 Open Access
Open Access
ARTICLE
Semantic Segmentation of Lumbar Vertebrae Using Meijering U-Net (MU-Net) on Spine Magnetic Resonance Images
1 Department of Computer Science and Engineering, College of Engineering Guindy, Anna University, Chennai, 600025, India
2 Department of Information Technology, Madras Institute of Technology, Anna University, Chrompet, Chennai, 600044, India
* Corresponding Authors: Shiloah Elizabeth Darmanayagam. Email: ,
Computer Modeling in Engineering & Sciences 2025, 142(1), 733-757. https://doi.org/10.32604/cmes.2024.056424
Received 22 July 2024; Accepted 30 September 2024; Issue published 17 December 2024
Abstract
Lower back pain is one of the most common medical problems in the world and it is experienced by a huge percentage of people everywhere. Due to its ability to produce a detailed view of the soft tissues, including the spinal cord, nerves, intervertebral discs, and vertebrae, Magnetic Resonance Imaging is thought to be the most effective method for imaging the spine. The semantic segmentation of vertebrae plays a major role in the diagnostic process of lumbar diseases. It is difficult to semantically partition the vertebrae in Magnetic Resonance Images from the surrounding variety of tissues, including muscles, ligaments, and intervertebral discs. U-Net is a powerful deep-learning architecture to handle the challenges of medical image analysis tasks and achieves high segmentation accuracy. This work proposes a modified U-Net architecture namely MU-Net, consisting of the Meijering convolutional layer that incorporates the Meijering filter to perform the semantic segmentation of lumbar vertebrae L1 to L5 and sacral vertebra S1. Pseudo-colour mask images were generated and used as ground truth for training the model. The work has been carried out on 1312 images expanded from T1-weighted mid-sagittal MRI images of 515 patients in the Lumbar Spine MRI Dataset publicly available from Mendeley Data. The proposed MU-Net model for the semantic segmentation of the lumbar vertebrae gives better performance with 98.79% of pixel accuracy (PA), 98.66% of dice similarity coefficient (DSC), 97.36% of Jaccard coefficient, and 92.55% mean Intersection over Union (mean IoU) metrics using the mentioned dataset.Keywords
Artificial Intelligence (AI) is undeniably revolutionizing medical research and patient care, with its multiple applications in several fields. Machine learning and artificial intelligence have the potential to give clear interpretations to experts and to compile important features that might help clinicians in accurate diagnosis, treatment planning, and disease monitoring [1]. A branch of machine learning called deep learning focuses on teaching artificial neural networks to learn and make predictions or judgments; it draws inspiration from the design and operation of the network of interconnected neurons in the human brain. Specifically, the use of AI in Computer-Aided Diagnosis may effectively improve the diagnostic process in patients affected by Lower Back Pain (LBP) [2].
Over the last two decades, Computer Aided Diagnosis (CAD) has made remarkable strides, surpassing traditional methods that involved laborious film digitization, Central Processing Unit (CPU)-intensive computations, and a limited scope. Currently, CAD serves as a potent tool, with thoroughly assessed methodologies applied to sizable and clinically relevant databases. The evolution involves creating methodologies to assess CAD performance, validating algorithms with relevant cases for accurate measurement and robustness, executing observer studies to evaluate radiologists in diagnostic tasks, both with and without computer aid, and culminating in performance assessment through clinical trials. This progress signifies the overcoming of traditional constraints through the integration of CAD technology [3].
World Health Organisation (WHO) report on 19 June 2023 stated that in 2020, low back pain (LBP) affected 619 million people globally, and it is estimated that the number of cases will increase to 843 million cases by 2050, driven largely by population expansion and aging.
The intricate structure of the spine referred to as the vertebral column or backbone, supports, stabilises, and protects the spinal cord while enabling mobility and flexibility. It is made up of several vertebrae, which are bones, piled on top of one another. Lumbar region consists of vertebrae L1, L2, L3, L4 and L5, intervertebral discs L1-L2, L2-L3, L3-L4, L4-L5 and L5-S1 where S1 is the sacral vertebra. Lumbar vertebrae L1 to L5 are the largest and sturdiest vertebrae that bear the majority of the upper body’s weight and endure a great deal of stress and pressure. Lower back discomfort is largely caused by the lumbar vertebrae and intervertebral discs of the spine anatomy [4]. Numerous conditions, including sciatica, herniated discs, degenerative disc disease, spinal stenosis, muscular strains, sprains, or spasms, can result in lower back pain. Herniated discs occur when the cushioning discs between the vertebrae rupture or bulge out of place, putting pressure on the nerves and causing pain. Spinal stenosis occurs when the spinal canal narrows. Degenerative disc diseases occur when the discs between the vertebrae break down or wear away, leading to pain and stiffness. Osteoarthritis is a condition that affects the joints, causing pain and stiffness. Spondylolisthesis occurs when one vertebra slips out of place onto the vertebra below it, causing pain and nerve compression [5,6].
Lumbar vertebrae segmentation for Computer-aided diagnosis of lumbar diseases can be a challenging task due to the complexity of the lumbar spine anatomy and the variability of the imaging modalities used for diagnosis, such as X-ray, Computed Tomography (CT), and Magnetic Resonance Imaging (MRI) [7]. For examining the spine’s soft tissues, MRI is thought to be the most effective imaging modality. T1 and T2-weighted MR imaging are two of the most often used techniques for MR imaging. The relaxation durations T1 and T2 are used to illustrate the different types of tissues in the body [8]. One of the primary challenges is the complexity of the MRI images, which typically consist of multiple structures, such as bones, muscles, organs, and soft tissue. These structures have different shapes, sizes, and appearances, making it challenging to distinguish them from each other accurately. Another challenge is the variability in the MRI image quality, such as variations in resolution, contrast, and artifacts, which can affect the segmentation accuracy. These variations can lead to partial volume effects, blurring of boundaries, and image noise, which can make it difficult for automated segmentation algorithms to distinguish between different structures accurately. Herniated discs, spinal cord compression, and spinal tumours are just a few of the ailments that an MRI is especially helpful for detecting [9].
Segmentation plays an important role in analyzing and diagnosing Lower Back Pain. The goal of segmentation is to separate the lumbar vertebrae from the surrounding anatomical structures, creating precise boundaries for each vertebra. Segmentation of the lumbar region can be performed using traditional image processing techniques or advanced deep-learning approaches. Traditional methods of segmentation rely on algorithms, such as region growing, thresholding, or active contour models, to differentiate the lumbar vertebrae from the rest of the region. Deep learning techniques, in particular Convolutional Neural Networks (CNNs), have achieved better results in automated segmentation tasks. These techniques make use of deep neural networks’ ability to directly extract complex features and spatial correlations from the data, leading to accurate and robust segmentations [10].
Ronneberger et al. [11] first suggested the U-Net in 2015, and it has since been widely used for a variety of medical image processing applications, including segmentation. Furthermore, the U-Net has been shown to achieve state-of-the-art performance on various MRI segmentation tasks, including brain tumour segmentation, liver segmentation, and prostate segmentation. The network can be divided into two paths: one is the contracting path, and the other is an expanding path. The contracting path performs down-sampling for feature extraction, constructed the same as a convolutional neural network but followed by an expanding path that performs up-sampling for precise localization of features in the higher resolution layers. Another important aspect that makes the network so special is taking the convolution layer feature maps that are trained in the down-sampling path and concatenating them to the corresponding de-convolution layers of the up-sampling path. In the down-sampling path, the input image runs through multiple convolutional layers, adding pooling in between to down-sample and reducing the size of the image, simultaneously increasing the number of layers by doubling the number of filters of convolutional layers on each convolution block. The up-sampling path remains symmetric to the down-sampling path, turning the network into a U-shaped neural network, named “U-Net”. The process of automatically labelling or segmenting various anatomical components or regions inside the lumbar spine in medical imaging is known as semantic segmentation of the lumbar region [12].
With the rapid development of deep learning and its wide application in the field of computer vision, a series of lightweight models for semantic segmentation have been proposed, such as the UNet series, SegNet, PSPNet, the DeepLab series and other classic variants of full convolutional networks (FCNs), each of which have achieved good results [13]. The complex characteristics and spatial correlations seen in the lumbar area are intended to be learned and captured by the convolutional neural network architecture. U-Net and Fully Convolutional Networks (FCNs) are frequently utilized for semantic segmentation. These architectures have an encoder for feature extraction and a decoder for creating segmentation maps.
The U-Net architecture has several advantages for the semantic segmentation of MRI images. It is designed to handle the challenges of medical image analysis tasks, such as the presence of small structures, limited training data, and inter-patient variability. The architecture is also relatively lightweight, making it suitable for deployment on limited computational resources as compared to FCN. U-Net image segmentation method investigates the MRI image features accurately and provides better assistance for doctors in their clinical practice [14,15]. The highlights of this work are as follows:
• In the dataset preparation phase, the selective Digital Imaging and Communications in Medicine (DICOM) slices of mid-sagittal MRI images have been used to generate the mask of the region of interest to localize and label it, effectively.
• The proposed MU-Net architecture consists of a modified convolutional layer that incorporates the Meijering filter for the semantic segmentation of vertebrae L1, L2, L3, L4, L5, and S1.
• Generated pseudo color mask images along with the corresponding slices of MRI have been used to train the network to detect the foreground of the lumbar vertebral column in the testing phase and to localize it by labelling the individual lumbar and sacral vertebrae.
• The performance of the work is tested against standard performance measures for semantic segmentation namely, pixel accuracy, dice similarity coefficient, Jaccard coefficient, and mean IoU. The obtained results are compared with the existing approaches and the efficiency of the proposed semantic segmentation system is proven.
The remainder of this paper has been organized according to the arrangement that is detailed below. The relevant research on semantic segmentation for the effective diagnosis of lumbar diseases is examined in Section 2. The computer-aided detection system that has been proposed for the identification, categorization, and localization of vertebrae for diagnosing lower back pain is discussed in Section 3. The performance of the proposed work is demonstrated in Section 4, and the study is brought to a close in Section 5.
This section studies and discusses the recent works on segmentation of the lumbar portion, labelling the vertebrae, and other forms of analysis. Mahdy et al. in [16] have proposed an automatic detection system that starts by visualizing the CT images in Digital Imaging and Communications in Medicine (DICOM) format in three views and then identifies regions for the lumbar or sacral. An adaptive threshold and modified region-growing technique have been used to separate the spine from the surrounding tissues, organs, and bones. The k-means clustering algorithm has been used to segment each vertebra in the spine. The intervertebral distance has been calculated to automatically detect the degenerative disc in the lumbar area. The proposed system has been evaluated on ten 3D Computed Tomography (CT) images downloaded from the Cancer Imaging Archive (TCIA). Zhang et al. in [17] have proposed the BN-U-Net network incorporates a specification layer, known as the Batch Normalization layer which allows for batch normalization of data in a specific layer of the convolutional network, thereby enhancing the performance of the U-Net network. The proposed model exhibits notable advancements in segmentation accuracy, sensitivity, and specificity for spinal MRI images compared to the FCN and U-Net algorithms. This improvement contributes to enhancing the overall diagnostic precision of spinal-related diseases in MRI images. Lu et al. in [18] have proposed a three-dimensional XUnet technique to accomplish automatic lumbar vertebrae segmentation. The public dataset VerSe 2020’s lumbar spine CT images have been used for experimentation.
Tang et al. in [19] have proposed a dual densely connected U-shaped neural network (DDU-Net) to segment the tissues with large-scale variant, inconspicuous edges like the spinal canal and extremely small sizes like the dural sac. 50% of the images have been fed into DDU-Net for training, 20% of the images have been used for hyper parameters optimization and prevention of over fitting, and 30% of the images have been used to evaluate the performance of the neural networks. The model has been evaluated using several semantic segmentation metrics like pixel accuracy (PA), mean pixel accuracy (MPA), mean intersection over union (MIoU), and frequency-weighted intersection over union (FWIoU). Ablation studies have been carried out with/without data augmentation, skip connections, dense blocks, and multi-branch networks. The Lumbar spinal CT image dataset used for experimentation contains 2393 axial CT images collected from 279 patients, with the ground truth of pixel-level segmentation labels.
A computational methodology capable of detecting the spinal cord has been proposed in [20]. It involves adaptive template matching for initial segmentation, intrinsic manifold simple linear iterative clustering (IMSLIC) for candidate segmentation, and convolutional neural networks for candidate classification. Template matching algorithm has been executed on each slice of the volume, where the similarity for each matching in each slice has been calculated. Initialization of the template has been defined as the one that has the greatest similarity. The number of the slice that has generated the best matching has been saved. IMSLIC is the technique that uses the least number of parameters. It will group neighboring pixels based on intensity and spatial distance. The dataset used for experimentation contains 36 patients’ CT images from 3 different institutes that have been clustered as 56,698 regions, where 4647 regions were the spinal cord and the rest all the non-spinal cord regions. The proposed methodology has been evaluated with an accuracy of 92.55%, specificity of 92.87%, and sensitivity of 89.23% with 0.065 false positives per image in the detection of the spinal cord.
Semi-automatic segmentation of the colon region on T2-weighted MRI scans has been proposed in [21]. In the first stage of segmentation, a custom novel tubularity filter using a set of points given by the specialists has been used to find an approximation to the colon medial path. In the second stage, a custom segmentation algorithm to detect the colon’s neighbouring regions and the fat capsule containing abdominal organs. Finally, segmentation has been performed via 3D graph cuts in a three-stage multigrid approach. Tubularity Detection Filter (TDF) has been built as a combination of two filters namely the ring filter and the directional filter. The non-colonic regions like the spinal cord have been discarded by segmenting and removed from the initial search space. The model has been evaluated on three groups of MRI scans that have been acquired in the scope of different clinical studies with Dice Similarity Coefficient of 0.92 and Sensitivity of 0.82.
A two-stage automated fracture detection system has been proposed in [22] to detect fractures in the human body. The Faster Region with Convolutional Neural Network (Faster R-CNN) has been used to detect the different types of bone regions in X-ray images. Crack-Sensitive Convolutional Neural Network (CrackNet) has been used to recognize the fractured bone region. The 20 different types of bones of the human anatomy, including vertebrae, have been analysed. Bone fracture identification using CrackNet starts with the Schmid convolutional layer which incorporates the Schmid filter into the convolution and connects to common convolutional layers to improve the recall rate of fractured images. The dataset used for experimentation consists of 3053 X-ray images, in which 2001 images have been used for training and 1052 images have been used for testing. Faster R-CNN and Crack-Sensitive CNN have achieved accuracy higher than 90%, an F-measure higher than 90%, 87.5% recall, and 89.09% precision, outperforming other methods on the bone fracture detection task. The study indicates that the usage of appropriate filters in the convolution operation results in better segmentation of structures.
He et al. have proposed a composite loss function with a dynamic weight, called the Dynamic Energy Loss function in [23] for spine MR image segmentation. The two datasets used for experimentation of the U-Net CNN model with the proposed loss function have achieved superior performance with Dice similarity coefficient values of 0.9484 and 0.8284 and also verified by the Pearson correlation, Bland-Altman, and intra-class correlation coefficient analysis. Li et al. have developed and validated a model for the simultaneous 3D semantic segmentation of multiple spinal structures at the voxel level named as the S3egANet in [24]. S3egANet explicitly solved the high variety and variability of complex 3D spinal structures through a multi-modality auto-encoder module which has been used for extracting the fine-grained structural information. For segmenting numerous spinal structures simultaneously with high accuracy and reliability, a multi-stage adversarial learning technique has been used. S3egANet has achieved the mean Dice coefficient of 88.3% and mean Sensitivity of 91.45% on MRI scans of 90 patients. Lessmann et al. in [25] have proposed an iterative instance segmentation. A fully convolutional neural network has been used to segment and label the vertebrae one by one, independently of the number of visible vertebrae. The network simultaneously carries out several tasks, including segmenting a vertebra, regressing its anatomical label, and predicting whether the vertebra is fully visible in the image. When all the detected vertebrae have been taken into consideration, the projected anatomical labels of the individual vertebrae have been further refined using a maximum likelihood approach.
The framework of U-Net architecture and the parametric level set have been proposed in [26] to extract the shape of bones and disks accurately. CNN has been trained using the dataset, as the level set is sensitive to initialization, training the CNN on training data and the output of the pre-trained network has been used as input to the level set. The level set works in iterations and refines the segmentation output to the desired level. Boundary and region-based optimization has been performed using the geodesic curve and Heaviside function. Two different datasets have been used for evaluation, namely 20 publicly available 3D spine MRI datasets to perform disc segmentation and 173 computed tomography scans for thoracolumbar (thoracic and lumbar) vertebrae segmentation. The dice score has been evaluated as
Zhang et al. in [27] have proposed a multi-task relational learning network (MRLN) that utilizes both the relationships between vertebrae and the relevance of the three tasks namely the accurate segmentation, localization, and identification of vertebrae. A dilation convolution group has been used to expand the receptive field, and Long Short-Term Memory (LSTM) to learn the prior knowledge of the order of the relationship between the vertebral bodies. A co-attention module has been used to learn the correlation information, localization-guided segmentation attention (LGSA), and segmentation-guided localization attention (SGLA), in the decoder stage of segmentation and localization tasks. To avoid the cumbersome weight adjustment for different task loss functions, a novel XOR loss has been formulated which provides a direct evaluation criterion for the localization relationship of the semantic location regression and semantic segmentation.
Li et al. in [28] have proposed a dual-branch multi-scale attention module in the U-Net architecture. The network contains multi-scale feature extraction based on three
Pang et al. in [30] have proposed a novel two-stage framework named SpineParseNet to achieve automated spine parsing for volumetric MR images. The network consists of a 3D graph convolutional segmentation network (GCSN) for 3D coarse segmentation and a 2D residual U-Net (ResUNet) for 2D segmentation refinement. The dataset used for experimentation consists of 215 subjects on T2-weighted volumetric MR images and has achieved performance with mean dice similarity coefficients of
Guinebert et al. in [33] have developed a Picture Archiving and Communication System (PACS) with a DICOM viewer to extract training data and implement two CNN networks, U-Net++ and Yolov5x to segment and detect discs and vertebrae. U-Net++ has been dedicated to performing the semantic segmentation task and Yolov5x for the analysis of degenerative disc disease, disc herniation, and vertebral fracture. Two hundred and forty-four T2 weighted sagittal MRI scans from the university hospital Pasteur 2 of Nice, France, used for experimentation have achieved accuracy of the order of 0.96 and 0.93 Dice index for intervertebral discs and vertebral bodies with an area under the precision-recall curve of 0.88 for fractures and 0.76 for degenerative disc disease. Saenz-Gamboa et al. in [34] have proposed a model for automatic semantic segmentation of lumbar spine MRI using variants of U-Net to assign the class label to each pixel of an image with vertebrae, intervertebral discs, nerves, blood vessels, and other tissues. Several complementary blocks, such as three types of convolutional blocks, spatial attention models, deep supervision, and multi-level feature extractor have been used to define the variants. The study showed that variants of U-Net yield better performance in semantic segmentation. Haq et al. in [35] have proposed a BTS-GAN system for breast tumor segmentation. The upgraded U-Net architecture utilized in the BTS-GAN system incorporates improvements like integrating skip connections between encoder and decoder layers and introducing a parallel dilated convolution (PDC) module. The PDC module in BTS-GAN improves tumor recognition across scales and enhances context sensitivity without adding parameters. It comprises three parallel branches with dilated convolutions for multi-scale feature fusion, preserving context while minimizing information loss. These enhancements aim to boost the network’s ability to retrieve features, resulting in more precise and efficient segmentation of breast tumors in MRI scans.
Luan et al. in [36] have developed a new deep model, namely Gabor Convolutional Networks (GCNs or Gabor CNN), with Gabor filters incorporated into deep CNNs. GCNs have been readily compatible with any well-liked deep learning architecture and implement the Gabor filter-based convolution operator so that the robustness of learned features against the orientation and scale changes can be reinforced. In medical image processing, many filters like tube filters, ridge filters, neurites, or vessel filters have been used in literature to segment the muscles, bones, nerves, or blood vessels. Meijering et al. in [37] have proposed a Meijering filter which is an improved steerable filter for computing local ridge strength and orientation. They have used a graph-searching algorithm with a novel cost function for exploiting the image features to obtain globally optimal tracings. In [38], the Meijering neuriteness ridge filter has been performed to show the ridges of the vessels in the frames. Based on the noise level, the Meijering filter was the most appropriate to show the ridge like structures namely intervertebral discs and spinal canal which helps to give the clear representation of vertebrae. The proposed work has been inspired by the literature and differs in the following ways:
• The bone fracture identification using CrackNet in [22] starts with the Schmid convolutional layer which incorporates the Schmid filter in the convolution process. Similarly, Gabor CNN incorporates Gabor filters into the convolutional layer in [34]. It is found from the literature that the Meijering filter is the best suitable ridge or vesselness filter used for the segmentation of lumbar portions than the other ridge filters or tubularity directional filter in [21]. Hence the MU-Net (Meijering U-Net) architecture is proposed which incorporates the Meijering filter in the convolutional layer as discussed in [35,36] to perform the segmentation of vertebrae.
• U-Net and its variants are one of the best-suited deep learning architectures for the segmentation or the semantic segmentation of medical images as discussed in [17,19,23,26,28–30,32,33]. Semantic segmentation of the vertebrae helps diagnosing the lumbar disorders certainly as discussed in [30–33]. In the proposed work, MU-Net is used for semantic segmentation of the lumbar and sacral vertebra.
This section discusses in detail about the dataset preparation, the pre-processing steps, and the proposed MU-Net architecture for the semantic segmentation of the vertebrae. The performance of semantic segmentation using MU-Net model as compared to the standard U-Net model has been contributed by the effectiveness of the Meijering convolutional layer incorporating the Meijering filter which enhances the vessel-like structures namely intervertebral discs and spinal canal that helps to give the clear representation of vertebrae. As depicted in Fig. 1, the work flow of the proposed scheme involves ground truth generation, pre-processing of input images, training phase and testing phase of the proposed MU-Net architecture and the performance evaluation of semantically segmented vertebrae image have been elaborated in detail in the further sections.
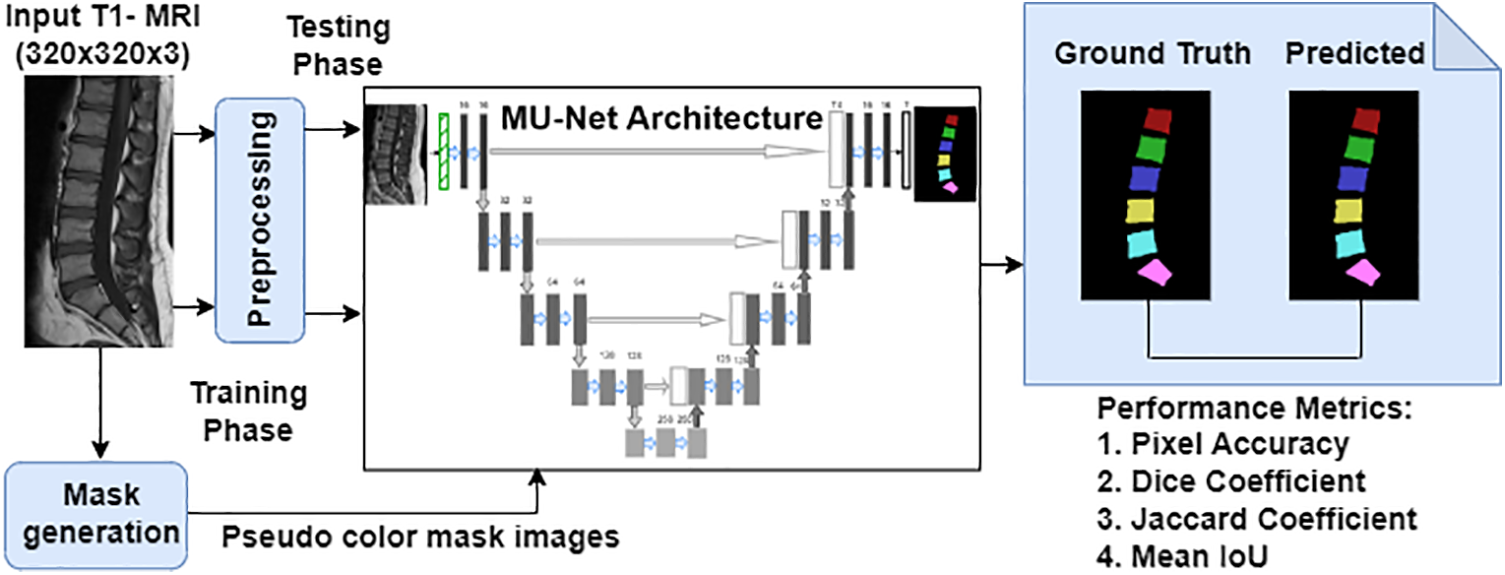
Figure 1: Overall architecture of the proposed scheme for semantic segmentation of the vertebrae
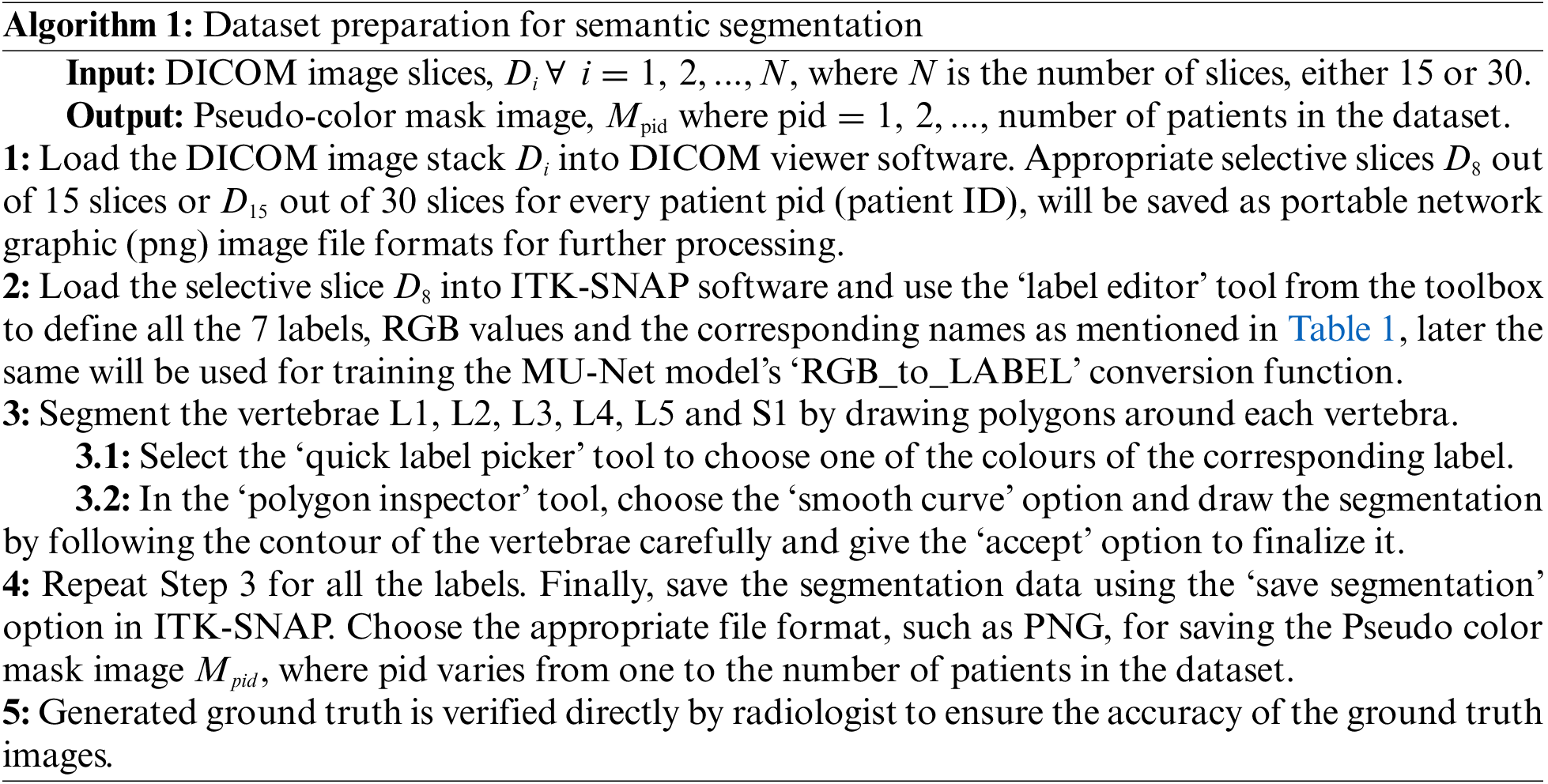
3.1 Dataset Preparation for Semantic Segmentation
Semantic segmentation of the lumbar vertebrae L1, L2, L3, L4, and L5 and sacral vertebra S1 is essential to diagnose the lower back pain disorders. To perform the segmentation, the ground truth information is generated initially before performing the segmentation task. Similarly, to perform the semantic segmentation task, the meaningful mask or ground truth to differentiate the segmented vertebrae must be generated clearly as depicted in Fig. 2. Dataset preparation provides the original selective slices using Algorithm 1 and their corresponding pseudo color ground truth mask images are generated as defined in Table 1. Red Green Blue (RGB) values represents the intensity values varying from 0 to 255, where 0 0 0 represents black and 255 255 255 represents white color. The ground truth generated and verified with radiologist and stored in directories for the further training of the deep learning model.
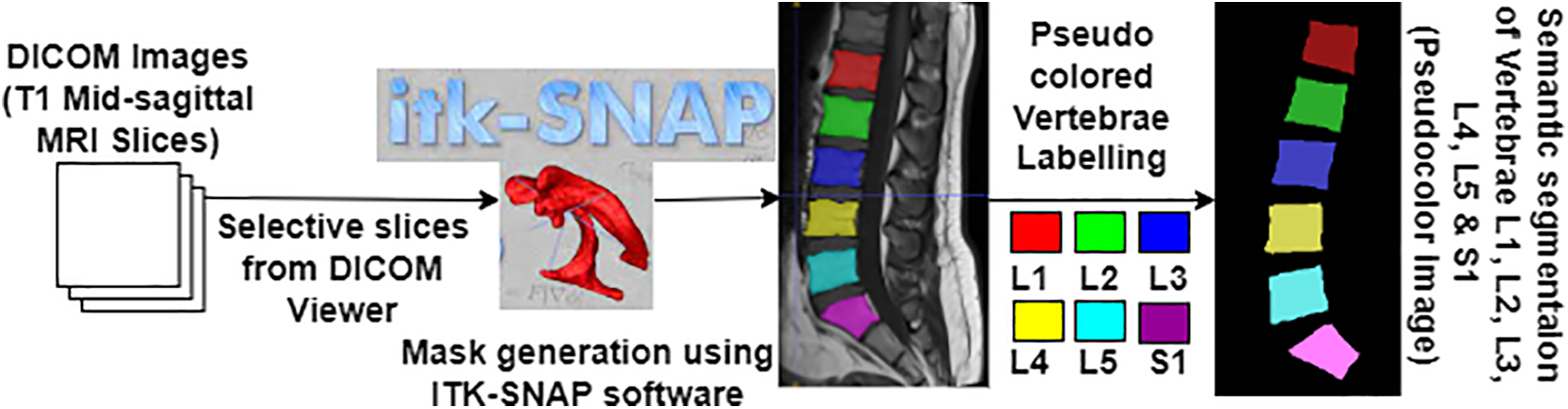
Figure 2: Pseudo color mask image generation

3.2 Pre-Processing Phase for MU-Net Model
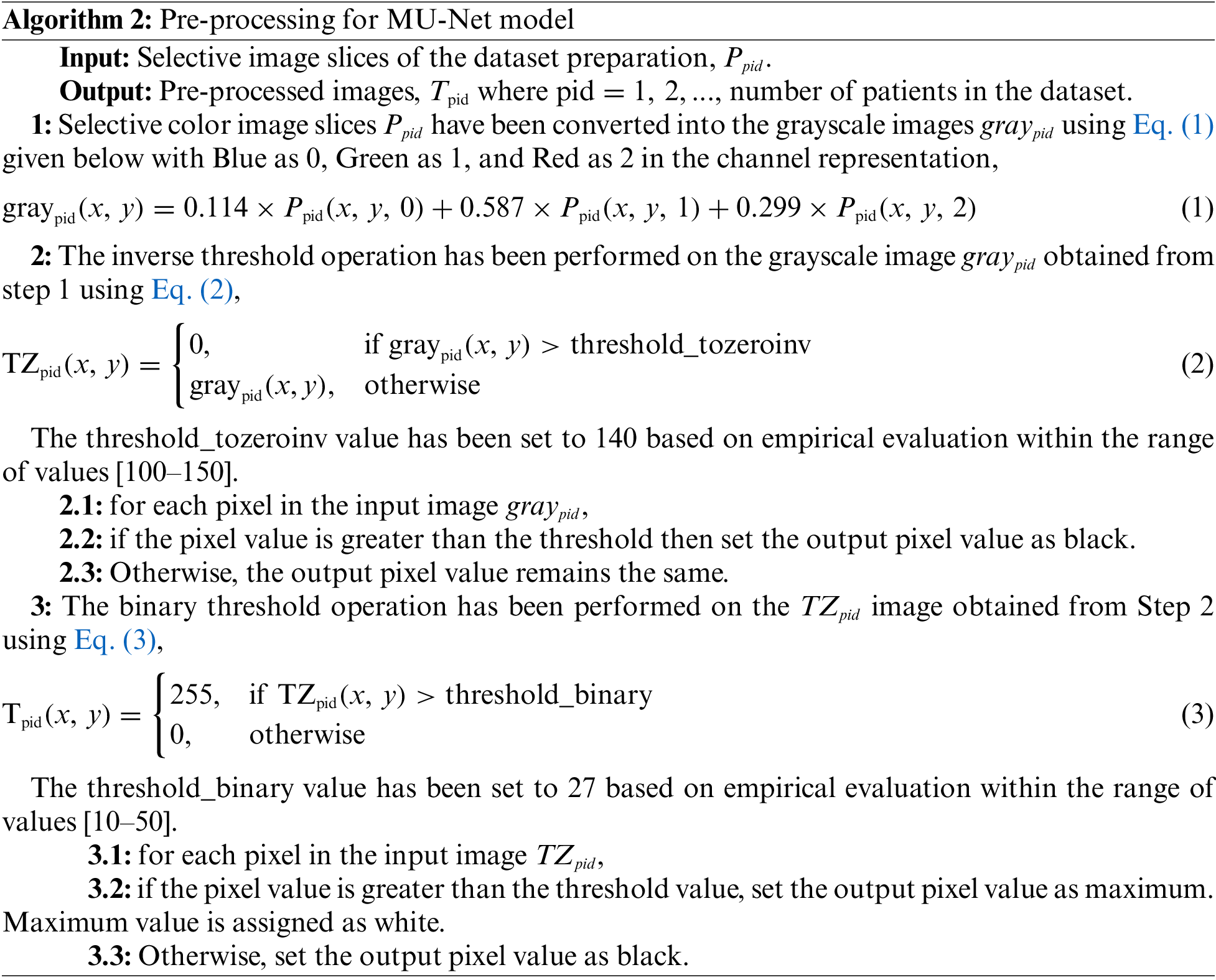
The selective DICOM slices obtained from the dataset preparation phase have been converted to.png image to make it comfortable to process in deep learning models. These slices have been (i) converted into grayscale and (ii) pre-processed using thresholding operations as described in Algorithm 2. T1-weighted MRI interpretation involves analyzing pixel intensity values, where bright pixels signify high signal intensity like vertebrae and dark pixels denote low signal intensity like spinal canal. This property of T1 weighted image has been utilized in binary thresholding to facilitate pre-processing that results in the effective segmentation of vertebrae. The Meijering filter can be applied then to the pre-processed image in the MU-Net model.
3.3 Proposed MU-Net Architecture for Semantic Segmentation
The pre-processed selective slice images and the corresponding pseudo mask images obtained from the dataset preparation phase and the pre-processing phase have been given for training the model with the downsampling path and the upsampling path of the proposed MU-Net model as depicted in Fig. 3.
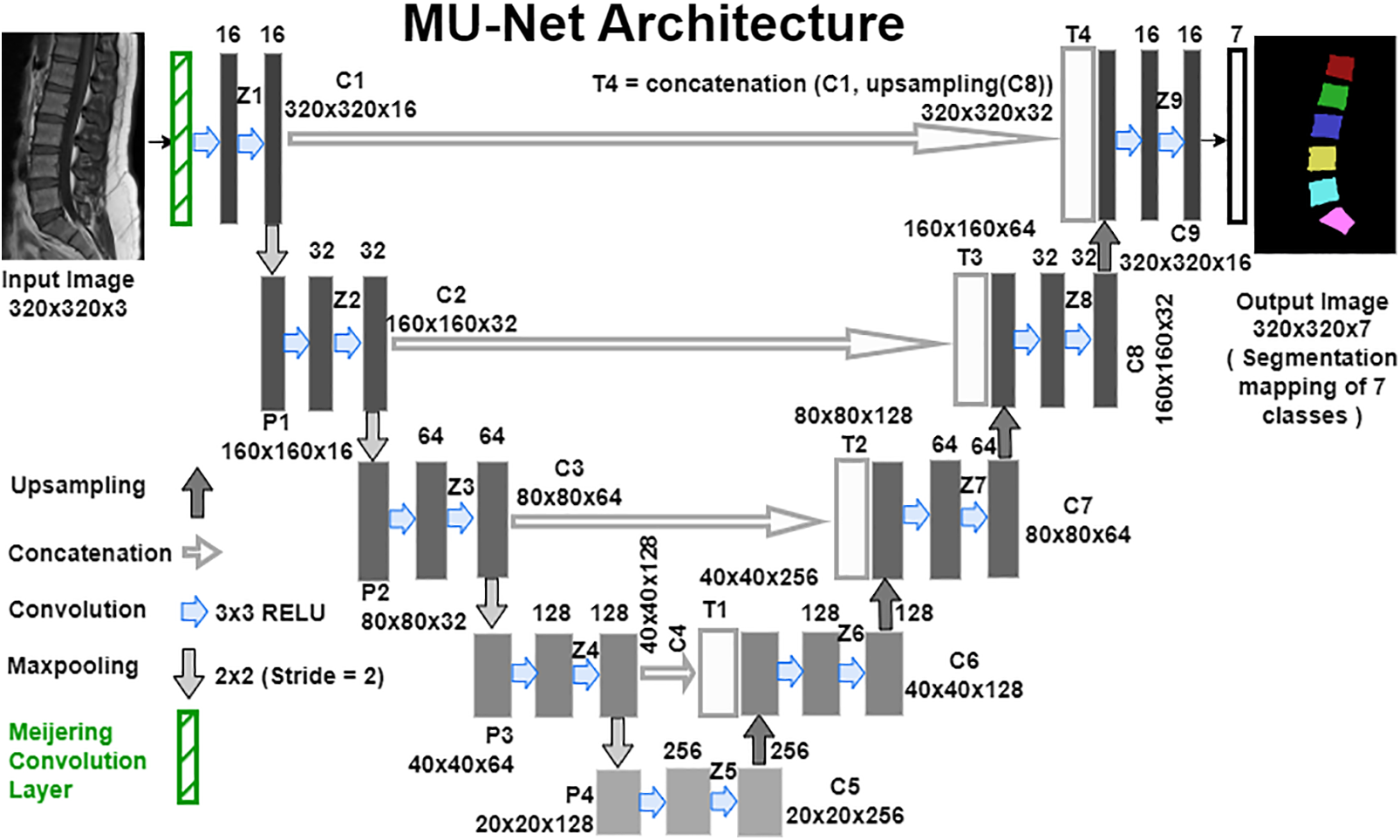
Figure 3: Meijering U-Net architecture for the semantic segmentation of vertebrae
The algorithm describes about the Meijering U-Net model creation to perform semantic segmentation of the vertebrae. Compile, train and save the model with defined inputs using the ‘categorical cross-entropy’ loss function and the ‘Adam’ optimizer. In the testing phase of the model, implement Steps 1 through 4 with the input test images to obtain the segmented feature map.
The Meijering convolutional layer is built by creating the Meijering filter [39,40] kernel matrix
where H(X) is defined in Eq. (5), the eigenvalues
Convolution operation is carried out between the input tensor X and
where

In the downsampling path of the MU-Net Architecture, the
where
where
In the up-sampling path of the MU-Net Architecture, on each level performs up-sampling using a
where C[i] is the output of the
For instance,
The last
where the resulting feature maps
4 Experimental Results and Discussion
The experiment has been carried out on a system with a 64-bit windows operating system (OS), 64 GB of random access memory (RAM), an NVIDIA Quadro P5000 GPU with 16 GB of memory, and an Intel Xeon processor with 3.60 GHz processing speed. The methods utilized in this work were implemented with the help of frameworks such as Keras (version 2.9.0), Tensorflow (version 2.7.0), Tensorflow-gpu (version 2.9.1), and OpenCV (version 4.5.5) from Python (version 3.9.7) libraries. The images from the original source [41] have been downloaded and the pseudo mask images have been created. This dataset has been utilized for experimentation of the proposed semantic segmentation technique.
The Lumbar Spine MRI Dataset from the Mendeley Data website in [42] is a publicly available standard dataset containing T1-weighted and T2-weighted mid-sagittal view images of 515 patients. T1-weighted mid-sagittal view comprises either 15 or 30 slices of size
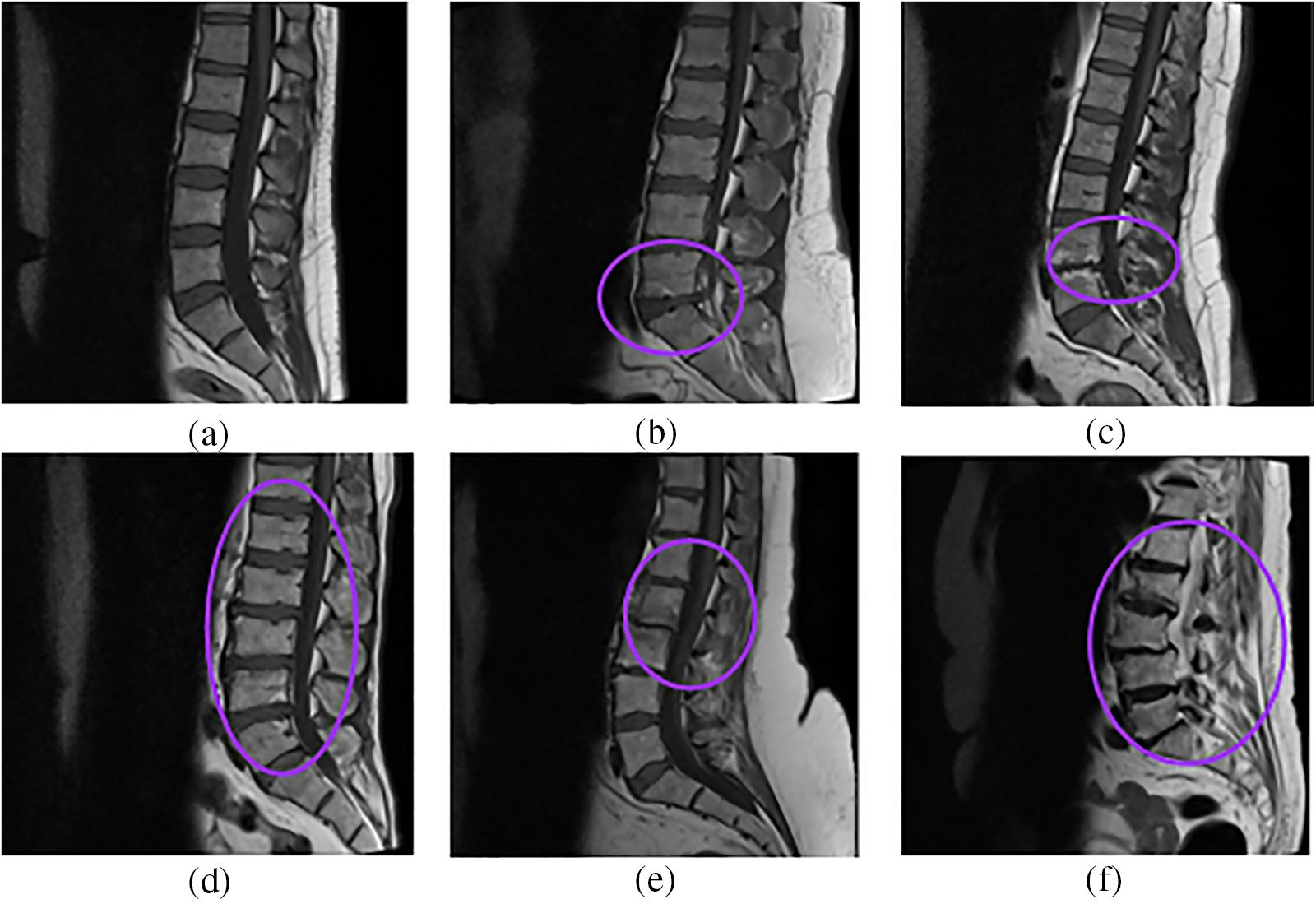
Figure 4: Sample images from the dataset (a) Normal, (b) Disc Herniation, (c) Degenerative disc disease, (d) Disc Bulging, (e) Spondylolisthesis, (f) Fractured
Digital Imaging and Communications in Medicine format and Neuroimaging Informatics Technology Initiative (NIfTI) format are commonly used file formats to store MRI images. To visualize the DICOM or NIfTI image files, DICOM viewer software or ImageJ software can be used. ImageJ is a Java-based graphic design program dedicated to analyzing images of different file formats. The appropriate slices of 1312 from all the patients are selected and saved in portable network graphic (png) format using DICOM viewer software for segmenting the lumbar and sacral vertebrae clearly.
The proposed models’ coefficients are optimized using Adam optimizer, and other specifications are kernel size, dropout, learning rate, activation function, loss function, and validation technique, as mentioned in Table 2. Dropout and learning rates have been optimized using the grid search method. The 5-fold validation technique gives better performance by reducing the overfitting problem.
From experimentation, it is found that the training time is comparatively less for the proposed MU-Net model, with an average of 86 s per epoch, as compared to the adopted U-Net, with an average of 102 s per epoch for the stated model specification. Due to the need to assess classification accuracy as well as localization correctness, evaluating semantic segmentation is fairly difficult [43]. The objective is to evaluate the performance of the proposed semantic segmentation method by finding the overlap and similarity between the semantic prediction and the ground truth. The parameters namely True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN) of the confusion matrix in the classification model are also evaluated in the segmentation model to measure the metrics namely Pixel Accuracy (PA), Intersection over Union (IoU), and Dice Similarity Coefficient (DSC).
True Positive (TP) Pixels that are correctly assigned as vertebrae.
True Negative (TN) Pixels that are correctly assigned as background.
False Positive (FP) Incorrect extraction of background pixels as vertebrae.
False Negative (FN) Incorrect extraction of vertebrae pixels as background.
Pixel Accuracy (PA):
Accuracy also known as Rand index or pixel accuracy (PA) is one of the most known evaluation metrics. The simplest metric computes the ratio between the number of pixels that are correctly classified to the total number of pixels for one class [44]. In semantic segmentation, PA is defined using Eq. (15) [45] that divides the number of accurate predictions (accurate positive and negative predictions) by the total number of predictions for one class. Mean Pixel Accuracy (MPA) is the average pixel accuracy for all the classes.
The splinter group of the pixels in a medical image is often taken up by a single ROI, with the background of the image taking up the remaining pixels. The accuracy metric will always yield an unjustified high score because of the genuine negative inclusion. Accuracy scores are frequently higher than 90% or very near to 100%, even while predicting the segmentation of an entire image as a background class. Due to class imbalance in segmentation, Pixel Accuracy is high, and then it doesn’t mean superior segmentation ability. So other metrics, namely DSC and IoU, which evaluate the similarity and overlap between the semantic prediction and the ground truth, play an important role in semantic segmentation comparatively.
Intersection over Union (IoU):
IoU or Jaccard Index or Jaccard similarity coefficient score measures the similarity between the predictions and the ground truth. Jaccard index score defined in Eq. (16) [43] varies from 0 to 1, where 0 represents no overlap and 1 represents the perfect overlap of segmented prediction with the mask image.
where A represents the ground truth and B represents the segmented images. Or it can also be represented with the confusion matrix parameters as defined in Eq. (17) [47],
IoU value or Jaccard index is evaluated for each segmented image and its corresponding mask for every input image separately and then the overall average IoU or Jaccard score is computed. IoU of the possible test cases in Fig. 4 have been calculated for the proposed model and compared with the adopted U-Net model. The outputs for sample images using the adopted model and proposed model are depicted in Fig. 5 and IoU variation with respect to the cases is depicted in Fig. 6 for better visualization. In Figs. 6 and 7, sample cases have been shown; 1(a) shows a healthy input sample, 2(a) shows a sample image indicating mild disc herniation, 3(a) shows the image depicting disc degeneration, 4(a) shows the image depicting disc bulging and 5(a) and 6(a) indicate the spondylolisthesis occurence on highly fractured or ruptured images.
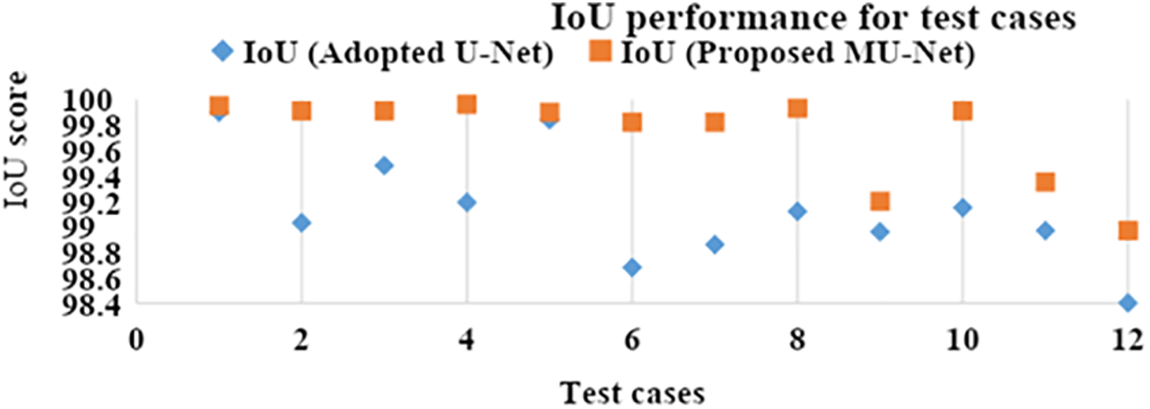
Figure 5: IoU performance of the adopted U-Net and proposed MU-Net models
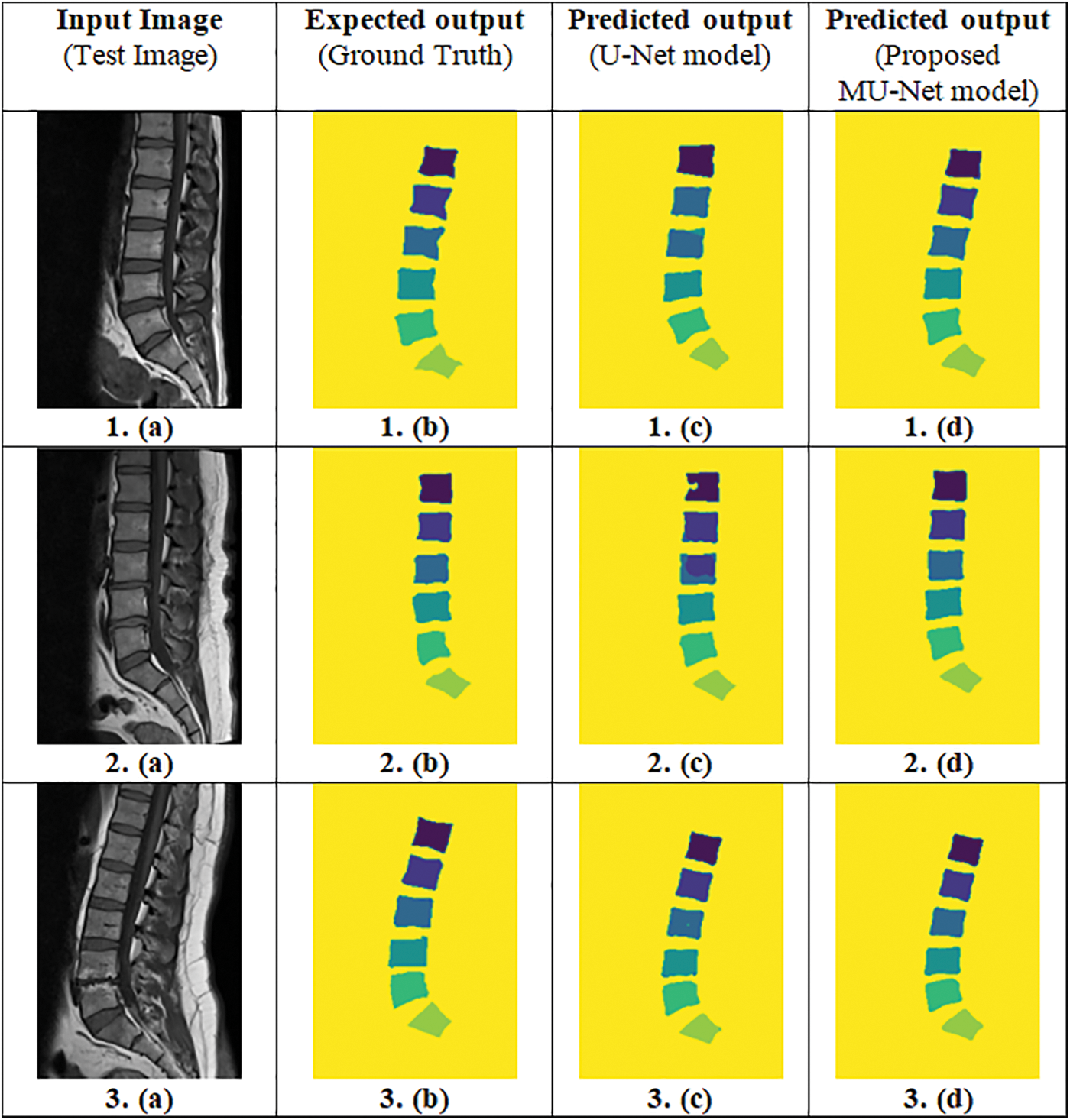
Figure 6: Predicted output of the proposed MU-Net model and the adopted U-Net model (Column (a)-Input image, Column (b)-Ground truth, Column (c)-Predicted output of the adopted U-Net model, Column (d)-Predicted output of the proposed MU-Net model)
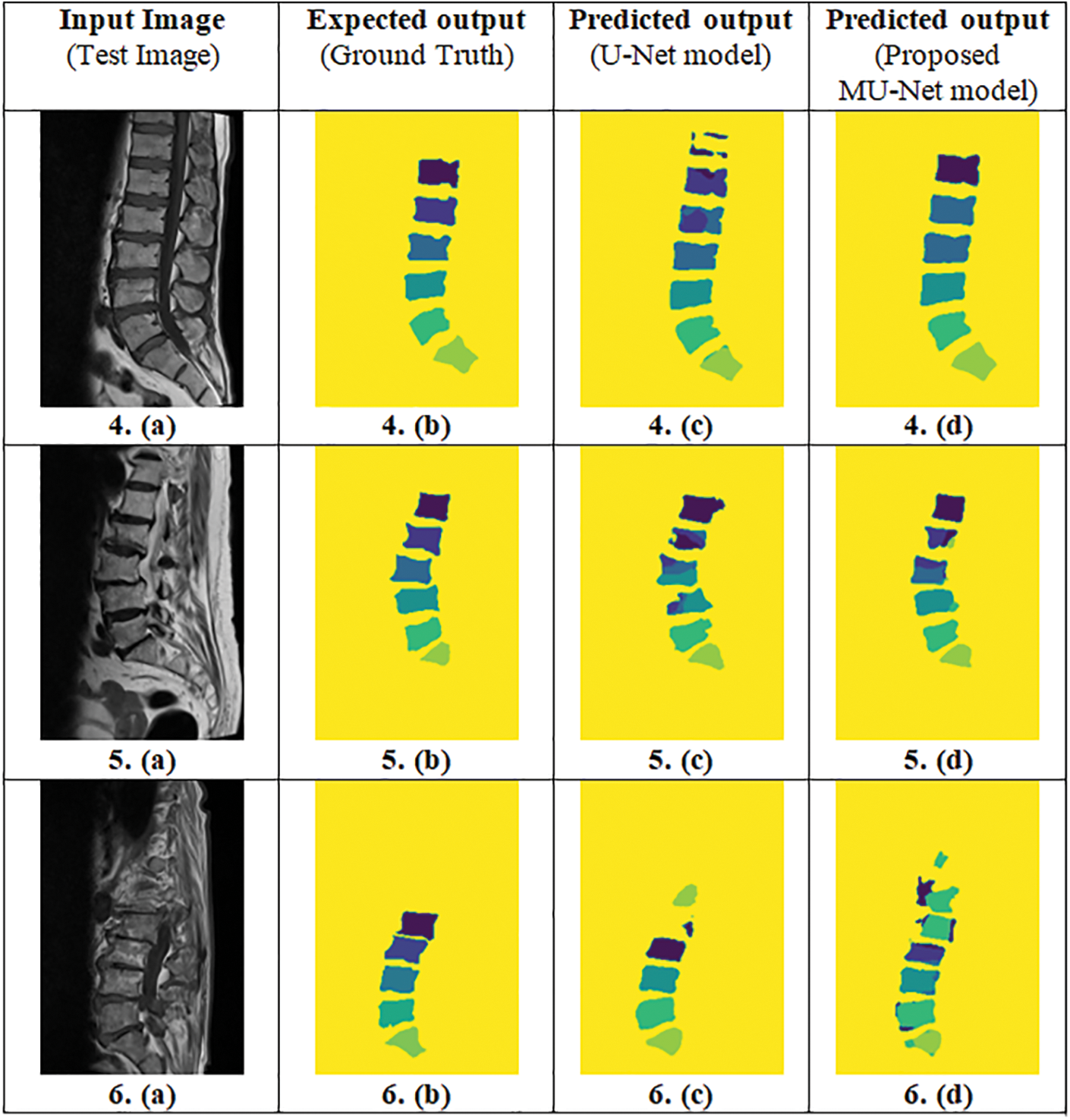
Figure 7: Predicted output of the proposed MU-Net model and the adopted U-Net model (Column (a)-Input image, Column (b)-Ground truth, Column (c)-Predicted output of the adopted U-Net model, Column (d)-Predicted output of the proposed MU-Net model)
Mean Intersection over Union (mIoU):
Mean IoU computes the IoU on a class basis and then averages it. Mean IoU calculates the overlap of every individual class in both the segmented mask and the actual ground truth one by one, and then calculates the sum of all the classes’ overlap value and divides it by the total number of classes as defined in Eq. (18),
where K = 7 represents all the 6 classes and one background class.
Dice Similarity Coefficient (DSC):
Dice Similarity Coefficient or SÃ-Dice index or F1 score is defined as two times the overlap area between the prediction and ground truth mask, divided by the sum of pixels in both the prediction and ground truth mask as defined in Eq. (19),
where A represents the ground truth and B represents the segmented images. Dice score or F1 score represented with the confusion matrix parameters as defined in Eq. (20) [43],
The proposed MU-Net model and the adopted U-Net model with pre-processed images as input are trained and tested for 60 epochs and the corresponding performance metrics are plotted in Fig. 8. The best performance for semantic vertebrae segmentation is achieved by MU-Net as depicted in Fig. 9 for better visualization in which Column 1 represents the performance metrics of the adopted U-Net model and Column 2 of the proposed MU-Net model.

Figure 8: Training and testing performance metrics for 60 epochs of the adopted U-Net and proposed MU-Net models
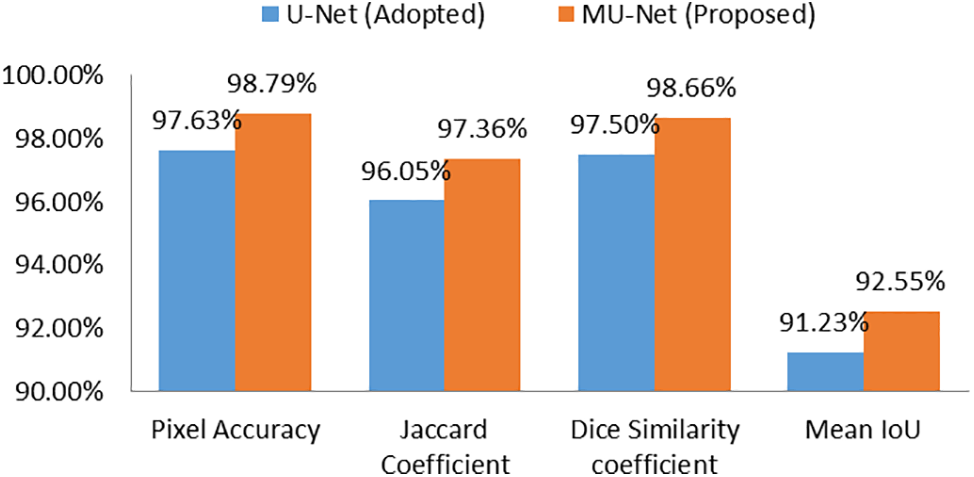
Figure 9: Performance metrics of the adopted U-Net and proposed MU-Net models
CAD systems in medical diagnostics raise ethical concerns to ensure that they serve as support tools rather than replacements for human expertise. CAD system improves the performance metrics over the past decades. Still, the final validation of the diagnosis remains with the radiologist.
The dataset used in our study has been sourced from Mendeley Data, which has been originally introduced and extensively validated by the authors mentioned in [31]. The authors explicitly considered a wide spectrum of variability in clinical settings. The MRI scanning parameters used in the scans can vary depending on the sequence and view plane types. The author ensures that the dataset is representative of diverse clinical conditions like image registration algorithm, patient age, patient movement, direction, and position of the image plane of axial view on sagittal view. The authors ensures good quality and also ensures avoiding the destroyed or fused lumbar spine elements getting included in the benchmark dataset.
Hyperparameters of the proposed model have been fine-tuned using the dropout regularization technique to enhance the model’s robustness by reducing overfitting, leading to better generalization across different imaging conditions. Data augmentation has been used to simulate variations in patient positioning, image noise, and lighting conditions to ensure that the model can handle variations in real-world clinical settings. The grid of three standard hyperparameter values [0.0001, 0.001, 0.01] has been assigned for learning rate and [0.1, 0.3, 0.5] assigned for dropout. Grid search has been performed using the defined grid, this gives the selective, optimized value for better performance of the model.
K-fold cross validation technique has been utilised with k value as 5. In 5-fold cross-validation, the data has been split into 5 subsets, trained on k−1 subsets, and tested on the kth subset, repeating the process k times. Grid search provides the hyperparameter combination with the best average performance across all the 5-folds and provides the optimal parameter values.
For the semantic segmentation of lumbar and sacral vertebrae, the proposed model gives better performance when compared to the state-of-art models, namely, UNet, SegNet, FCN, DDUNet, FCN-UNet, 6-FFN, and ResNet-UNet as mentioned in Table 3. All the models utilize the MRI modality on corresponding available datasets mentioned by authors using the variations of U-Net. The proposed MU-Net model adopted the U-Net model and the adopted SegNet model have been compared by the proposed sequence of methodology only by differing the corresponding U-Net architecture.
The integration of CAD as a support tool in clinical workflows is highlighted, with final diagnoses remaining the responsibility of trained professionals. Additionally, the importance of continuous education is stressed to ensure radiologists understand both the benefits and limitations of CAD systems for certain exceptional cases. The potential limitations of the proposed method are on different image types other than DICOM images, different imaging modalities like CT or X-ray, and other imaging conditions of highly fractured or ruptured vertebrae images. The hardware and software specifications used for the experiment have been comparatively high, which may not be accessible to all medical facilities to experiment with the proposed model for the diagnostic process. The limitation can be overcome by performing the necessary changes as mentioned in future work.
5 Conclusion and Future Enhancement
Lower back pain (LBP) is a widespread and significant health issue that affects people of all ages, from childhood to adulthood, and the elderly. To enhance outcomes and general well-being, lower back pain sufferers should seek an early diagnosis and proper treatment. The proposed model produced better results for the semantic segmentation of the lumbar portion comprising of lumbar vertebrae L1 through L5 and the sacral vertebra S1 from T1-weighted spine MRI. Semantic segmentation provides not only the segmented vertebrae but also the identification of which vertebrae it is. In the dataset preparation process, the pseudo color mask images representing each vertebra to the corresponding pixel intensity have been generated to train the MU-Net model. The pre-processing phase of the input image performs image enhancement with thresholding operations to make the image representation more clear for further segmentation process. In this work, a modified U-Net architecture called Meijering U-Net (MU-Net) has been proposed, which leverages the power of U-Net to tackle the complexities of medical spine image analysis tasks. The key addition in MU-Net is the Meijering convolutional layer, which incorporates the Meijering filter to enhance the vessel-like structures, namely intervertebral discs and spinal canal, which helps to give the clear representation of vertebrae in MU-Net as compared to U-Net for the effective semantic segmentation process. The ground truth masks and the input images were given as input to the MU-Net model. The MU-Net model has been used to extract the features from the images and perform pixel-wise classification based on the ground truth information used for training the model. The output is the predicted mask image that has been used to diagnose the vertebrae.
A dataset comprising 1312 images extracted from T1-weighted mid-sagittal MRI scans from the Lumbar Spine MRI Dataset publicly available from Mendeley Data is used for experimentation. By utilizing the Meijering filter in the convolutional layer, MU-Net has achieved a pixel accuracy of 98.79%, dice similarity coefficient of 98.66%, Jaccard coefficient or IoU of 97.36%, and 92.55% mean IoU metrics for the dataset, thereby aiding in the diagnostic process of lumbar diseases. Based on experimentation, it is found that the suggested method exhibits good performance across a variety of performance criteria, demonstrating its effectiveness. The proposed CAD system that generates diagnostic results from MRI would not only reduce the burden on a radiologist but also boost confidence in a diagnosis. Occasionally, the CAD system might also detect a disorder that a radiologist could have missed due to insufficient time to analyse a case. The proposed method allows for early diagnosis of disc bulging, disc herniation, and slipped vertebrae, which avoids leading to spinal stenosis, severe disc disorders, and spondylolisthesis that can be beneficial for a better patient outcome by avoiding surgery or prolonged treatment.
In the future, the proposed framework can be extended to objectively segment the cervical, thoracic, and pelvic regions as well as to give a more in-depth understanding of human spinal diseases on different imaging modalities. Another direction that may be explored is spinal canal detection along with vertebrae and intervertebral discs by fine-tuning the MU-Net model to diagnose Lumbar Spinal Stenosis (LSS) in guiding surgeons towards less invasive methods for improving patient care and optimizing surgical outcomes in the field of spinal surgery. The standard diagnostic machines used traditionally by radiologists can also deploy the model as software. Apparently by using the cloud environment to make the model into a SaaS (software as a service) with required configuration for execution on any system containing minimal specification with proper internet facility.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Study conception, design, material preparation, data collection and analysis were performed by Lakshmi S V V. The first draft of the manuscript was written by Lakshmi S V V. Shiloah Elizabeth Darmanayagam and Sunil Retmin Raj Cyril commented on previous versions of the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The Lumbar Spine MRI Dataset that supports the findings of this study is publicly available from https://data.mendeley.com/datasets/k57fr854j2/2 (accessed on 29 September 2024) [42]. The radiologist report for the mentioned dataset is publicly available from https://data.mendeley.com/datasets/s6bgczr8s2/2 (accessed on 29 September 2024).
Ethics Approval: Written formal consent was obtained from each patient prior to the data collection.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Galbusera F, Casaroli G, Bassani T. Artificial intelligence and machine learning in spine research. Hoboken, New Jersey, NY, USA: John Wiley and Sons Inc.; 2019. [Google Scholar]
2. Haque IRI, Neubert J. Deep learning approaches to biomedical image segmentation. Amsterdam, Netherlands: Elsevier Ltd.; 2020. [Google Scholar]
3. Giger ML, Chan HP, Boone J. Anniversary paper: history and status of CAD and quantitative image analysis: the role of medical physics and AAPM. Hoboken, New Jersey, USA: John Wiley and Sons Ltd.; 2008. [Google Scholar]
4. Saravagi D, Agrawal S, Saravagi M, Chatterjee JM, Agarwal M. Diagnosis of lumbar spondylolisthesis using optimized pretrained CNN models. Comput Intell Neurosci. 2022;2022:7459260. doi:10.1155/2022/7459260. [Google Scholar] [PubMed] [CrossRef]
5. Athertya JS, Kumar GS. Classification of certain vertebral degenerations using MRI image features. Biomed Phys Eng Express. 2021;7(4):45013. doi:10.1088/2057-1976/ac00d2. [Google Scholar] [PubMed] [CrossRef]
6. Lee J, Chung SW. Deep learning for orthopedic disease based on medical image analysis: present and future. Appl Sci. 2022 Jan 11;12(2):681. doi:10.3390/app12020681. [Google Scholar] [CrossRef]
7. Kim GU, Chang MC, Kim TU, Lee GW. Diagnostic modality in spine disease: a review. Asian Spine J. 2020;14:910–20. doi:10.31616/asj.2020.0593. [Google Scholar] [PubMed] [CrossRef]
8. Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Zeitschrift für Medizinische Physik. 2019 May 1;29(2):102–27. doi:10.1016/j.zemedi.2018.11.002. [Google Scholar] [PubMed] [CrossRef]
9. Hesamian MH, Jia W, He X, Kennedy P. Deep learning techniques for medical image segmentation: achievements and challenges. J Digit Imaging. 2019;8,32:582–96. doi:10.1007/s10278-019-00227-x. [Google Scholar] [PubMed] [CrossRef]
10. Andrew J, Divyavarshini M, Barjo P, Tigga I. Spine magnetic resonance image segmentation using deep learning techniques. In: 2020 6th International Conference on Advanced Computing and Communication Systems, ICACCS 2020, 2020; Coimbatore, TamilNadu, India: Institute of Electrical and Electronics Engineers Inc.; p. 945–50. [Google Scholar]
11. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. Springer Verlag, Germany: Springer Verlag; 2015. vol. 9351, p. 234–41. [Google Scholar]
12. Liu L, Cheng J, Quan Q, Wu FX, Wang YP, Wang J. A survey on U-shaped networks in medical image segmentations. Neurocomputing. 2020;10,409:244–58. doi:10.1016/j.neucom.2020.05.070. [Google Scholar] [CrossRef]
13. He H, Sun YD, Yang BX, Xie MX, Li SL, Zhou B, et al. Building extraction based on hyperspectral remote sensing images and semisupervised deep learning with limited training samples. Comput Electr Eng. 2023;110:108851. doi:10.1016/j.compeleceng.2023.108851. [Google Scholar] [CrossRef]
14. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2015 Jun 7–12; Boston, MA, USA; p. 3431–40. [Google Scholar]
15. Huang SY, Hsu WL, Hsu RJ, Liu DW. Fully convolutional network for the semantic segmentation of medical images: a survey. Diagnostics. 2022 Nov 11;12(11):2765. doi:10.3390/diagnostics12112765. [Google Scholar] [PubMed] [CrossRef]
16. Mahdy LN, Ezzat KA, Hassanien AE. Automatic detection system for degenerative disk and simulation for artificial disc replacement surgery in the spine. ISA Trans. 2018;10,81:244–58. doi:10.1016/j.isatra.2018.07.006. [Google Scholar] [PubMed] [CrossRef]
17. Zhang Q, Du Y, Wei Z, Liu H, Yang X, Zhao D. Spine medical image segmentation based on deep learning. J Healthc Eng. 2021;2021(1):1917946. doi:10.1155/2021/1917946. [Google Scholar] [PubMed] [CrossRef]
18. Lu H, Li M, Yu K, Zhang Y, Yu L. Lumbar spine segmentation method based on deep learning. J Appl Clin Med Phys. 2023 Jun;24(6):e13996. doi:10.1002/acm2.13996. [Google Scholar] [PubMed] [CrossRef]
19. Tang H, Pei X, Huang S, Li X, Liu C. Automatic lumbar spinal CT image segmentation with a dual densely connected U-Net. IEEE Access. 2020;8:89228–38. doi:10.1109/ACCESS.2020.2993867. [Google Scholar] [CrossRef]
20. Diniz JOB, Diniz PHB, Valente TLA, Silva AC, Paiva AC. Spinal cord detection in planning CT for radiotherapy through adaptive template matching, IMSLIC and convolutional neural networks. Comput Methods Programs Biomed. 2019;3,170:53–67. doi:10.1016/j.cmpb.2019.01.005. [Google Scholar] [PubMed] [CrossRef]
21. Orellana B, Monclus E, Brunet P, Navazo I, Ãlvaro B, Azpiroz F. A scalable approach to T2-MRI colon segmentation. Med Image Anal. 2020;7:63. doi:10.1016/j.media.2020.101697. [Google Scholar] [CrossRef]
22. Ma Y, Luo Y. Bone fracture detection through the two-stage system of crack-sensitive convolutional neural network. Inform Med Unlocked. 2021 Jan 1;22:100452. doi:10.1016/j.imu.2020.100452. [Google Scholar] [CrossRef]
23. He S, Li Q, Li X, Zhang M. An optimized segmentation convolutional neural network with dynamic energy loss function for 3D reconstruction of lumbar spine MR images. Comput Biol Med. 2023;6:160. doi:10.1016/j.compbiomed.2023.106839. [Google Scholar] [CrossRef]
24. Li T, Wei B, Cong J, Li X, Li S. S3egANet: 3D spinal structures segmentation via adversarial nets. IEEE Access. 2020;8:1892–901. doi:10.1109/ACCESS.2019.2962608. [Google Scholar] [CrossRef]
25. Lessmann N, Van Ginneken B, De Jong PA, Išgum I. Iterative fully convolutional neural networks for automatic vertebra segmentation and identification. Med Image Anal. 2019 Apr 1;53:142–55. doi:10.1016/j.media.2019.02.005. [Google Scholar] [CrossRef]
26. Rehman F, Shah SIA, Riaz N, Gilani SO. A robust scheme of vertebrae segmentation for medical diagnosis. IEEE Access. 2019;7:120387–98. doi:10.1109/ACCESS.2019.2936492. [Google Scholar] [CrossRef]
27. Zhang R, Xiao X, Liu Z, Li Y, Li S. MRLN: Multi-task relational learning network for MRI vertebral localization, identification, and segmentation. IEEE J Biomed Health Inform. 2020;10,24:2902–11. doi:10.1109/JBHI.2020.2969084. [Google Scholar] [PubMed] [CrossRef]
28. Li H, Luo H, Huan W, Shi Z, Yan C, Wang L, et al. Automatic lumbar spinal MRI image segmentation with a multi-scale attention network. Neural Comput Appl. 2021;9,33:11589–602. doi:10.1007/s00521-021-05856-4. [Google Scholar] [PubMed] [CrossRef]
29. Wang S, Jiang Z, Yang H, Li X, Yang Z. Automatic segmentation of lumbar spine MRI images based on improved attention U-net. Comput Intell Neurosci. 2022;2022(1):4259471. doi:10.1155/2022/4259471. [Google Scholar] [PubMed] [CrossRef]
30. Pang S, Pang C, Zhao L, Chen Y, Su Z, Zhou Y, et al. SpineParseNet: spine parsing for volumetric MR image by a two-stage segmentation framework with semantic image representation. IEEE Trans Med Imaging. 2021;1,40:262–73. doi:10.1109/TMI.2020.3025087. [Google Scholar] [PubMed] [CrossRef]
31. Al-Kafri AS, Sudirman S, Hussain A, Al-Jumeily D, Natalia F, Meidia H, et al. Boundary delineation of MRI images for lumbar spinal stenosis detection through semantic segmentation using deep neural networks. IEEE Access. 2019;7:43487–501. doi:10.1109/ACCESS.2019.2908002. [Google Scholar] [CrossRef]
32. Atli I, Gedik OS. Sine-Net: a fully convolutional deep learning architecture for retinal blood vessel segmentation. Eng Sci Technol. 2021;24(2):271–83. doi:10.1016/j.jestch.2020.07.008. [Google Scholar] [CrossRef]
33. Guinebert S, Petit E, Bousson V, Bodard S, Amoretti N, Kastler B. Automatic semantic segmentation and detection of vertebras and intervertebral discs by neural networks. Comput Methods Programs Biomed Update. 2022;2:100055. doi:10.1016/j.cmpbup.2022.100055. [Google Scholar] [CrossRef]
34. Sáenz-Gamboa JJ, Domenech J, Alonso-Manjarrés A, Gómez JA, de la Iglesia-Vayá M. Automatic semantic segmentation of the lumbar spine: clinical applicability in a multi-parametric and multi-center study on magnetic resonance images. Artif Intell Med. 2023 Jun 1;140:102559. doi:10.1016/j.artmed.2023.102559. [Google Scholar] [PubMed] [CrossRef]
35. Haq IU, Ali H, Wang HY, Cui L, Feng J. BTS-GAN: computer-aided segmentation system for breast tumor using MRI and conditional adversarial networks. Eng Sci Technol, Int J. 2022;36:101154. doi:10.1016/j.jestch.2022.101154. [Google Scholar] [CrossRef]
36. Luan S, Chen C, Zhang B, Han J, Liu J. Gabor convolutional networks. IEEE Trans Image Process. 2018;9,27:4357–66. doi:10.1109/TIP.2018.2835143. [Google Scholar] [PubMed] [CrossRef]
37. Meijering E, Jacob M, Sarria JCF, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytom Part A. 2004;58:167–76. doi:10.1002/cyto.a.20022. [Google Scholar] [PubMed] [CrossRef]
38. Gharaibeh M, Almahmoud M, Ali MZ, Al-Badarneh A, El-Heis M, Abualigah L, et al. Early diagnosis of alzheimer’s disease using cerebral catheter angiogram neuroimaging: a novel model based on deep learning approaches. Big Data Cogn Comput. 2021 Dec 28;6(1):2. doi:10.3390/bdcc6010002. [Google Scholar] [CrossRef]
39. Wang A, Yan X, Wei Z. ImagePy: an open-source, Python-based and platform-independent software package for bioimage analysis. Bioinformatics. 2018;9,34:3238–40. doi:10.1093/bioinformatics/bty313. [Google Scholar] [PubMed] [CrossRef]
40. Survarachakan S, Pelanis E, Khan ZA, Kumar RP, Edwin B, Lindseth F. Effects of enhancement on deep learning based hepatic vessel segmentation. Electronics. 2021 May 13;10(10):1165. doi:10.3390/electronics10101165. [Google Scholar] [CrossRef]
41. Al Kafri A, Sudirman S, Hussain A, Fergus P, Al-Jumeily Obe D, Al-Jumaily M, et al. A framework on a computer assisted and systematic methodology for detection of chronic lower back pain using artificial intelligence and computer graphics technologies. In: Intelligent computing theories and application. Cham, Switzerland: Springer International Publishing; 2016. vol. 9771, p. 843–54. [Google Scholar]
42. Sudirman S, Al Kafri A, Natalia F, Meidia H, Afriliana N, Al-Rashdan W, et al. Lumbar spine MRI dataset. In: Mendeley Data; 2019. [Google Scholar]
43. Muller D, Soto-Rey I, Kramer F. Towards a guideline for evaluation metrics in medical image segmentation. BMC Res Notes. 2022;15:210. doi:10.1186/s13104-022-06096-y. [Google Scholar] [PubMed] [CrossRef]
44. Rajalakshmi TS, Senthilnathan R. Dataset and performance metrics towards semantic segmentation. Int J Eng Manage Res. 2023;2,13:40–9. doi:10.31033/ijemr.13.1.5. [Google Scholar] [CrossRef]
45. Krithika Alias AM, Suganthi K. Review of semantic segmentation of medical images using modified architectures of UNET. Diagnostics. 2022 Dec 6;12(12):3064. doi:10.3390/diagnostics12123064. [Google Scholar] [PubMed] [CrossRef]
46. Kingma DP. Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980. 2014. [Google Scholar]
47. Masood RF, Taj IA, Khan MB, Qureshi MA, Hassan T. Deep learning based vertebral body segmentation with extraction of spinal measurements and disorder disease classification. Biomed Signal Process Control. 2022 Jan 1;71:103230. doi:10.1016/j.bspc.2021.103230. [Google Scholar] [CrossRef]
48. Lu JT, Pedemonte S, Bizzo B, Doyle S, Andriole KP, Michalski MH, et al. Deep spine: automated lumbar vertebral segmentation, disc-level designation, and spinal stenosis grading using deep learning. In: Machine Learning for Healthcare Conference, 2018 Nov 29; USA: PMLR; p. 403–19. [Google Scholar]
49. Rak M, Steffen J, Meyer A, Hansen C, Tonnies KD. Combining convolutional neural networks and star convex cuts for fast whole spine vertebra segmentation in MRI. Comput Methods Programs Biomed. 2019;177:47–56. doi:10.1016/j.cmpb.2019.05.003. [Google Scholar] [PubMed] [CrossRef]
50. Huang J, Shen H, Wu J, Hu X, Zhu Z, Lv X, et al. Spine explorer: a deep learning based fully automated program for efficient and reliable quantifications of the vertebrae and discs on sagittal lumbar spine MR images. Spine J. 2020;20(4):590–9. doi:10.1016/j.spinee.2019.11.010. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

 Downloads
Downloads
 Citation Tools
Citation Tools