 Open Access
Open Access
REVIEW
Advancements in Liver Tumor Detection: A Comprehensive Review of Various Deep Learning Models
1 Deptartment of Artificial Intelligence and Data Science, Vardhaman College of Engineering, Hyderabad, 501218, India
2 Deptartment of CSE, V.S.B College of Engineering, Karur, 639111, India
3 Deptartment of Electrical and Electronics Engineering, Vardhaman College of Engineering, Hyderabad, 501218, India
4 Centre for Research Impact & Outcome, Chitkara University, Rajpura, 140401, Punjab, India
5 Department of Computer Science and Engineering, School of Engineering and Technology, Central University of Haryana, Mahendergarh, 123031, Haryana, India
6 Department of Computer Science and Information Engineering, National Yunlin University of Science and Technology, Yunlin, 64002, Taiwan
7 Intelligence Recognition Industry Service Research Center, National Yunlin University of Science and Technology, Yunlin, 64002, Taiwan
* Corresponding Author: Shih-Yu Chen. Email:
Computer Modeling in Engineering & Sciences 2025, 142(1), 91-122. https://doi.org/10.32604/cmes.2024.057214
Received 11 August 2024; Accepted 18 November 2024; Issue published 17 December 2024
Abstract
Liver cancer remains a leading cause of mortality worldwide, and precise diagnostic tools are essential for effective treatment planning. Liver Tumors (LTs) vary significantly in size, shape, and location, and can present with tissues of similar intensities, making automatically segmenting and classifying LTs from abdominal tomography images crucial and challenging. This review examines recent advancements in Liver Segmentation (LS) and Tumor Segmentation (TS) algorithms, highlighting their strengths and limitations regarding precision, automation, and resilience. Performance metrics are utilized to assess key detection algorithms and analytical methods, emphasizing their effectiveness and relevance in clinical contexts. The review also addresses ongoing challenges in liver tumor segmentation and identification, such as managing high variability in patient data and ensuring robustness across different imaging conditions. It suggests directions for future research, with insights into technological advancements that can enhance surgical planning and diagnostic accuracy by comparing popular methods. This paper contributes to a comprehensive understanding of current liver tumor detection techniques, provides a roadmap for future innovations, and improves diagnostic and therapeutic outcomes for liver cancer by integrating recent progress with remaining challenges.Keywords
Abbreviations
| AI | Artificial Intelligence |
| CSSM | Conditional-Statistical Shape Model |
| CT | Computed Tomography |
| DT | Decision Tree |
| ELM | Extreme Learning Machine |
| FCN | Fully Convolution Network |
| FE | Feature Extraction |
| FMLS | Fast-Marching Level-Set |
| FSVM | Fuzzy Support Vectors Machine |
| GLCM | Gray-level co-occurrence matrix |
| HFF | Hierarchical feature fusion |
| LC | Liver Cancer |
| LS | Liver Segmentation |
| MI | Medical Images |
| MLP | Multi-Layer Perceptions |
| MRI | Magnetics Resonance Imaging |
| NN | Neural Networks |
| PET | Positron emissions tomography |
The human body is composed of numerous cell types. In a controlled and orderly manner, these cells grow and split to generate new cells. Therefore, the tumor can develop, and a mass of additional tissue can be formed. It can be benign (that is not cancerous) or malignant (cancerous). Abnormal cells are present in malignant tumors [1]. The body’s largest glandular organ is the liver [2]. Liver Cancer (LC) is a lethal disease around the globe. The best treatment available is surgical resection. However, the conditions applied i.e., tumor sizes should be met. Therefore, it is critical to early diagnosis and accurate appraisal of tumors [3]. In the medical field, LT diagnosing has a broad scope. It is challenging for researchers to segment LT as Computed Tomography (CT) images [4]. Presently, Ultrasound Imaging (US), Magnetic Resonance Imaging (MRI), computer axial tomography, and Positron Emissions Tomography (PET) images are some Medical Images (MI) that are extensively utilized for detecting cancers [5]. Precise and robust lesion segmentation is the primary concern in examining liver pathologies, radiotherapy, and surgery planning [6]. An impact is shown by the progression of medical technology and artificial intelligence (AI) on the comprehension of MI. Computer-Aided Diagnosis (CAD) is one of the fields of main research focus, wherein computer technologies are utilized to detect and characterize malfunction of MI [7].
The research field of MI analysis was dominated by the automatic detection and segmentation of tumors. It is also a vital pre-processing step in CAD [8]. The recent advancements in computer vision spurred the resurgence and refinement of deep Neural Networks (NN). It can now surpass mankind’s performance in object classification as natural images [8,9]. Next, classification techniques are applied to segmented images to classify tissue into two types classified as normal and abnormal [10,11]. Additional investigation is performed on the abnormal tissue image to extract helpful information from a segmented image with some noises [12]. Some comprehensive reviews were done on LS [13,14]. The LT detection techniques are reviewed in this paper. As per the image features it works on, a segmentation technique is categorized as centered on a methodical study of disparate LC detection methods and a systematic summary of the methods. Thus, the performance of every category is summarized, and an optimum solution is found for a specific segmentation task [15,16]. The fundamental architecture for LT detection is exhibited in Fig. 1.
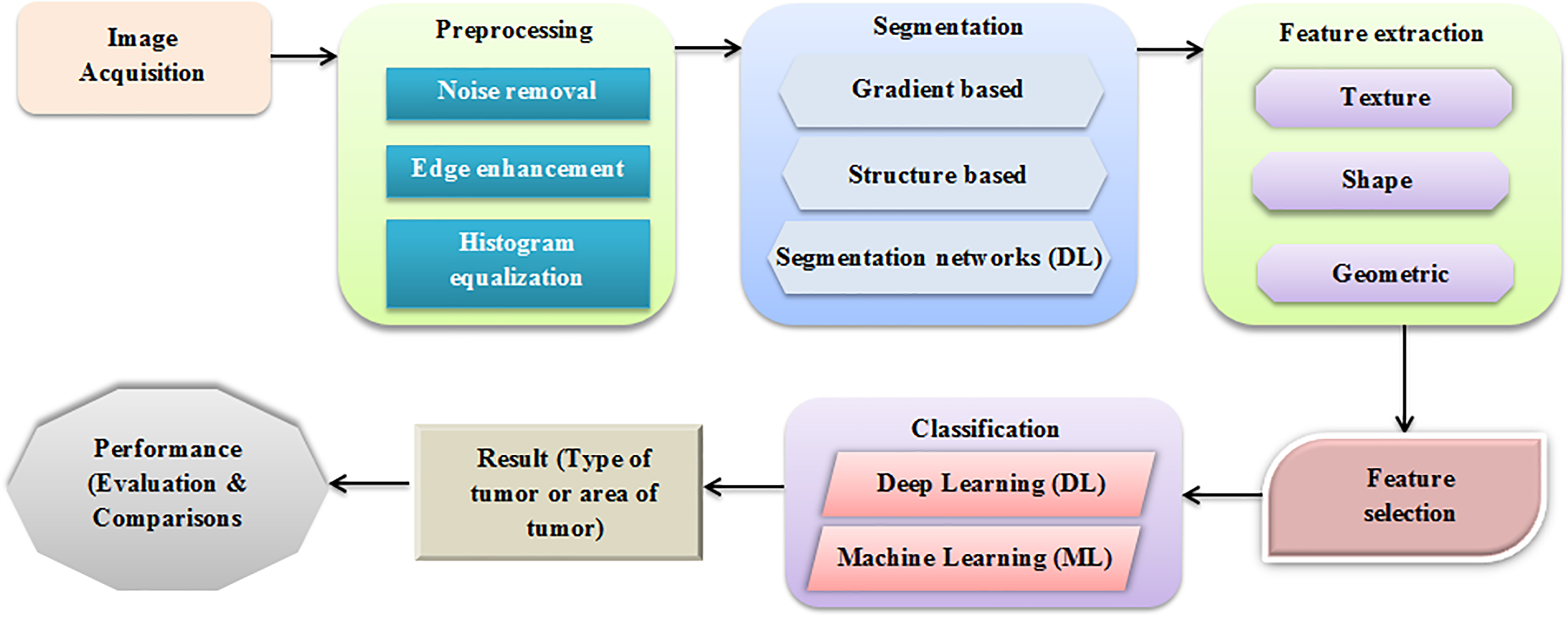
Figure 1: Basic architecture of LT detection
The working pipeline utilized to detect LT is depicted in Fig. 1. There are several cancers in the liver. Predicting whether the tumor is present or not and identifying diverse tumor stages are vital steps in the treatment and cancer evaluation. Pre-processing is vital to diminish the available noise and elevate the CT image’s edges to segment the liver and tumor effectively. Through several features, such as texture, shape, and size, CT and LT images can be detected and categorized [17]. Tumors are classified based on the chosen feature using diverse classifiers. In the evaluation, the performance of several models for LT detection is discussed. This paper is organized as follows: Section 2 is devoted to the literature review, and Section 3 describes the results from reviewed papers, irrespective of the input dataset, methods, and illustrations. Section 4 is for discussion, and Section 5 exhibits the conclusions and future work. Fig. 2 presents the generalized steps for data pre-processing of images [18,19].
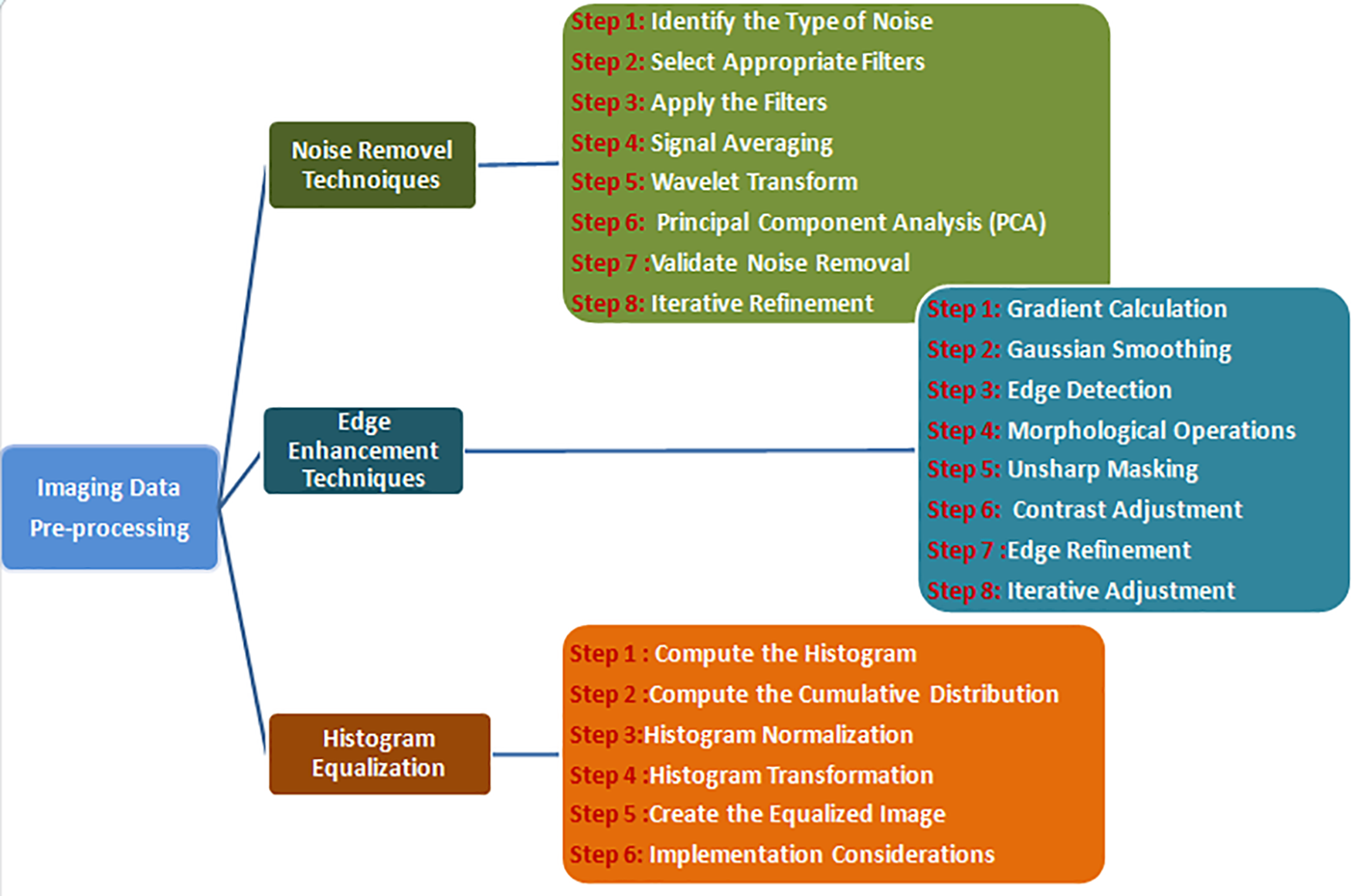
Figure 2: Different types image data pre-processing steps
A. Noise removal techniques
Noise is an unavoidable aspect of digital images, emerging during the acquisition, coding, transmission, and processing phases. Removing noise from digital images is difficult without a solid grasp of filtering techniques. This review offers a brief overview of various noise filtering methods, where the selection of filters is guided by analyzing noise characteristics and segmentation from CT volumes [20,21]. A thorough and quantitative assessment of noise and the most suitable filters is presented in this discussion [22,23]. The generalized expression of the noise model is illustrated in Eq. (1).
The position of the noise in the image is referred to as ENoisy(x,y). The original image is denoted by A(x, y) and its noise pixel is r. The Gaussian noise can be represented by N(x, y)–N(0, S2). The expression for the mean filter model is presented in Eq. (2). The Gaussian filter is outlined in Eq. (3). The mean squared error of the model can be expressed by Eq. (4) [24,25].
B. Edge Enhancement Techniques
Edge enhancement is essential in image processing because the human visual system depends on edges to understand image content. Edges in different directions can be specifically identified and enhanced. These enhanced edges can be blended with the original image to preserve the context. Horizontal edges and lines, in particular, are often accentuated during this process [26,27].
The Prewitt operator is another gradient-based edge detection method, akin to the Sobel operator but using simpler coefficients. The horizontal and vertical kernels are shown in Eqs. (5) and (6).
C. Histogram equalization
Histogram equalization is an image processing technique to enhance contrast by redistributing intensity levels. It aims to publish the most common intensity values, producing a more balanced image and revealing details in regions with low contrast, as depicted in Eqs. (7) and (8).
Eq. (7) is the Cumulative Distribution Function (CDF) in the context of histogram equalization, where CDF(I) is the cumulative sum of the histogram values up to intensity level and h(j) is the histogram count for intensity from j [28].
where T(I) is the transformed intensity for the pixel value, and CDF(I) is the cumulative distribution function value at the intensity within the max and min range. The total number of pixels in the image is (M*N). The number of possible intensity levels (for an 8-bit image, Q = 256).
Based on the working pipeline, the state-of-the-art reports, modern research developments, present trends, and recommendations for further enhancement in the automatic LT detection field are depicted in this section. (i) Image pre-processing, (ii) TS, (iii) Feature Extraction (FE), and (iv) classification utilizing standard datasets are the four noteworthy stages discussed in this paper [29,30]. Section 2.1 reviews the pre-processing and segmentation techniques. The methods are discussed in Section 2.2. The Deep Learning (DL) and Machine Learning (ML) classification for LT detection are briefly explained in Section 2.3. Also, the paper reviews the various research perspectives on the proposed topic of study, Liver Tumor Segmentation [31,32].
2.1 Pre-Processing and Segmentation Methods
This section reviews the modern achievements in autonomous LS with pre-processing. Before the data are forwarded to the segmentation stage, it typically undergoes pre-processing. Pre-processing is employed to eradicate noise and preserve the edges. The pre-processing and segmentation techniques involved in LT detection are elaborated. Abdominal CT images are pre-processed to increase LS’s speed and accuracy [33–35]. Centered upon an amalgamation of region growing and threshold algorithms, a technique for LS was developed. All the schemes are determined in one among the three categories incorporating Grey Level centered, Structure centered, and using Segmentation networks [36–40].
2.1.1 Grey-Scale-Based Approach
Grey level-based pre-processing procedure with a hybridized semi-automatic technique for LS centered upon level-set techniques utilizing manifold seed points [41]. The proposed hybridized technique encompassed a modified Fast Marching Level Set (FMLS) and a Threshold Centered Level Set (TCLS). A customized FMLS was employed to detect an optimum initial liver region as of manifold seed points chosen by the user. A TLS was utilized to extract the actual liver centered upon the initial liver region. The hybridized technique was favored for LS in pre-operative virtual liver surgery planning. Nevertheless, the automatic techniques sacrificed the liver extraction’s accuracy as their algorithm can distinguish the liver from the neighboring organs based on the resemblance of image intensity betwixt the organs [42].
A similar work was done on hybridized watershed segmentation for LT diagnosis. The nose was eliminated by a median filter [43]. The watershed segmentation ameliorated the region, indicating the presence of the needed objects. A solitary intensity threshold was returned, separating pixels into ‘2’ classes: foreground and background. The Wavelet transform determined the threshold values. It was simple and took very little time. An effectual system for detecting LC was deemed. The input was considered in the image or video form, and the output was the same as the inputted image. However, the method encompassed an over-segmentation issue.
Lakshmi et al. [44] introduced Kernelized Fuzzy C-Mean (KFCM) clustering with an adaptive LT segmentation threshold. A succession of test images on MICCAI 2008 LT segmentation datasets was taken to extract the tumor area. The tumor area was gauged. To reduce the effect of noise and enhance clustering, the KFCM introduced a kernel function on FCM. The result exhibited positive outcomes for the algorithm. Segmenting the LC area was effective, and it achieved a high peak signal-to-noise ratio (PSNR), lower Mean Squared Error (MSE) values, and higher consistency for detecting smaller changes in images. Nevertheless, the technique was not appropriate for noisy images.
The FCM approach for CT and LT segmentation was proposed, which analyzes altered intensity values and higher frequencies eradicated utilizing histogram equalization and a median filter scheme to increase the contrast of liver CT images [45]. The approach centered on neutrosophy handled indeterminacy, better uncertainty, lessened over-segmentation, and better accuracy and performance on uninformed and noisy images. The accuracy attained by the suggested approach was nearly 95% better than LS. However, the system was tested with limited CT images.
The Geodesic Actives Contour algorithm was aimed at generating pre-processing of the LT images [46,47]. After pre-processing, the tumor’s Region of Interest (ROI) was determined. The abnormal region’s boundary in a single slice was signified. Next, a generative model processed the remaining slices and was ameliorated by incorporating a restraint. A probabilistic scheme was employed to search for the tumor boundary, and the solution was attained by Bayesian inference. The Kullback-Leiblers divergence was employed to measure the outcomes’ consistency with the model’s restraint. However, simple training datasets were employed.
A novel algorithm recommended for identifying a unified level set model for integrating image gradient, area competition, and prior information was intended for CT-LT segmentation [47]. Unsupervised fuzzy clustering helped to assess LT’s probabilistic distribution [48,49]. It was employed to ameliorate the object indication function, state the directional balloon force, and regulate region competition. An inclusive and flexible platform was provided for balancing various forces, such as the dynamic interface, the image in the examination, and other previous information. The unified level set model was an effectual solution for LT segmentation on contrast-ameliorated CT images. Nevertheless, the model encompassed restrictions for these images with lower contrast, weaker boundaries, and field homogeneity.
2.1.2 Structure-Based Approach
Work analyzing structure-based approaches develops Principal Components Analysis for automatic liver image pre-processing [48–50]. Based on the created atlas, the utmost probable liver area of the test image was additionally determined via a probability map’s posterior classification approach, resulting in rough segmentation. Shapes intensity before the level set produced the final LS. ‘25’ test CT datasets as of the partner site were considered. Its outcomes were contrasted with two top-notch LS methods. Nevertheless, a smaller dataset was utilized for training [51–53].
Image rendered with hierarchical local region-centered Sparse Shape Composition (SSC) for LS on CT scans was found to be significant and promising [54,55]. In the initial training step, to augment the flexibility of shape prior models and devotedly capture the comprehensive local shape information, a multi-level local region-centered SSC model termed Multi Linear Regression (MLR)-SSC was described. The liver shapes were disintegrated into manifold areas in a multi-level fashion. The segmentation work was more effective and robust to local minima. The method rendered an inferior initial shape, which brought about a larger segmentation error on the last outcomes.
Generic affine in-variance shape parameterized together with graph cuts intended for the segmentation utilizes training sets with regions of atypical local shape that were determined as a preliminary segmentation. The geodesics active contour locally corrected the organ segmentation on abnormal images [56–58]. Utilizing shape and amelioration constraints, optimized graph cuts were employed for segmenting the vasculature in addition to hepatic tumors. A significant reduction in the LS errors could be seen. All tumors were detected, and the trouble was anticipated with a 0.9% error. The technique’s robustness in examining livers as complex clinical cases to permit temporal monitoring of patients having hepatic cancer was demonstrated. Nevertheless, a high false-positive rate was present [59–61].
2.1.3 Segmentation Based Approach
Image segmenting cascaded with Enhanced Deep Convolution Neural Network (EDCNN) aims at effective LT segmentation. For cascade segmenting of the liver in addition to lesions on CT images with restricted image quantity, deep EDCNN was built and trained. The liver image was segmented using the EDCNN and rendered for the EDCNN training [62]. Next, the EDCNN segmented the tumor areas on the liver ROI areas as envisaged utilizing the EDCNN. The false positives were significantly reduced by partitioning the hepatic tumor on the liver ROI. A public dataset was taken for testing. In addition, many metrics were utilized to quantitatively evaluate its performance. A DICE score of 95.22% was produced aimed at the test set of CT images. However, the system was trained with restricted datasets [63–65].
Li et al. [66] presented probability in addition to the local restraint level set model intended for LT segmentation as of CT volumes. The target’s Density Distributions (DD) and the multi-modal DD of the backdrop can minimize the probability energy approximated manifold areas. The ramp related to the edges for weak boundaries was preserved via the edge detector. The scheme to the Chans-Vese and the geodesics level set models and the manual segmentation performed by specialists were compared. Concerning segmenting the hepatictumors, the Chan-Vese model was not triumphant. The model trounced the geodesic level set model. However, the system was trained with restricted image features [67–69].
Another work with gradient ameliorated level-set centered segmentation is intended for LT as of CT Images [70]. It was truncated to keep the CT image intensity values in a fixed range to enhance the image contrast surrounding the liver and LT. These were done during the pre-processing step. Firstly, the Convolutional Neural Network (CNN) segmented the liver in a coarse-to-fine manner intended for removing non-liver tissues aimed at succeeding TS. For roughly localizing the liver [71,72], a 2D slice-centered U-net was utilized. 3D patch-centered full CNN was employed to refine the LS and roughly localize the LT. The better segmentation performance of the pipeline was identified over top-notch methods has illustrated in Fig. 3. Nevertheless, the system encompassed a high computational expense.
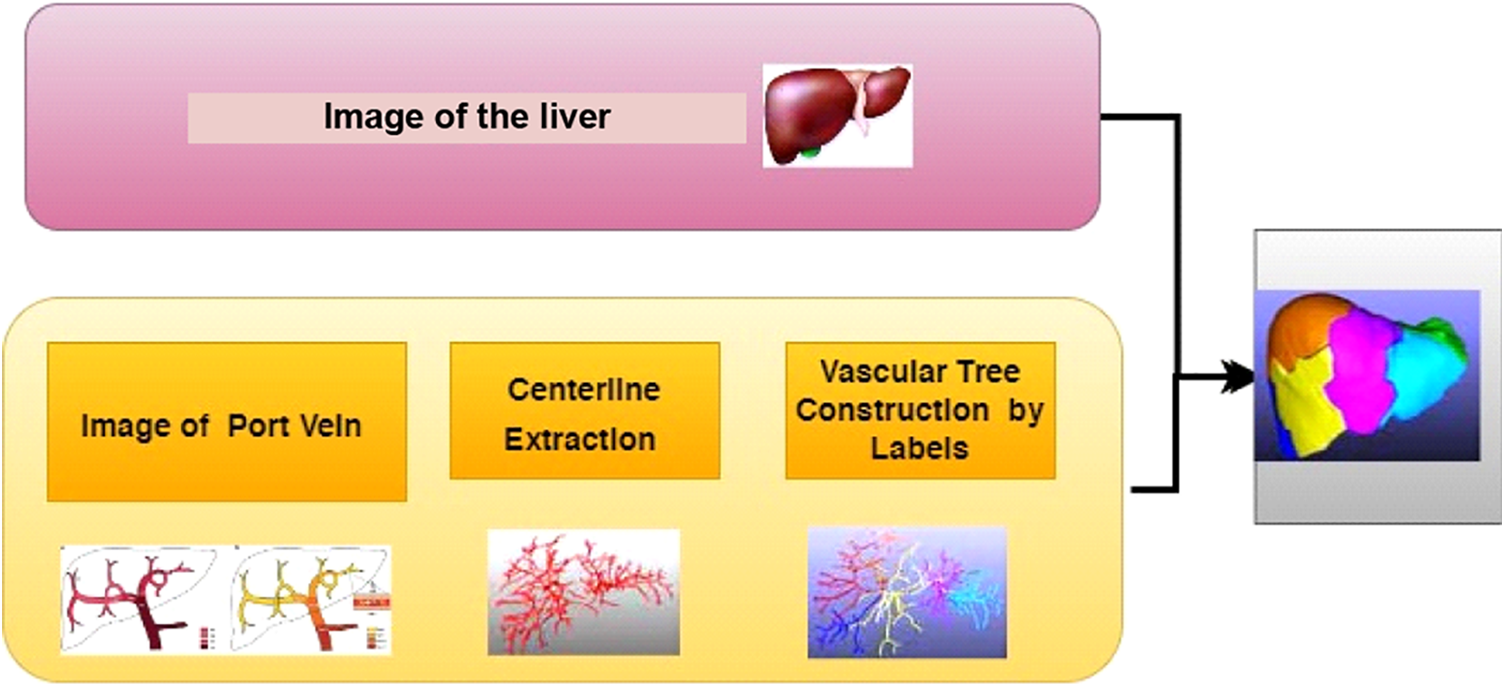
Figure 3: Basic segmentation process for LT detection
Lu et al. [73] formed a 3D CNN for automatic LS. In addition, a liver probability map was attained to generate a preliminary segmentation. Next, the learned probability map was incorporated into the graph cut energy function for additional segmentation alteration. The detiled image segmentation process has been illustated in the Fig. 4. It does not need any user interaction aimed at initialization, which was the main benefit. Therefore, non-experts could perform the method. In addition, it was an early endeavor to engage DL algorithms for 3D-LS. Nevertheless, poor classification accuracy was present.
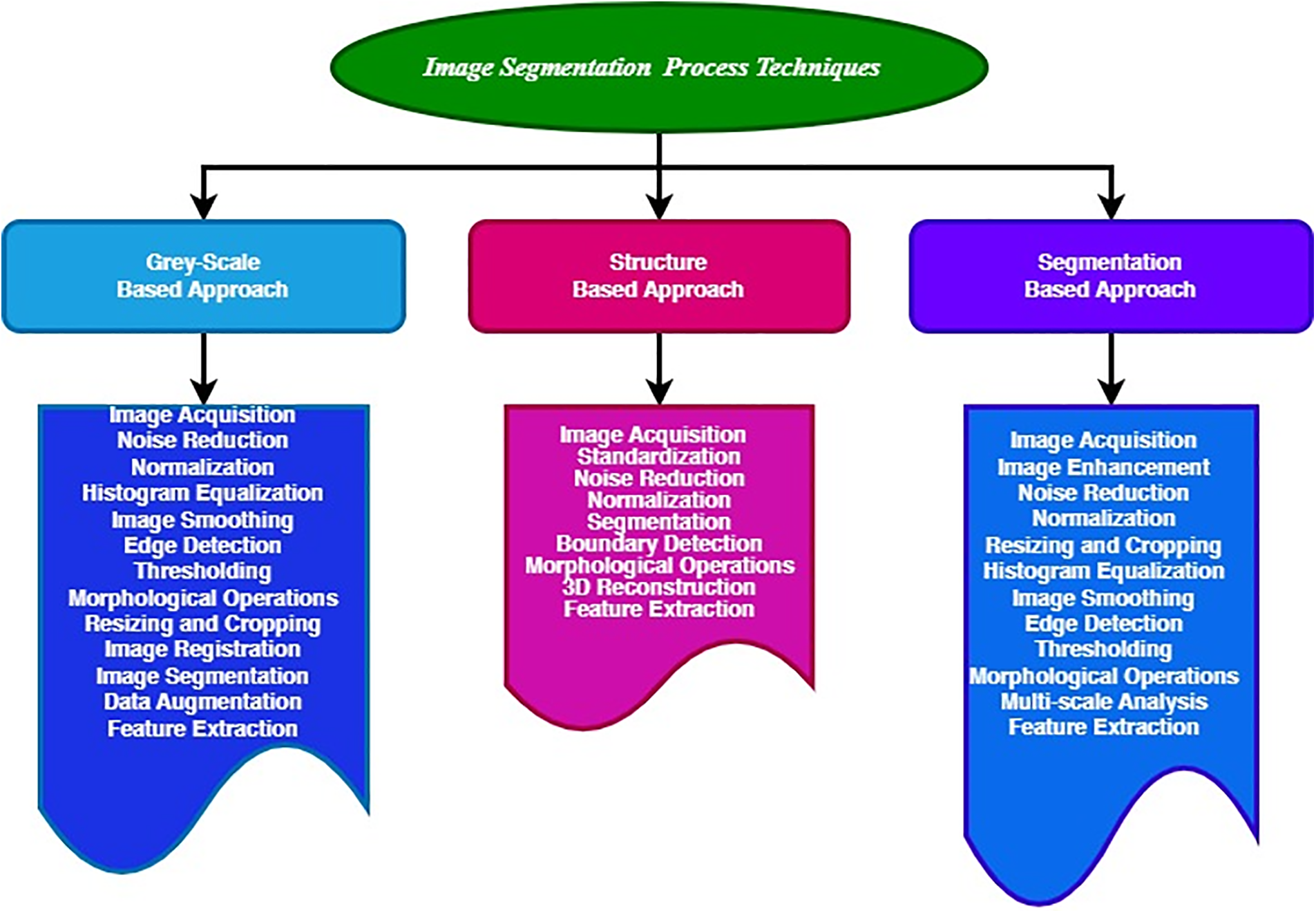
Figure 4: Different types of image segmentation process
Chi et al. [74] aimed at automatic liver vasculature segmentation on CT images centered on the vessel context. Voxels were grouped hierarchically to liver vasculature. Initially, they were locally grouped into vessel branches with the benefit of a vessel junction gauge. Next, utilizing manifold feature point voting mechanisms, they were globally grouped into vasculature. ‘10’ clinical CT datasets were estimated. The below 3 min are taken by segmenting 3rd-order vessel trees as CT images of the portal venous phase on a PC with a 2.0 GHz dual-core processor. The average segmentation accuracy was 98%. However, the system encompassed higher segmentation errors.
A fully Convolutional Network (FCN) was developed to address the LS task and detect liver metastases on CT examinations [75]. Although considering semantic segmentation, FCN was demonstrated to be an enormously powerful tool. A relatively smaller dataset was taken where the FCN performance was explored and contrasted with patch-centered CNN and sparsity-centered classification schemes. True positive rates of 0.86 and 0.6 false positives per case were achieved utilizing the fully automatic algorithm, resulting in propitious and clinically pertinent outcomes. Nevertheless, the system needed a huge dataset.
2.2 Feature Extraction Techniques
The various FE techniques (Fig. 5) and their achievements for LT detection by features like texture, shape, and size are listed in Table 1.
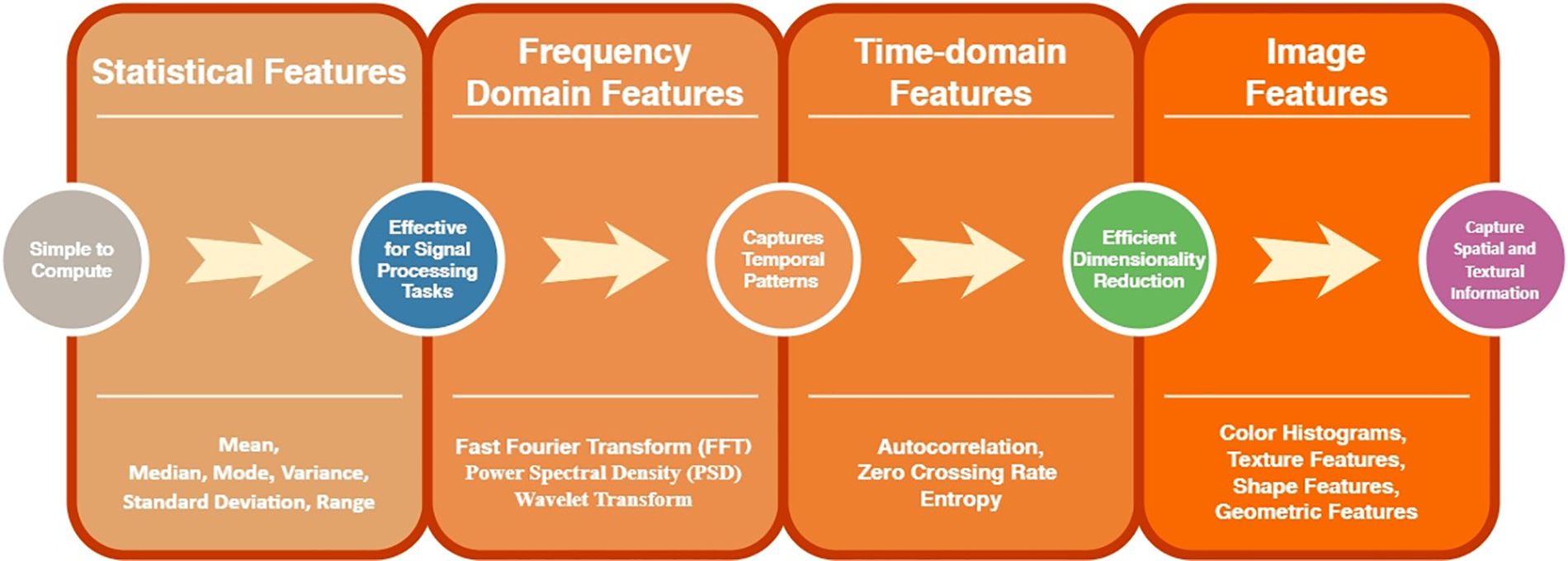
Figure 5: Various feature extraction techniques

Studies [26–28] look into different elements of liver image analysis. Cheng et al. [26] analyzed ultrasonic liver tissue using wavelet sub-images and HFF, which successfully selects discriminative features but has a restricted dynamic range. Shyam et al. [27] focused on liver segmentation utilizing high-level semantic features and NucleiSegNet, which achieves higher accuracy with the KMC Liver dataset but encounters false-positive predictions. Guangming [28] employed FSVM and textural characteristics such as energy and contrast to identify liver tissue using B-Mode Sonography and the GLCM. It delivers excellent specificity but is challenging to understand. Studies [29–31] used a variety of approaches for segmenting liver tissue and identifying lesions. Ramin et al. [29] employed a complicated method for tissue segmentation in CT images, concentrating on edge detection using a Kirsch filter on a non-public CT dataset. Although they enhanced the identification of concave and convex locations, they added complexity. Sho et al. [30] used a statistical shape model for automated liver segmentation utilizing conditional features on abdominal CT volumes, which improves classification accuracy but struggles to discriminate between healthy and sick patients. Laszlo et al. [31] created a model to identify liver lesions using geometric characteristics and GLCM using the Ground Truth lesion database. This is appropriate for all hepatic lesions; however, the contouring is less exact.
Studies [32,33] use modern methods to diagnose liver conditions and segment lesions. Chin et al. [32] identified liver diseases utilizing a CAD system with 3D form, texture features, kinetic properties, GLCM, and an elliptic model on CT images. It has good accuracy and sensitivity but requires manual segmentation. In a study [33], liver lesions were segmented using an adaptive local window approach using global and local statistics on CT and MRI datasets, displaying resilience to baseline contour position but struggling with diverse hepatic lesions. The MICCAI 2007 dataset is used in a study [34] to develop a liver segmentation approach that uses local characteristics and the liver border with Sparse a priori Skin Sparing Mastectomy (SPSSM). Although it can efficiently accommodate deformation, it necessitates a high sample size. These studies present a variety of techniques, emphasizing benefits such as effective feature selection and high accuracy while addressing shortcomings such as complexity and difficulties in discriminating between healthy and sick tissues.
2.3 Classification Techniques for Liver Tumor Detection
Classification is the final stage in an autonomous CAD system. The extracted set of feature vector(s) as of the former stage is acquired as the input. The classification phase aims to implement a learning-centered methodology regarding its inputted feature vector(s) for disease diagnosis. The classification techniques for LT detection are briefly explained here.
SVM was found to be highly influential for the automated liver as well as TS [35]. Utilizing training sets, the regions of a characteristic local shape were ascertained as of a liver’s initial segmentation. The liver segmentation in abnormal images was locally corrected by the geodesic active contour. Utilizing shape and enhancement constraints, graph cuts segmented the hepatic tumors. Significant reduction in the LS errors and also every tumor were detected. SVM and feature selection were utilized to reduce the total false tumor detection. The tumor trouble was anticipated with a 0.9% error. Nevertheless, the technique was computationally costly.
SVM aims at automated LT detection [36]. An automated CAD of LT as of CT images was generated. Markov Random Fields (MRF) embedded level set technique segmented the liver. Robustness was rendered to noise as well as fast segmentation. Shape analysis techniques found the shape ambiguities of the liver (segmented), which utilized a training set intended for correction. The graph cut technique detected hepatic tumors as of the corrected LS. FE was employed to classify them utilizing an SVM. However, the system encompassed poor classification accuracy.
Das et al. [37] presented a fuzzy clustering with a DT classifier for LC detection. With adaptive thresholding, the liver and other body parts were separated initially. Next, with spatial fuzzy clustering, the cancer-affected lesions of the liver were segmented. As for segmented cancerous areas, the informative features were extracted. It was classified into Hepato Cellular Carcinoma (HCC) and MET utilizing MLP and C4.5 DT classifiers. An effectual approach intended to automatically recognize LC was the SFCM-centered segmentation with a C4.5 DT classifier. Nevertheless, a lower recognition rate was obtained.
Huang et al. [38] rendered an arbitrary feature subspace ensemble-centered Extreme Learning Machine (ELM) intended for LT detection and segmentation. The ELM autoencoder was executed as a pre-training step to increase testing accuracy. ELM was trained as a 1-class classifier with merely healthy liver samples in automatic LT detection. The performance was contrasted with ‘2’-class ELM. A semi-automatic approach was utilized to train the classifier to choose samples in 3D space in LT segmentation. The technique was tested and evaluated using a cluster of patients’ CT data, and the experiment showed propitious outcomes. Nevertheless, the segmentation process was tedious.
Kernel-centered ELM for LT detection together with segmentation trained as a 1-class classifier with merely healthy liver samples in training [39]. It brought about a technique of tumor detection centered upon novelty detection. It was contrasted with ‘2’-class ELM. The semi-automatic approach was adopted using arbitrarily choosing samples in 3D space inside a restricted ROI intended for classifier training to extract the tumor boundary. A cluster of patients’ CT data was considered. The experimentation showed better detection and encouraged segmentation outcomes. However, it limited its applications to higher dimensional and larger data.
DL models are utilized to diagnose the LT by disparate image acquisitions with the progression of AI. Disparate works are done for lever segmentation and LT detection utilizing DL approaches. The DL techniques for LT detection are listed in Table 2. The summary in Table 2 outlines several techniques that provide insight into the range of accuracy reported in several methods discussed in the study. Image pre-processing is crucial for a precise outcome of succeeding steps. Generally, noise elimination, contrast enhancement, and edge enhancement are the ‘3’ kinds of pre-processing. As the Wiener filter can polish the boundary and conserve the image’s interior information well, it was utilized by the models to perform image pre-processing. The cumulative distribution function was utilized by histogram equalization to normalize image intensity, thereby elevating image brightness. The pre-processing schemes utilized in the above models executed comparatively well in network training for automated LS.
The entire ‘3’ sorts of LS methods have their advantages. Generally, to handle the complex segmentation issue, the GL-centered models are often utilized together. Using prior knowledge, the structure-centered models handled the liver’s unclear boundary. This means that some issues that the GL-based methods cannot handle can be dealt with through this model. Owing to the capacity to resolve computer vision tasks, a primary focus for the research community in previous years is deep convolutional NN [74–77]. Vital attributes for detecting tumor regions as of the image are termed Features. A statistical approach and a structured approach are the two ways it is done. Major researchers utilized a statistical approach. Co-occurrence matrix, Fractals, Gabor filters, and wavelet transform were the several statistical techniques for gauging texture. GLCM, utilized by major researchers, captures numerical feature values by utilizing spatial relationships among neighborhood pixel features. Several autonomous approaches for brain tumor detection, namely NN and SVM, have been popular in the prior decades. As DL can signify complicated structures, own-learning, and process enormous MRI-centered image data efficiently, it has reached a central tract regarding automation of Brain tumor detection. It can present the current trends and achievements in ML and DL techniques as shown in Fig. 6. Due to their automated FE techniques, DL models have been identified as being able to execute better and make the system more effective and intelligent [78].

Figure 6: Variety of dataset usage on diagnosing the LT
Several models can encounter a few challenges in TS and detection even though they were introduced with various benefits. These models are found to be rapid. However, they can miss their effectiveness when the target’s GL varies. Many false positive regions requiring post-processing are encompassed in their outcomes. Requiring an enormous quantity of training data to cover the whole liver condition is difficult for those models. It is even more critical with the liver’s non-standard shape, which makes it challenging to define the liver utilizing a unified model. TS grounded on DL has become the standard of autonomous TS, and outstanding achievements have been made in the image segmentation domain. In order to deliver great performance, a few challenges of DL are derived. The deep learning classifier is grounded mainly on the available dataset’s magnitude and quality. The success ratio of DL can be directly hampered mainly in the medical domains if restricted data or information is present. The model’s performance was primarily affected by the slight alteration in the values of hyperparameters. Accurate outcomes for real-time issues cannot be rendered by the default value of parameters. DL models need neuron training repeatedly for diverse issues as they cannot adapt to another domain. For the scope of enhancements and alterations, infinite opportunities are available. Noteworthy enhancements in the accuracy of analysis and prediction in diagnosing disease by DL cannot be ignored.
The disparate outcomes of the ML and DL techniques show the classifiers’ performance. Centered on some performance metrics, such as sensitivity, accuracy, and specificity, together with NPV and PPV, the performance of the techniques is gauged. The below figures are utilized to comprehend the performance of the classifiers involved in LT detection. Datasets used by the different methods are tabulated as follows in Table 3.

The accuracy of DL classifiers is illustrated in Fig. 7. 85.7% accuracy was obtained by the Deep Convolutional Generative Adversarial Network (DCGAN) [60]. CNN [62] and 3D U-net [63] encompassed 99.94% and 73.6% accuracy. CNN [64] reaches 92%. Next, 96% accuracy is attained by the Adaptive Neural Network (ANN) [66]. CBAM [67] and DCNN [68] encompassed 90.93% and 79.7% accuracy. CNN [70] rendered 75% accuracy. PNN [50] attained 96.7% accuracy.

Figure 7: Accuracy of different DL classifiers with PNN, DCGAN, CNN, CNN-R, 3D U Net, ANN, CBAM, DCNN and CNN-N
The sensitivity of DL classifiers is exhibited in Fig. 8. DCGAN [60] renders 78.6% of sensitivity. Graph convolution Embedded LSTM Long Short Term Memory (GCLSTM) [61], in addition to CNN [64], achieved 77.06% and 92% of sensitivity was attained by the Artificial Neural Network (ANN) [66] and 85.7% of sensitivity was rendered by the FCNet (Fully Complex) Network [44]. PNN [67] and FCM [68] encompassed 97.3% and 91% of 98.64% sensitivity.

Figure 8: Sensitivity of different DL classifiers with FCNet, PNN, FCM, DCGAN, GCLSTM, CNN, ANN and CBAM
The specificity of DL classifiers is depicted in Fig. 9. DCGAN [60] reaches 88.4%, and CNN [64] encompasses 98% of specificity. Next, 98.11% of specificity renders ANN [66]. CBAM [67] achieves 91% of specificity. FCNet [68] and NN [69] encompass 92.4% and 72%. Then, PNN [70] has 94% specificity, and PNN [74] achieves 96% specificity. FCM attained 92% of specificity [78].

Figure 9: Specificity of different DL classifiers with FCNet, PNN, FCM, DCGAN, GCLSTM, CNN, ANN and CBAM
Fig. 10 analyses the NPV of the various DL classifiers.It indicates that NPV of 96% and 91.66% are attained by the PNN and CBAM [67], which are higher than PNN [70], BEDM, and ANN. The NPV of PNN, BEDM, and ANN are 91.4%, 83.51%, and 78.45%.
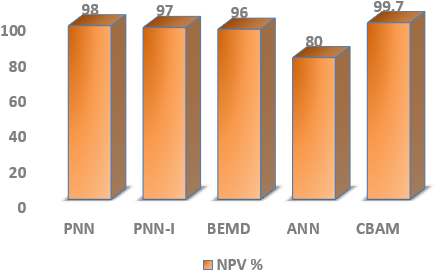
Figure 10: SNPV of DL classifiers with PNN, PNN-I, BEMD, ANN and CBAM
The PPV of different DL classifiers is analyzed in Fig. 11. The CBAM [67] has a higher PPV value. BEMD attains 98.67%, and PNN achieves 94% of PPV. In addition, PNN and ANN have a PPV of 87.83% and 68.62%.
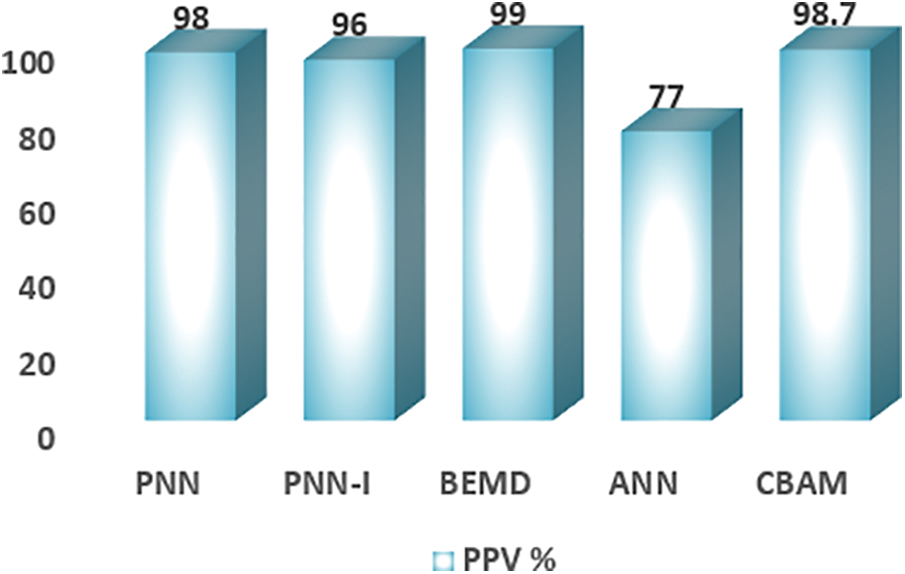
Figure 11: SPPV of DL classifiers with PNN, BEMD, ANN and CBAM
The accuracy of the ML classifiers is exhibited in Fig. 7. BEMD achieves 92.95% of accuracy. After that, SVM attains 98.9%. SVM and SVM [36] have 87.6% and 77% accuracy. The SFCM-DT reaches 95.02% accuracy. Lastly, ELM attained 74.75% accuracy (the same has illustrated in Fig. 12).
Figure 12: Accuracy of different ML classifiers with SVM, SFCM-DT, ELM and BEMD
3.1 Research Attainments of Existing Approaches
Researchers have developed several systems to classify LT. The outcomes are compared using the figures given above, and the classifier’s performance is analyzed. DL techniques, say [63–65], are superior to ML techniques [78–82]. Semi-automatic methods offer no advantages over recent automatic methods. Automatic methods certainly attain some of the best outcomes for LT segmentation. From the study, automatic segmentation offers notable advantages compared to a semi-automatic technique. The classification accuracy is improved with DL techniques, and the training time is lessened. If liver lesion segmentation is performed inside a liver envelope, it seems more accurate, especially for automatic segmentation. Lastly, DL techniques seem to augment tumor detection and segmentation accuracy.
3.2 Research Problems and Trends in Tumor Analysis
The study analyzed the research problem on liver tumors, including properties like brightness, contrast characteristics, surrounding tissues, larger sample size, and others. The same characteristics are widely used in experiments that take any of these characteristics. However, the research does not consider multiple characteristics. Likewise, the most critical factor to focus on is irregular tumor shapes that vary significantly at different cancer stages. These issues are highlighted earlier by several research communities in Tables 1 and 2, respectively. All the approaches mainly focus on tumor segmentation and identification, which strongly relies on the attributes of the study. Though several studies exist on tumor analysis, no strongly dependent approach can more effectively identify liver tumors, leading to effective tumor detection and classification. The research trends mainly focus on analyzing the model using machine learning, leading to prominent results. Though the results were promising from the existing approach, it faced some time constraints. The study presented an outline of these algorithms and the key factors that lead to possible improvements in any parametric analysis.
4 Research Impact on Existing Systems, Proposed Solution, and Experimental Analysis
4.1 Research Impact on Liver Tumor Detection
The proposed study mainly focuses on research aspects of analyzing the pros and cons of existing systems by looking into the depth of several algorithms and in wider, several broad categories, Tables 1 and 2, which highlight the significance of methods on feature extraction and strategies adopted for different works on liver tumor analysis and deep learning techniques in the proposed context of study. Therefore, research impact on liver tumor analysis is deemed challenging, as inferred from several studies highlighted in the survey (presented in Sections 1 and 2). Table 4 summarizes the key research findings and narrows the future research prospects. In this view, the identified research gap is presented, and the following section provides an improved solution to the gap and narrows the significance of the proposed research illustration.
Compared to the existing systems, an improved model is suggested, focusing on multi-variate analysis and the dimensions of the existing systems. Fig. 13 presents the proposed model, analyzing the drawbacks highlighted in Table 3. The classifier presents the image analysis and passes through several layers for improved outcomes compared to fine-tuning the expected outcomes. The results mainly aim to classify liver tumors [123–126]. However, the study can also be well applied to several diseases, as shown in the image. In our model, the criteria for each category are based on specific anatomical and pathological characteristics relevant to liver tumor analysis [127–129], such as (i) Liver—The categorization focuses on the presence, size, and location of the tumor within the liver, as well as any associated changes in liver texture or shape [130–134]. (ii) Bladder—This is primarily included to differentiate between abdominal organs and to exclude areas irrelevant to liver analysis, based on spatial positioning [135–137]. (iii) Lung—The inclusion of the lung is primarily to assess any secondary findings or metastasis, as liver tumors can sometimes spread to nearby organs [138–140]. These categories were determined based on clinical relevance and typical patterns observed in liver tumor cases [141–143].
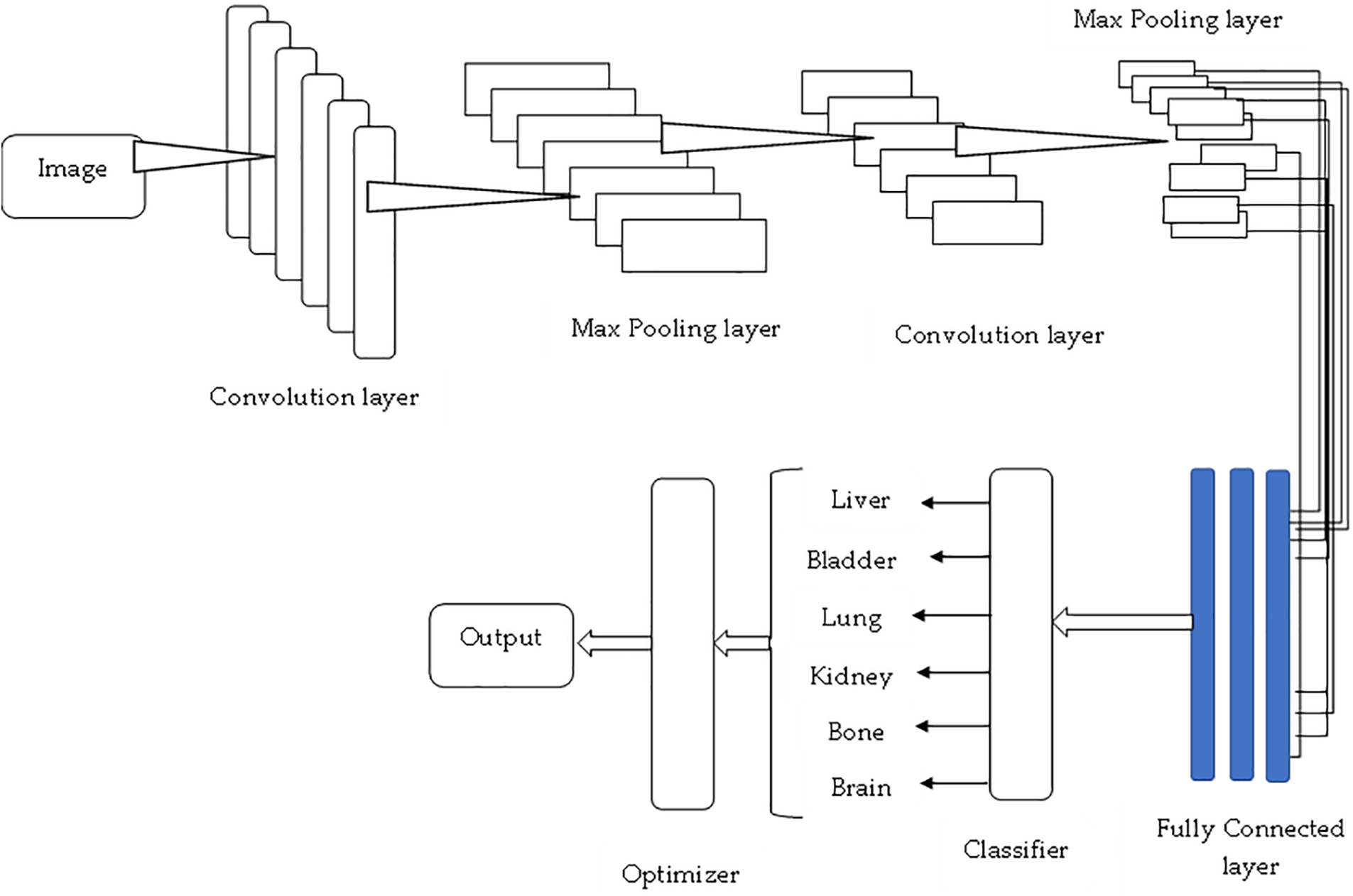
Figure 13: Proposed system model for liver tumor analysis
Collaborative Fusion Convolutional Neural Network (CFCNN) for tumor detection is developed to address the implications of liver and lesion segmentation. The detection of lesions from the input data is considered very small, especially the automatic segmentation, and if it is done manually, then it can only be applied to 2D. Fig. 14 shows the original images, which are considered for testing purposes. In order to produce the 1 to 1.2 scales of the samples, the size of the lesions can be modified. The measurements are sampled evenly, and the photos are resampled using neighboring methods as shown in Fig. 15. Using the CFCNN method, various measurements and scales are generated while considering available datasets.

Figure 14: Tumor segmentation samples

Figure 15: Training vs. validation of study results
The study compares performance evaluations’ similarity with several metrics such as precision, recall, accuracy, and F1-score. The proposed method is found to be more efficient for the existing model. The similarity coefficient evaluation has illustrated through Table 5, and convergence has shown in Fig. 16.

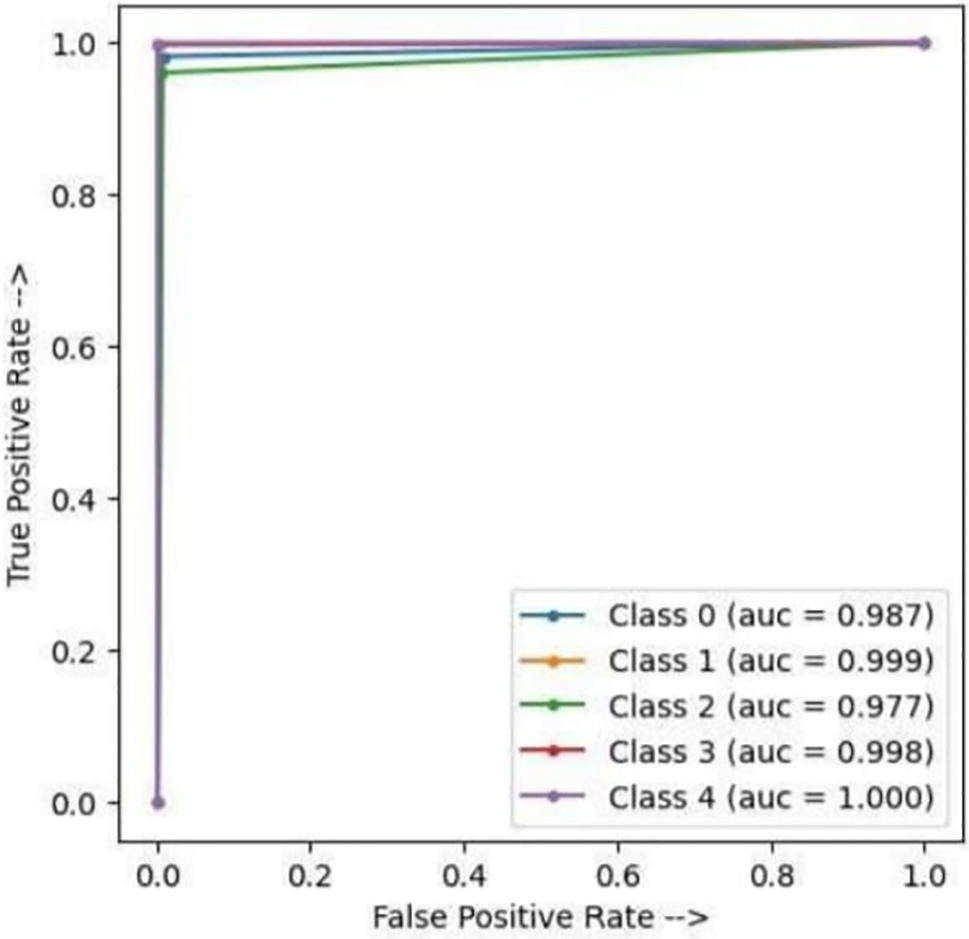
Figure 16: AUC-ROC curve
This study emphasizes the crucial role of efficient pre-processing and segmentation in liver tumor identification, highlighting recent advancements and innovative computational approaches. By examining a probabilistic distribution strategy for liver tumor analysis, we shed light on the advantages of various feature extraction and diagnostic techniques. Among these, the Gray Level Co-occurrence Matrix (GLCM) stands out as one of the most effective feature extraction methods, widely used for its ability to enhance processing speed, handle data inputs, and maximize diagnostic performance in terms of accuracy and precision. As shown in Table 1, GLCM-based methods consistently deliver reliable results across diverse datasets, underscoring their utility in tumor feature analysis. Table 2 presents a comparison of twenty diagnostic techniques, detailing the strengths and limitations of each approach. Combinational methods incorporating transfer learning have shown particular promise, often outperforming traditional approaches by leveraging pre-trained models to enhance accuracy in complex, image-based diagnostics. However, despite these benefits, transfer learning is computationally intensive, which can limit its practicality in real-time or resource-constrained settings. A comprehensive analysis of sensitivity, accuracy, specificity, and predictive values (NPV and PPV) across ML and DL methods reveals that certain ML algorithms remain highly effective in liver tumor diagnosis, especially when tailored to specific data needs. Section 3 summarizes two decades of research, tracing the evolution of ML and DL algorithms and their impact on clinical diagnostic capabilities. While DL approaches generally offer higher accuracy for processing large, complex datasets, ML techniques provide reliable, lower-cost options suitable for smaller datasets and limited-resource environments. In addition to offering a detailed review of current diagnostic methods, this paper highlights key challenges, including the need for diverse, high-quality datasets, computational efficiency, and strategies to prevent overfitting in smaller datasets. Addressing these limitations will be essential for improving the clinical applicability and diagnostic accuracy of liver tumor detection. In conclusion, this research reveals substantial potential for continued innovation in liver tumor diagnostics.
Future research should focus on refining these techniques to reduce computational demands without sacrificing diagnostic precision. Also studies should focus on developing computationally efficient adaptive models, exploring hybrid approaches that combine the strengths of machine learning and deep learning, and expanding the use of unlabeled datasets to drive advancements in unsupervised learning. By harnessing these developments, the field can advance toward implementing faster, more accurate, and resource-efficient liver tumor diagnostics in clinical settings, ultimately improving patient outcomes. This study underscores the importance of effective pre-processing and segmentation in liver tumor identification, highlighting recent advancements and creative computational techniques. Through our examination of a probabilistic distribution strategy for liver tumor analysis illuminating the benefits of various feature extraction and diagnostic methods.
Acknowledgement: The authors would like to thank the National Yunlin University of Science and Technology, Taiwan, and Vardhaman College of Engineering, Hyderabad, India, for providing the necessary facilities and resources for conducting this research.
Funding Statement: This work received financial support from the “Intelligent Recognition Industry Service Center” as part of the Featured Areas Research Center Program under the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan, and the National Science and Technology Council, Taiwan, under grants 113-2221-E-224-041 and 113-2622-E-224-002. Additionally, partial support was provided by Isuzu Optics Corporation.
Author Contributions: Shanmugasundaram Hariharan: data collection, draft the article; D Anandan: interpretation of results; Murugaperumal Krishnamoorthy: diagrams and draft preparation; Vinay Kukreja: manuscript validation; Nitin Goyal: supervising and result validation; Shih-Yu Chen: supervising and draft checking. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Jayanthi M, Kanmani B. Extracting the liver and tumor from abdominal CT images. In: 5th International Conference on Signals and Image Processing, 2014 Jan 8–10; Bangalore, India: IEEE. [Google Scholar]
2. Bing NL, Chee KC, Stephen C, Sim HO. A new unified level set method for semi automatic liver tumor segmentation on contrast enhanced CT images. Expert Syst Appl. 2012;39(10):9661–8. doi:10.1016/j.eswa.2012.02.095. [Google Scholar] [CrossRef]
3. Daniel P, Nikos P, Stephane C. Automatic detection of liver tumors. In: 5th IEEE International Symposium on Biomedical Imaging From Nano to Macro, 2008 May 14–17; Paris, France: IEEE. [Google Scholar]
4. Yugander P, Raghotham RG. Liver tumor segmentation in noisy CT images using distance regularized level set evolution based on fuzzy C-means clustering. In: 2nd IEEE International Conference on Recent Trends in Electronics Information and Communication Technology, 2017 May 19–20; Bangalore, India. [Google Scholar]
5. Nader H, Abdel M, Mohiy MH, Khalid MA. Fully automatic liver tumor segmentation from abdominal CT scans. In: International Conference on Computer Engineering & Systems, 2010 Nov 30–Dec 2; Cairo, Egypt: IEEE. [Google Scholar]
6. Chethan KS, Nataraj KR. Automated identification of hemangioma based on CT-scan tumor edge detection. In: 2nd International Conference on Trends in Electronics and Informatics, 2018 May 11–12; Tirunelveli, India. [Google Scholar]
7. Ryo H, Yutaro I, Lanfen L, Hongjie H, Yen WC. Automatic segmentation of liver tumor in multiphase CT images by mask R-CNN. In: 2nd Global Conference on Life Sciences and Technologies, 2020 Mar 10–12; Kyoto, Japan. [Google Scholar]
8. Eugene V, An T, Chris P, Samuel K. Liver lesion segmentation informed by joint liver segmentation. In: 15th International Symposium on Biomedical Imaging, 2018 Apr 4–7; Washington, DC, USA. [Google Scholar]
9. Vinita D, Jyotika P. Review of image processing techniques for automatic detection of tumor in human liver. Int J Comput Sci Mobile Comput. 2014;13(3):371–8. [Google Scholar]
10. Luo SH, Li XC, Li JM. Review on the methods of automatic liver segmentation from abdominal images. J Comput Commun. 2014;2(2):1–7. doi:10.4236/jcc.2014.22001. [Google Scholar] [CrossRef]
11. Yang X, Yu HC, Choi Y, Lee W, Wang B, Yang J, et al. A hybrid semi-automatic method for liver segmentation based on level-set methods using multiple seed points. Comput Methods Programs Biomed. 2014;113(1):69–79. doi:10.1016/j.cmpb.2013.08.019. [Google Scholar] [PubMed] [CrossRef]
12. Thulasidass S, Soundari DV, Chinnapparaj S, Naveen R. Liver tumor diagnosis by using hybrid watershed segmentation method. Mater Today: Proc. 2021;37:2848–57. doi:10.1016/j.matpr.2020.08.660. [Google Scholar] [CrossRef]
13. Amita D, Sukanta KS. Kernelized fuzzy C-means clustering with adaptive thresholding for segmenting liver tumors. Procedia Comput Sci. 2016;92(10):389–95. doi:10.1016/j.procs.2016.07.395. [Google Scholar] [CrossRef]
14. Ahmed MA, Aboul EH. CT liver tumor segmentation hybrid approach using neutrosophic sets, fast fuzzy c-means and adaptive watershed algorithm. Artif Intell Med. 2019;97(19):105–17. doi:10.1016/j.artmed.2018.11.007. [Google Scholar] [PubMed] [CrossRef]
15. Nasim N, Amir HF, Yen WC. Integration of a knowledge-based constraint into generative models with applications in semi-automatic segmentation of liver tumors. Biomed Signal Process Control. 2020;57:1–11. [Google Scholar]
16. Jia YZ, Damon WKW, Feng D, Sudhakar K, Venkatesh M, Qi T, et al. Liver tumour segmentation using contrast-enhanced multi-detector CT data performance benchmarking of three semiautomated methods. Eur Radiol. 2010;20(7):1738–48. doi:10.1007/s00330-010-1712-z. [Google Scholar] [PubMed] [CrossRef]
17. Wang JK, Cheng YZ, Guo CY, Wang YD, Tamura S. Shape intensity prior level set combining probabilistic atlas and probability map constrains for automatic liver segmentation from abdominal CT images. Int J Comput Assist Radiol Surg. 2016;11(5):817–26. doi:10.1007/s11548-015-1332-9. [Google Scholar] [PubMed] [CrossRef]
18. Shi C, Cheng Y, Liu F, Wang Y, Bai J, Tamura S, et al. A hierarchical local region-based sparse shape composition for liver segmentation in CT scans. Pattern Recognit. 2016;50(2):88–106. doi:10.1016/j.patcog.2015.09.001. [Google Scholar] [CrossRef]
19. Marius GL, William JR, Jeremy MW, Vivek P, Ronald MS. Liver and tumor segmentation and analysis from CT of diseased patients via a generic affine invariant shape parameterization and graph cuts. 1st ed. Berlin, Heidelberg: Springer; 2012. [Google Scholar]
20. Umit B, Guo YH, Erkan T, Abdulkadir Ş. Cascaded deep convolutional encoder-decoder neural networks for efficient liver tumor segmentation. Med Hypotheses. 2019;134:1–20. [Google Scholar]
21. Li C, Wang X, Eberl S, Fulham M, Yin Y, Chen J, et al. A likelihood and local constraint level set model for liver tumor segmentation from CT volumes. IEEE Trans Biomed Eng. 2013;60(10):2967–77. doi:10.1109/TBME.2013.2267212. [Google Scholar] [PubMed] [CrossRef]
22. Zhang Y, Jiang B, Wu J, Ji D, Liu Y, Chen Y, et al. Deep learning initialized and gradient enhanced level-set based segmentation for liver tumor from CT images. IEEE Access. 2020;8:76056–68. doi:10.1109/ACCESS.2020.2988647. [Google Scholar] [CrossRef]
23. Lu F, Wu F, Hu P, Peng Z, Kong D. Automatic 3D liver location and segmentation via convolutional neural network and graph cut. Int J Comput Assist Radiol Surg. 2016;12(2):171–82. [Google Scholar] [PubMed]
24. Chi Y, Liu J, Venkatesh SK, Huang S, Zhou J, Tian Q, et al. Segmentation of liver vasculature from contrast enhanced CT images using context-based voting. IEEE Trans Biomed Eng. 2010;58(2):2144–53. [Google Scholar]
25. Avi BC, Idit D, Eyal K, Michal A, Hayit G. Fully convolutional network for liver segmentation and lesions detection. 1st ed. Cham: Springer; 2016. [Google Scholar]
26. Wu CC, Lee WL, Chen YC, Hsieh KS. Evolution-based hierarchical feature fusion for ultrasonic liver tissue characterization. IEEE J Biomed Health Inform. 2013;17(5):967–76. doi:10.1109/JBHI.2013.2261819. [Google Scholar] [PubMed] [CrossRef]
27. Shyam L, Devikalyan D, Kumar A, Anirudh K, Aman K, Jyoti K. NucleiSegNet robust deep learning architecture for the nuclei segmentation of liver cancer histopathology images. Comput Biol Med. 2020;128:1–16. [Google Scholar]
28. Xian G. An identification method of malignant and benign liver tumors from ultrasonography based on GLCM texture features and fuzzy SVM. Expert Syst Appl. 2010;37(10):6737–41. doi:10.1016/j.eswa.2010.02.067. [Google Scholar] [CrossRef]
29. Ramin R, Soroush BS. Automated liver and tumor segmentation based on concave and convex points using fuzzy C-means and mean shift clustering. Measurement. 2019;150:1–39. [Google Scholar]
30. Sho T, Elco O, Akinobu S, Hidefumi W, Shigeru N. A conditional statistical shape model with integrated error estimation of the conditions application to liver segmentation in non-contrast CT images. Med Image Anal. 2014;18(1):130–43. doi:10.1016/j.media.2013.10.003. [Google Scholar] [PubMed] [CrossRef]
31. Laszlo R, Adam P. Automated liver lesion detection in CT images based on multi-level geometric features. Int J Comput Assist Radiol Surg. 2013;9(4):577–93. [Google Scholar]
32. Chin CC, Hong HC, Yeun CC, Ming YY, Chung ML, Wei CK, et al. Computer-aided diagnosis of liver tumors on computed tomography images. Comput Methods Programs Biomed. 2017;145:45–51. doi:10.1016/j.cmpb.2017.04.008. [Google Scholar] [PubMed] [CrossRef]
33. Assaf H, Christopher FB, Guilherme MC, Elhamy H, Claude BS, Sandy N, et al. Adaptive local window for level set segmentation of CT and MRI liver lesions. Med Image Anal. 2017;37(3):46–55. doi:10.1016/j.media.2017.01.002. [Google Scholar] [PubMed] [CrossRef]
34. Xuehu W, Yongchang Z, Lan G, Xuan W, Xinting S, Xiangfeng K, et al. Liver segmentation from CT images using a sparse priori statistical shape model (SP-SSM). PLoS One. 2017;12(10):1–23. [Google Scholar]
35. Marius GL, William JR, Jianfei L, Jeremy MW, Vivek P, Shijun W, et al. Tumor burden analysis on computed tomography by automated liver and tumor segmentation. IEEE Trans Med Imaging. 2012;31(10):1965–76. doi:10.1109/TMI.2012.2211887. [Google Scholar] [PubMed] [CrossRef]
36. Amitha R, Jayasree M. Automated liver tumor detection using Markov random field segmentation. Procedia Technol. 2016;24(10):1305–10. doi:10.1016/j.protcy.2016.05.126. [Google Scholar] [CrossRef]
37. Amita D, Priti DS, Panda S, Sukanta S. Detection of liver cancer using modified fuzzy clustering and decision tree classifier in CT images. Pattern Recognit Image Anal. 2019;29(2):201–11. doi:10.1134/S1054661819020056. [Google Scholar] [CrossRef]
38. Weimin H, Yongzhong Y, Zhiping L, Guang BH, Jiayin Z, Yuping D, et al. Random feature subspace ensemble based extreme learning machine for liver tumor detection and segmentation. In: 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 2014 Aug 26–30; Chicago, IL, USA. [Google Scholar]
39. Huang WM, Li N, Lin ZP, Huang GB, Zong WW, Zhou YP. Liver tumor detection and segmentation using kernel-based extreme learning machine. In: 35th Annual International Conference of the IEEE EMBS, 2013 Jul 3–7; Osaka, Japan. [Google Scholar]
40. Banafsheh AH, Rahil H. An improved fuzzy differential evolution approach applied to classification of tumors in liver CT scan images. Medic Biol Eng Comput. 2019;57(10):2277–87. doi:10.1007/s11517-019-02009-7. [Google Scholar] [PubMed] [CrossRef]
41. Eleftherios T, Georgios CM, Katerina N, Konstantinos D, Manos C, Antonios D, et al. Extending 2-D convolutional neural networks to 3-D for advancing deep learning cancer classification with application to MRI liver tumor differentiation. IEEE J Biomed Health Inform. 2019;23(3):923–30. doi:10.1109/JBHI.6221020. [Google Scholar] [CrossRef]
42. Dong X, Zhou Y, Wang L, Peng J, Fan Y. Liver cancer detection using hybridized fully convolutional neural network based on deep learning framework. IEEE Access. 2019;(8):129889–98. [Google Scholar]
43. Sun C, Xu A, Liu D, Xiong Z, Ding W. Deep learning-based classification of liver cancer histopathology images using only global labels. IEEE J Biomed Health Inform. 2019;24(6):1643–51. [Google Scholar] [PubMed]
44. Lakshmi PB, Jayanthi K, Biju P, Ramkumar G. GoogLeNet based ensemble FCNet classifier for focal liver lesion diagnosis. IEEE J Biomed Health Inform. 2020;24(6):1686–94. doi:10.1109/JBHI.2019.2942774. [Google Scholar] [PubMed] [CrossRef]
45. Refael V, Leo J, Naama LC, Ariel E, Jacob S. Patient-specific and global convolutional neural networks for robust automatic liver tumor delineation in follow-up CT studies. Med Biol Eng Comput. 2018;56(9):1699–713. doi:10.1007/s11517-018-1803-6. [Google Scholar] [PubMed] [CrossRef]
46. Deepti M, Vinod K, Suresh CS, Niranjan K, Naveen K. Neural network based focal liver lesion diagnosis using ultrasound images. Comput Med Imaging Graph. 2011;35(4):315–23. doi:10.1016/j.compmedimag.2011.01.007. [Google Scholar] [PubMed] [CrossRef]
47. Mala K, Sadasivam V, Alagappan S. Neural network based texture analysis of CT images for fatty and cirrhosis liver classification. Appl Soft Comput. 2015;32:80–6. doi:10.1016/j.asoc.2015.02.034. [Google Scholar] [CrossRef]
48. Dirk S, Dirk L, Bert S, Bart DD, Dirk V, Paul S. Semi automatic level set segmentation of liver tumors combining a spiral scanning technique with supervised fuzzy pixel classification. Med Image Anal. 2010;14(1):13–20. doi:10.1016/j.media.2009.09.002. [Google Scholar] [PubMed] [CrossRef]
49. Sun C, Guo S, Zhang H, Li J, Chen M, Ma S, et al. Automatic segmentation of liver tumors from multiphase contrast-enhanced CT images based on FCNs. Artif Intell Med. 2017;83(5):58–66. doi:10.1016/j.artmed.2017.03.008. [Google Scholar] [PubMed] [CrossRef]
50. Kumar SS, Moni RS, Rajeesh J. An automatic computer-aided diagnosis system for liver tumours on computed tomography images. Comput Elect Eng. 2013;39(5):1516–26. doi:10.1016/j.compeleceng.2013.02.008. [Google Scholar] [CrossRef]
51. Gaurav S, Saini BS, Dilbag S. Segmentation of cancerous regions in liver using an edge based and phase congruent region enhancement method. Comput Electr Eng. 2015;53:244–62. [Google Scholar]
52. Patrick FC, Mohamed E, Elshaer A, Florian E, Sunil T, Marc B, et al. Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. Cham: Springer; 2016. [Google Scholar]
53. Sangman K, Jusung P. Hybrid feature selection method based on neural networks and cross-validation for liver cancer with microarray. IEEE Access. 2018;6:78214–24. doi:10.1109/ACCESS.2018.2884896. [Google Scholar] [CrossRef]
54. Bai Z, Jiang H, Li S, Yao YD. Liver tumor segmentation based on multi-scale candidate generation and fractal residual network. IEEE Access. 2019;7:82122–33. doi:10.1109/ACCESS.2019.2923218. [Google Scholar] [CrossRef]
55. Xiao H. Automatic liver lesion segmentation using a deep convolutional neural network method. Med Phys. arXiv:1704.07239. 2017. [Google Scholar]
56. Masuda Y, Tateyama T, Xiong W, Zhou J, Wakamiya M, Kanasaki SM, et al. Liver tumor detection in CT images by adaptive contrast enhancement and the Em/Mpm algorithm. In: 18th IEEE International Conference on Image Processing, 2011 Sep 11–14; Brussels, Belgium. [Google Scholar]
57. Chung M, Lee J, Lee M, Lee J, Shin YG. Deeply self-supervised contour embedded neural network applied to liver segmentation. Comput Methods Programs Biomed. 2020;192(4):1–11. doi:10.1016/j.cmpb.2020.105447. [Google Scholar] [PubMed] [CrossRef]
58. Hussein A, Amr A. Computer-aided classification of liver lesions from CT images based on multiple ROI. Procedia Comput Sci. 2016;90:80–6. doi:10.1016/j.procs.2016.07.027. [Google Scholar] [CrossRef]
59. Lei B, Jinman K, Ashnil K, Dagan F. Automatic liver lesion detection using cascaded deep residual networks. arXiv:1704.02703. 2017. [Google Scholar]
60. Maayan FA, Idit D, Eyal K, Michal A, Jacob G, Hayit G. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing. 2018;321(5):321–31. doi:10.1016/j.neucom.2018.09.013. [Google Scholar] [CrossRef]
61. Liang D, Lin L, Chen X, Hu H, Zhang Q, Che Q, et al. Multi-stream scale insensitive convolutional and recurrent neural networks for liver tumor detection in dynamic CT images. In: IEEE International Conference on Image Processing (ICIP), 2019 Sep 22–25; Taipei, Taiwan. [Google Scholar]
62. Wang J, Xu Z, Pang ZF, Huo Z, Luo J. Tumor detection for whole slide image of liver based on patch-based convolutional neural network. Multimed Tools Appl. 2020;80:17429–40. [Google Scholar]
63. Hong Y, Mao XW, Hui QL, Ouyang X, Peng Z, Kong D. Automatic liver and tumor segmentation based on deep learning and globally optimized refinement. Appl Math-J Chin Univ. 2021;36(2):304–16. doi:10.1007/s11766-021-4376-3. [Google Scholar] [CrossRef]
64. Charlie AH, Clinton JW, Lynn JS, Marc F, Isabel S, Todd S, et al. Deep learning for liver tumor diagnosis part I development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29(7):3338–47. doi:10.1007/s00330-019-06205-9. [Google Scholar] [PubMed] [CrossRef]
65. Rajendra AU, Joel EWH, Yuki H, Jen HT, Arkadiusz G, Anushya V, et al. Automated diagnosis of focal liver lesions using bidirectional empirical mode decomposition features. Comput Biol Med. 2018;94(7):11–8. doi:10.1016/j.compbiomed.2017.12.024. [Google Scholar] [PubMed] [CrossRef]
66. Yoo N, Ju HL, Ga YK, Yuan YJ, Sung MK. Classification of focal liver lesions on ultrasound images by extracting hybrid textural features and using an artificial neural network. Biomed Mater Eng. 2015;26(1):1599–611. [Google Scholar]
67. Anirudh AA, Kumar A, Shyam L, Jyoti KPU, Prakash S. LiverNet efficient and robust deep learning model for automatic diagnosis of sub-types of liver hepatocellular carcinoma cancer from H&E stained liver histopathology images. Int J Comput Assist Radiol Surg. 2021;16(9):1549–63. doi:10.1007/s11548-021-02410-4. [Google Scholar] [PubMed] [CrossRef]
68. Liang C, George P, Iasonas K, Kevin M, Alan LY. DeepLab semantic image segmentation with deep convolutional nets, atrous convolution and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2018;40(4):834–48. doi:10.1109/TPAMI.2017.2699184. [Google Scholar] [PubMed] [CrossRef]
69. Simranjeet R, Abeer A, Prasad PWC, Thair AD, Ahmed D, Ahmad A. Deep learning for liver tumour classification enhanced loss function. Multimed Tools Appl. 2020;80:4729–50. [Google Scholar]
70. Omar IA, Ashrani AAR, Ehsan G. An automated liver tumour segmentation from abdominal CT scans for hepatic surgical planning. Int J Comput Assist Radiol Surg. 2018;13(8):1169–76. doi:10.1007/s11548-018-1801-z. [Google Scholar] [PubMed] [CrossRef]
71. Deepika R, Swathi K, Mythilee KL. “Liver tumor detection using fast fuzzy C-Means clustering”. In: 2022 1st International Conference on Computational Science and Technology (ICCST), 2022; Chennai, India; p. 1–5. [Google Scholar]
72. Pathak K, Singh DK. Convolutional neural networks based liver tumor classification. In: 2022 International Conference on Augmented Intelligence and Sustainable Systems (ICAISS), 2022; Trichy, India; p. 171–7. [Google Scholar]
73. Ramgopal NC, Gantela R, Thankam T, SenthamilSelvan R. Automatic liver cancer detection in abdominal liver images using soft optimization techniques. In: 2022 International Conference on Knowledge Engineering and Communication Systems (ICKES), 2022; Chickballapur, India; p. 1–5. [Google Scholar]
74. Zheng R, Wang Q, Lv S, Li C, Wang C, Chen W, et al. Automatic liver tumor segmentation on dynamic contrast enhanced MRI using 4D information: deep learning model based on 3D convolution and convolutional LSTM. IEEE Trans Med Imaging. 2022;41(10):2965–76. doi:10.1109/TMI.2022.3175461. [Google Scholar] [PubMed] [CrossRef]
75. Kiruthiga R, Abbas MAS, Ashok KM, Raj RA, Balaji N. Gradient-driven texture-normalized liver tumor detection using deep learning. In: 2022 International Conference on Power, Energy, Control and Transmission Systems (ICPECTS), 2022; Chennai, India; p. 1–5. [Google Scholar]
Jain RK, Sato T, Watasue T, Nakagawa T, Iwamoto Y, Han X, et al. Unsupervised domain adaptation using adversarial learning and maximum square loss for liver tumors detection in multi-phase CT images. In: 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 2022 Jul 11–15; Glasgow, UK; IEEE; p. 1536–9. [Google Scholar]
77. Sridhar K, Kavitha C, Wen CL, Balasubramanian PK. Detection of liver tumour using deep learning based segmentation with coot extreme learning model. Biomedicines. 2023;11(3):800. [Google Scholar]
78. Tosaki T, Yamakawa M, Shiina T. A study on the optimal condition of ground truth area for liver tumor detection in ultrasound images using deep learning. J Med Ultrasonics. 2023;50(2):167–76. doi:10.1007/s10396-023-01301-2. [Google Scholar] [PubMed] [CrossRef]
79. Geetha C, Arunachalam AR. Prediction of parameters of liver tumor using feature extraction and supervised function. Measurem Sens. 2022;22:1000386. [Google Scholar]
80. Eigo H, Keisuke D, Yoshito M, Ni Ni, Masatoshi K. Liver tumor detection and classification from abdominal ultrasound images with CenterNet using contrastive learning. In: International Workshop on Advanced Imaging Technology (IWAIT), 2023, Jan 9–11; Jeju, Republic of Korea. doi:10.1117/12.2662969. [Google Scholar] [CrossRef]
81. Chi J, Han X, Wu C, Wang H, Ji P. X-Net: multi-branch UNet-like network for liver and tumor segmentation from 3D abdominal CT scans. Neurocomputing. 2021;459:81–96. doi:10.1016/j.neucom.2021.06.021. [Google Scholar] [CrossRef]
82. Rahman H, Tanvir F, Naik B, Azhar I, Junaid T, Shanshan T, et al. A deep learning approach for liver and tumor segmentation in CT images using ResUNet. Bioengineering. 2022;9(8):368. doi:10.3390/bioengineering9080368. [Google Scholar] [PubMed] [CrossRef]
83. Ayalew Y, Yodit A, Kinde AF, Mohammed AM. Modified U-Net for liver cancer segmentation from computed tomography images with a new class balancing method. BMC Biomed Eng. 2021;3:1–13. [Google Scholar]
84. Sabir MW, Zia K, Naufal MS, Danish MK, Mahmoud A, Khasawneh A, et al. Segmentation of liver tumor in CT scan using ResU-Net. Appl Sci. 2022;12(17):8650. doi:10.3390/app12178650. [Google Scholar] [CrossRef]
85. Jensen CT, Gupta S, Saleh MM, Liu X, Wong VK, Salem U, et al. Reduced-dose deep learning reconstruction for abdominal CT of liver metastases. Radiology. 2022;303(1):90–8. doi:10.1148/radiol.211838. [Google Scholar] [PubMed] [CrossRef]
86. Almotairi S, Ghada K, Mohamed A, Badr A, Mohammed AMS. Liver tumor segmentation in CT scans using modified SegNet. Sensors. 2020;20(5):1516. doi:10.3390/s20051516. [Google Scholar] [PubMed] [CrossRef]
87. Altini N, Berardino P, Giacomo DC, Antonio B, Gioacchino B, Vito T, et al. Liver, kidney and spleen segmentation from CT scans and MRI with deep learning: a survey. Neurocomputing. 2022;490:30–53. doi:10.1016/j.neucom.2021.08.157. [Google Scholar] [CrossRef]
88. Gul S, Muhammad SK, Asima B, Amith K, Mohamed AA, Muhammad EHC. Deep learning techniques for liver and liver tumor segmentation: a review. Comput Biol Med. 2022;147:105620. doi:10.1016/j.compbiomed.2022.105620. [Google Scholar] [PubMed] [CrossRef]
89. Araújo J, Denes L, Luana B, Cruz D, João O, Bandeira D, et al. Liver segmentation from computed tomography images using cascade deep learning. Comput Biol Med. 2022;140:105095. doi:10.1016/j.compbiomed.2021.105095. [Google Scholar] [PubMed] [CrossRef]
90. Ichikawa Y, Yoshinori K, Akio Y, Naoki N, Motonori N, Masaki I, et al. Deep learning image reconstruction for improvement of image quality of abdominal computed tomography: comparison with hybrid iterative reconstruction. Jpn J Radiol. 2021;39:598–604. doi:10.1007/s11604-021-01089-6. [Google Scholar] [PubMed] [CrossRef]
91. Bhayana R, Avik S, Matthew DL, Denston EC, Mark A, Anderson M, et al. Abdominal imaging findings in COVID-19: preliminary observations. Radiology. 2020;297(1):E207–15. doi:10.1148/radiol.2020201908. [Google Scholar] [PubMed] [CrossRef]
92. Elayan A, Hamzeh B, Moath B, Ahmad S, Sufian AH, Ahmad HE. Primary hepatic neuroendocrine tumor: a case report and literature review. Cureus. 2022;14(2):e22370. [Google Scholar]
93. Ansari MY, Yin Y, Shidin B, Julien A, Abdulla AA, Mohamed W, et al. A lightweight neural network with multiscale feature enhancement for liver CT segmentation. Sci Rep. 2022;12(1):14153. doi:10.1038/s41598-022-16828-6. [Google Scholar] [PubMed] [CrossRef]
94. Li J, Liu K, Hu Y, Zhang H, Heidari AA, Chen H, et al. Eres-UNet++: liver CT image segmentation based on high-efficiency channel attention and Res-UNet++. Comput Biol Med. 2023;158:106501. doi:10.1016/j.compbiomed.2022.106501. [Google Scholar] [PubMed] [CrossRef]
95. Starekova J, Diego H, Perry JP, Scott BR. Quantification of liver fat content with CT and MRI: state of the art. Radiology. 2021;301(2):250–62. doi:10.1148/radiol.2021204288. [Google Scholar] [PubMed] [CrossRef]
96. Liu T, Liu J, Ma Y, He J, Han J, Ding X, et al. Spatial feature fusion convolutional network for liver and liver tumor segmentation from CT images. Med Phys. 2021;48(1):264–72. doi:10.1002/mp.14585. [Google Scholar] [PubMed] [CrossRef]
97. Liu J, Zhang Y, Chen JN, Xiao J, Lu Y, Landman BA, et al. Clip-driven universal model for organ segmentation and tumor detection. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, 2023 Oct 1–6; Paris, France; p. 21152–64. [Google Scholar]
98. Higashigaito K, André E, Matthias E, Thomas GF, Bernhard S, Hatem A. Contrast-enhanced abdominal CT with clinical photon-counting detector CT: assessment of image quality and comparison with energy-integrating detector CT. Acad Radiol. 2022;29(5):689–97. doi:10.1016/j.acra.2021.06.018. [Google Scholar] [PubMed] [CrossRef]
99. Hotta M, Angela CR, Mahbod GJ, Nandakumar M, Andrea F, Matthias RB, et al. Non-oncologic incidental uptake on FAPI PET/CT imaging. British J Radiol. 2023;96(1142):20220463. doi:10.1259/bjr.20220463. [Google Scholar] [PubMed] [CrossRef]
100. Zan Q, Fan L, Ma L, Yang Q, Zhao K, Huang Y, et al. Dual-channel fluorescent probe for simultaneously detecting H2S and viscosity/polarity and its application in non-alcoholic fatty liver, tumor tissue, and food spoilage. Sens Actuat B: Chem. 2023;(397):134596. doi:10.1016/j.snb.2023.134596. [Google Scholar] [CrossRef]
101. Zhou J, Sun H, Wang Z, Cong W, Zeng M, Zhou W, et al. Guidelines for the diagnosis and treatment of primary liver cancer. Liver Cancer. 2023;12(5):405–44. doi:10.1159/000530495. [Google Scholar] [PubMed] [CrossRef]
102. Aruna S, Saranya A, Guru PD, Kavya SP, Piyush KP. Machine learning approach for detecting liver tumours in CT images using the gray level co-occurrence metrix. In: 2023 International Conference on Applied Intelligence and Sustainable Computing (ICAISC), 2023 Jun 16–17; Dharwad, India; IEEE; p. 1–5. [Google Scholar]
103. Hu C, Xia T, Cui Y, Zou Q, Wang Y, Xiao W, et al. Trustworthy multi-phase liver tumor segmentation via evidence-based uncertainty. Eng Appl Artif Intell. 2024;133:108289. doi:10.1016/j.engappai.2024.108289. [Google Scholar] [CrossRef]
104. Luo L, Wang T, Cheng M, Ge X, Song S, Zhu G, et al. Rare benign liver tumors that require differentiation from hepatocellular carcinoma: focus on diagnosis and treatment. J Cancer Res Clin Oncol. 2023;149(7):2843–54. doi:10.1007/s00432-022-04169-w. [Google Scholar] [PubMed] [CrossRef]
105. Bakrania A, Joshi N, Zhao X, Zheng G, Bhat M. Artificial intelligence in liver cancers: decoding the impact of machine learning models in clinical diagnosis of primary liver cancers and liver cancer metastases. Pharmacol Res. 2023;189:106706. doi:10.1016/j.phrs.2023.106706. [Google Scholar] [PubMed] [CrossRef]
106. Chen X, Chang Y, Wu J, Xu J, Zhao H, Nie Z, et al. Outcomes of radiofrequency ablation for liver tumors in patients on hemodialysis: results from the US Nationwide Inpatient Sample 2005–2020. Eur J Radiol. 2024;178:111640. doi:10.1016/j.ejrad.2024.111640. [Google Scholar] [PubMed] [CrossRef]
107. Bilic P, Christ P, Li HB, Vorontsov E, Ben-Cohen A, Kaissis G, et al. The liver tumor segmentation benchmark (LiTS). Med Image Anal. 2023;84:102680. doi:10.1016/j.media.2022.102680. [Google Scholar] [PubMed] [CrossRef]
108. Khan RA, Fu M, Burbridge B, Luo Y, Wu FX. A multi-modal deep neural network for multi-class liver cancer diagnosis. Neural Netw. 2023;165:553–61. doi:10.1016/j.neunet.2023.06.013. [Google Scholar] [PubMed] [CrossRef]
109. Wang X, Wang S, Zhang Z, Yin X, Wang T, Li N. CPAD-Net: contextual parallel attention and dilated network for liver tumor segmentation. Biomed Signal Process Control. 2023;79:104258. doi:10.1016/j.bspc.2022.104258. [Google Scholar] [CrossRef]
110. Chen J, Niu C, Yang N, Liu C, Zou S, Zhu S. Biomarker discovery and application—an opportunity to resolve the challenge of liver cancer diagnosis and treatment. Pharmacol Res. 2023;189:106674. doi:10.1016/j.phrs.2023.106674. [Google Scholar] [PubMed] [CrossRef]
111. Haverkamp T, Olivia B, Thomas K, Carolin M, Wilko W, Thomas S, et al. Heterogeneous molecular behavior in liver tumors (HCC and CCA) of two patients with acute intermittent porphyria. J Cancer Res Clin Oncol. 2023;149(6):2647–55. doi:10.1007/s00432-022-04384-5. [Google Scholar] [PubMed] [CrossRef]
112. Zeng S, Wang Y, Chen C, Kim H, Liu X, Jiang M, et al. An ER-targeted, viscosity-sensitive hemicyanine dye for the diagnosis of nonalcoholic fatty liver and photodynamic cancer therapy by activating pyroptosis pathway. Angew Chem. 2024;136(9):e202316487. doi:10.1002/ange.v136.9. [Google Scholar] [CrossRef]
113. Sawhney G, Aditya RB, Kumar S, Diotima B, Munendra S, Daljeet SD, et al. Nanotechnology at the forefront of liver cancer diagnosis. In: Nanophototherapy. 2025. p. 575–93. [Google Scholar]
114. Jung EM, Yi D, Friedrich J. Current aspects of multi-modal ultrasound liver diagnostics using contrast-enhanced ultrasonography (CEUSfat evaluation, fibrosis assessment, and perfusion analysis–an update. Clin Hemorheol Microcirc. 2023;83(2):181–93. doi:10.3233/CH-239100. [Google Scholar] [PubMed] [CrossRef]
115. Deng Y, Min Z, Shi YL. Isolated liver gastrointestinal stromal tumor: a case report. J Clin Ultrasound. 2024;52(8):1188–92 [Google Scholar]
116. Mauro E, Joana FF, Tamara S, Alexandre S, Amparo C, Marta B, et al. New challenges in the management of cholangiocarcinoma: the role of liver transplantation, locoregional therapies, and systemic therapy. Cancers. 2023;15(4):1244. doi:10.3390/cancers15041244. [Google Scholar] [PubMed] [CrossRef]
117. Omar MA, Mohamed MO, Khaled F, Ashraf AT, Yasser ES, Tarek ME, et al. Biomarkers for hepatocellular carcinoma: from origin to clinical diagnosis. Biomedicines. 2023;11(7):1852. doi:10.3390/biomedicines11071852. [Google Scholar] [PubMed] [CrossRef]
118. Takahashi Y, Erdenetsogt D, Hiroyuki K, Toshio F. Pathology and pathogenesis of metabolic dysfunction-associated steatotic liver disease-associated hepatic tumors. Biomedicines. 2023;11(10):2761. doi:10.3390/biomedicines11102761. [Google Scholar] [PubMed] [CrossRef]
119. Souza D, Melroy A, Mahesh G, Shraddha P. Liver cancer. In: Tata memorial centre textbook of oncology. Singapore: Springer Nature Singapore; 2024. p. 535–47. [Google Scholar]
120. Albrecht T, Annik R, Jana DA, Jan PN, Beate KS, Tiemo SG, et al. Deep learning-enabled diagnosis of liver adenocarcinoma. Gastroenterology. 2023;165(5):1262–75. doi:10.1053/j.gastro.2023.07.026. [Google Scholar] [PubMed] [CrossRef]
121. Yang L, Tian Y, Cao X, Wang J, Luo B. Identification of novel diagnostic biomarkers associated with liver metastasis in colon adenocarcinoma by machine learning. Discover Oncology. 2024;15(1):1–15. [Google Scholar]
122. Vaz J, Peter J, Strömberg U, Patrik M, Berne E, David B. Metabolic dysfunction-associated steatotic liver disease has become the most common cause of hepatocellular carcinoma in Sweden: a nationwide cohort study. Int J Cancer. 2024;156(1):40–51. [Google Scholar]
123. Houssein EH, Diego O, Nagwan AS, Noha FM, Marwa ME. Liver cancer algorithm: a novel bio-inspired optimizer. Comput Biol Med. 2023;165:107389. doi:10.1016/j.compbiomed.2023.107389. [Google Scholar] [PubMed] [CrossRef]
124. Guan MC, Shi YZ, Qian D, Na L, Ting TF, Gui XZ, et al. The performance of GALAD score for diagnosing hepatocellular carcinoma in patients with chronic liver diseases: a systematic review and meta-analysis. J Clin Med. 2023;12(3):949. doi:10.3390/jcm12030949. [Google Scholar] [PubMed] [CrossRef]
125. Khan SU, Ibrar MK, Munir UK, Muhammad AUD, Muhammad ZK, Nazir MK, et al. Role of LGMN in tumor development and its progression and connection with the tumor microenvironment. Front Mol Biosci. 2023;10:1121964. doi:10.3389/fmolb.2023.1121964. [Google Scholar] [PubMed] [CrossRef]
126. Wu B, Gilbert M. Application of digital pathology and machine learning in the liver, kidney and lung diseases. J Pathol Inf. 2023;14:100184. doi:10.1016/j.jpi.2022.100184. [Google Scholar] [PubMed] [CrossRef]
127. Radak M, Haider YL, Hossein M. Machine learning and deep learning techniques for breast cancer diagnosis and classification: a comprehensive review of medical imaging studies. J Cancer Res Clin Oncol. 2023;149(12):10473–91. doi:10.1007/s00432-023-04956-z. [Google Scholar] [PubMed] [CrossRef]
128. Saeedi S, Sorayya R, Hamidreza K, Sharareh R, Niakan K. MRI-based brain tumor detection using convolutional deep learning methods and chosen machine learning techniques. BMC Med Inform Decis Mak. 2023;23(1):16. doi:10.1186/s12911-023-02114-6. [Google Scholar] [PubMed] [CrossRef]
129. Joshi AA, Rabia MA. Deep learning approach for brain tumor classification using metaheuristic optimization with gene expression data. Int J Imaging Syst Technol. 2024;34(2):e23007. doi:10.1002/ima.v34.2. [Google Scholar] [CrossRef]
130. Rai HM. Cancer detection and segmentation using machine learning and deep learning techniques: a review. Multimed Tools Appl. 2024;83(9):27001–35. [Google Scholar]
131. Bouamrane A, Makhlouf D. Enhancing lung cancer detection and classification using machine learning and deep learning techniques: a comparative study. In: 2023 International Conference on Networking and Advanced Systems (ICNAS), 2023 Oct 21–23; Algiers, Algeria; IEEE; p. 1–6. [Google Scholar]
132. Sarkar S, Kong M, Waleed I, Ryan WT, Irbaz BR, Alvin CS, et al. Performing automatic identification and staging of urothelial carcinoma in bladder cancer patients using a hybrid deep-machine learning approach. Cancers. 2023;15(6):1673. doi:10.3390/cancers15061673. [Google Scholar] [PubMed] [CrossRef]
133. Mohanty BC, Subudhi PK, Ratnakar D, Bidyadhar M. Feature-enhanced deep learning technique with soft attention for MRI-based brain tumor classification. Int J Inform Technol. 2024;16(3):1617–26. [Google Scholar]
134. Khandakar S, Mohd AAM, Islam MM, Kaosar H, Melon MMM, Javed MS. Unveiling early detection and prevention of cancer: machine learning and deep learning approaches. Educ Adm Theory Pract. 2024;30(5):14614–28. [Google Scholar]
135. Sahu A, Pradeep KD, Sukadev M. Recent advancements in machine learning and deep learning-based breast cancer detection using mammograms. Phys Med. 2023;114:103138. doi:10.1016/j.ejmp.2023.103138. [Google Scholar] [PubMed] [CrossRef]
136. Jose P, Shanmugasundaram H, Vimaladevi M, Sujaudeen N, Murugaperumal K, Chekuri AK. Enhanced QSVM with elitist non-dominated sorting genetic optimisation algorithm for breast cancer diagnosis. IET Quantum Commun. 2024 Oct 23;23(3):31. doi:10.1049/qtc2.12113. [Google Scholar] [CrossRef]
137. Asif S, Zhao M, Tang F, Zhu Y. An enhanced deep learning method for multi-class brain tumor classification using deep transfer learning. Multimed Tools Appl. 2023;82(20):31709–36. doi:10.1007/s11042-023-14828-w. [Google Scholar] [CrossRef]
138. Liu X, Wang Z. Deep learning in medical image classification from mri-based brain tumor images. arXiv:2408.00636. 2024. [Google Scholar]
139. Zebari NA, Ahmed AHA, Ridwan BM, Merdin SS. Enhancing brain tumor classification with data augmentation and DenseNet121. Acad J Nawroz Univ. 2023;12(4):323–34. doi:10.25007/ajnu.v12n4a1985. [Google Scholar] [CrossRef]
140. Ullah MS, Muhammad AK, Anum M, Olfa M, Oumaima S, Nazik A. Brain tumor classification from MRI scans: a framework of hybrid deep learning model with Bayesian optimization and quantum theory-based marine predator algorithm. Front Oncol. 2024;14:1335740. doi:10.3389/fonc.2024.1335740. [Google Scholar] [PubMed] [CrossRef]
141. Yadav A, Rakesh K, Meenu G. An analysis of convolutional neural network and conventional machine learning for multiclass brain tumor detection. In: AIP Conference Proceedings, 2024; Melville, NY, USA: AIP Publishing. [Google Scholar]
142. Malik MH, Hamid G, Tahir R, Bibi M, Zhang H, Qasim U. Feature extraction-based liver tumor classification using machine learning and deep learning methods of computed tomography images. Cogent Eng. 2024;11(1):2338994. doi:10.1080/23311916.2024.2338994. [Google Scholar] [CrossRef]
143. Tejaswi VSD, Venubabu R. Liver cancer diagnosis: enhanced deep maxout model with improved feature set. Cancer Investig. 2024;42(8):710–25. doi:10.1080/07357907.2024.2391359. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

 Downloads
Downloads
 Citation Tools
Citation Tools