Mader KS. CT Medical Images | Kaggle. 2017. Available from: https://www.kaggle.com/datasets/kmader/&break;siim-medical-images?select=full_archive.npz. [Accessed 2024]. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools