 Open Access
Open Access
ARTICLE
Three-Stage Transfer Learning with AlexNet50 for MRI Image Multi-Class Classification with Optimal Learning Rate
1 School of Computing, Sastra Deemed to be University, Thanjavur, 613401, India
2 Department of Computational Intelligence, SRM Institute of Science and Technology, Kattankulathur, 603203, India
3 Department of AI and Software Engineering, Kangwon National University, Samcheok, 25913, Republic of Korea
4 Department of Electronics, Information and Communication Engineering, Kangwon National University, Samcheok, 25913, Republic of Korea
* Corresponding Author: Woong Cho. Email:
Computer Modeling in Engineering & Sciences 2025, 142(1), 155-183. https://doi.org/10.32604/cmes.2024.056129
Received 15 July 2024; Accepted 11 October 2024; Issue published 17 December 2024
Abstract
In radiology, magnetic resonance imaging (MRI) is an essential diagnostic tool that provides detailed images of a patient’s anatomical and physiological structures. MRI is particularly effective for detecting soft tissue anomalies. Traditionally, radiologists manually interpret these images, which can be labor-intensive and time-consuming due to the vast amount of data. To address this challenge, machine learning, and deep learning approaches can be utilized to improve the accuracy and efficiency of anomaly detection in MRI scans. This manuscript presents the use of the Deep AlexNet50 model for MRI classification with discriminative learning methods. There are three stages for learning; in the first stage, the whole dataset is used to learn the features. In the second stage, some layers of AlexNet50 are frozen with an augmented dataset, and in the third stage, AlexNet50 with an augmented dataset with the augmented dataset. This method used three publicly available MRI classification datasets: Harvard whole brain atlas (HWBA-dataset), the School of Biomedical Engineering of Southern Medical University (SMU-dataset), and The National Institute of Neuroscience and Hospitals brain MRI dataset (NINS-dataset) for analysis. Various hyperparameter optimizers like Adam, stochastic gradient descent (SGD), Root mean square propagation (RMS prop), Adamax, and AdamW have been used to compare the performance of the learning process. HWBA-dataset registers maximum classification performance. We evaluated the performance of the proposed classification model using several quantitative metrics, achieving an average accuracy of 98%.Keywords
Transfer learning has become a crucial component in computer vision, addressing the limitations of traditional machine learning [1], which typically relies on training and testing on datasets with the same feature space. However, in real-world applications, unseen cases often differ significantly from the training data, leading to challenges in model generalization [2]. Transfer learning involves two disjoint domains, the source domain (training) and the target domain (testing). The source domain can consist of multiple datasets, whereas the target domain is used exclusively for testing. Unlike conventional methods [3] where training and testing share the same feature space [4], transfer learning operates with distinct feature spaces for training and testing, reflecting real-world complexities more accurately.
The advantages of transfer learning [5] are listed below:
• Knowledge Transfer: Transfer learning emulates the human visual system by applying knowledge from past experiences to new, unseen cases. This enhances the model’s ability to generalize.
• Versatility: It allows the model to handle diverse scenarios without the need to relearn feature spaces for each new instance, making it adaptable to various applications.
The disadvantages of transfer learning are listed below:
• Complexity: The need to determine which features to learn, when to apply transfer learning, and where it will be most effective introduces complexity.
• Domain Dependence: The success of transfer learning can heavily depend on the similarity between the source and target domains. If they are too dissimilar, the transfer may not be effective.
The approach is superior in real-world scenarios where datasets are often diverse and do not share the same feature space. Transfer learning provides a more robust and flexible solution than traditional methods, which may struggle with unseen cases. By leveraging existing knowledge, transfer learning reduces the need for extensive retraining, saving time and computational resources.
For those focused on model performance, transfer learning offers a way to improve accuracy and generalization in complex, real-world applications. From a research perspective, the challenges of selecting the right features, timing, and application make transfer learning a rich field for exploration and innovation. In practical applications, transfer learning’s ability to adapt to new data with minimal retraining makes it an attractive choice for industries that require flexible and scalable solutions.
In summary, transfer learning stands out as a powerful tool in computer vision, offering significant advantages in handling diverse and unseen cases, though it comes with its own set of challenges that need careful consideration.
To find tumors, the feature analysis on MRI is used in the neurology field [6,7]. In a diagnosis using MRI, a large skilled human interpretation is required. The diagnosis can be made with small size new feature set. This proposed method aims to apply transfer learning to classify the MRI images with different sources and target domains with different feature spaces. So, it is not required to apply feature selection and feature reduction processes on the source domain.
Tumors from MRI images can be segmented using an automated technique proposed by Alhassan et al. [8]. This technique has preprocessing and segmentation techniques for clustering and extending the range of data to separate benign from malignant tumors or tissue. This work proposes the clustering approach of the Bat Algorithm with Fuzzy C-ordered means (BAFCOM) as a current learning-based approach for processing the automatic segmentation in multimodal MRI images to find brain cancers. Using the Bat technique, the BAFCOM clustering technique finds the starting centroids and distances inside the pixels. By measuring the distance between the tumor Region of Interest (RoI) and the non-tumor RoI, this technique also obtains the tumor. Using the Enhanced Capsule Networks (ECN) processing method, the MRI image was next categorized as either normal or a brain tumor.
Glory et al. [9] presented a Deep Neural Network (DNN) architecture combining MobilenetV2 and U-net. It takes advantage of both the local and universal contextual features of 2D MRI FLAIR images. Encoders and decoders are used in the proposed network design. Various performance indicators have been calculated, such as accuracy, Intersection Over Union (IOU), and dice loss. Brain tumor identification, evaluation, and treatment rely heavily on automated 3D MRI segmentation. This project aims to use Brain Tumor Image Analysis (BraTumIA) to do 3D volumetric segmentation.
Cristin et al. [10] introduced a reliable classification technique called fractional-chicken swarm optimization (fractional-CSO) to classify the severity degree of malignancies. This study integrates the derivative factor with the behavior of the chicken swarm to improve the precision of severity level classification. Optimal outcomes are achieved by fine-tuning the positioning of the rooster, which is determined by a superior measure of physical prowess. The classification of cancer is performed following the preprocessing of brain images and the effective extraction of characteristics. In addition, the proposed fractional-CSO algorithm is utilized to train a deep recurrent neural network, which is employed to classify the severity level of malignancies.
Using 3D MR and 2D ultrasound (US) data, Qiao et al. [11] proposed the MRI-US multi-modality network (MUM-Net) to categorize breast tumors into various categories. Our explicit distillation of modality-agnostic variables for tumor classification is the main finding of MUM-Net. To be more precise, the authors use min-max training techniques and a discrimination-adaption module to separate features into modality-specific and modality-agnostic ones. Then, using an affinity matrix and the selection of the nearest neighbor, they presented a feature fusion module to improve the compactness of the modality-agnostic features. They created a paired MRI-US breast tumor classification dataset of 502 samples and three medical indications to validate the suggested strategy.
An improved strategy for classifying Schizophrenia (SCZ) and Healthy Controls (HC) using individual hierarchical brain networks created from structural MRI images was put forth by Chawla et al. [12]. Individual hierarchical networks are built using this technique, with each node and edge signifying an ROI and the correlation between two ROIs, respectively. The authors show that edge features significantly improve SCZ/HC classification performance compared to node characteristics. Classification performance is further examined by merging edge characteristics with node features using a multiple kernel learning architecture.
Khaliki et al. [13] examined the classification process of transfer learning methods on brain pictures to determine the transfer learning method with the most superior performance. The researchers examined the efficacy of Convolutional Neural Network (CNN) and CNN based models such as Inception V3, EfficientNet B4, and VGG19 on brain pictures. They also explored the use of transfer learning using CNN as a multilayer without employing transfer learning.
Ahmmed et al. [14] developed and optimized two robust frameworks, ResNet 50 and Inception V3, specifically tailored for the categorization of brain MRI images. Expanding on the past achievements of ResNet 50 and Inception V3 in accurately categorizing various medical imaging datasets, our study includes datasets with unique features, such as one with four distinct categories and another with two. The authors have incorporated crucial strategies, such as Early Stopping and ReduceLROnPlateau, to enhance the model by optimizing hyperparameters. This entailed augmenting the model with more layers, exploring different loss functions and learning rates, and integrating dropout layers and regularization techniques to guarantee the convergence of predictions. In addition, the implementation of strategic improvements, such as tailored pooling and regularization layers, has greatly increased the precision of our models, leading to exceptional classification accuracy.
Majeed et al. [15] and his colleagues utilized a MobileNetV3 model that is optimized for mobile CPU usage to extract features and transfer information. The authors proceed to develop a model for classifying brain lesions by integrating lightweight DepthWise and PointWise blocks. The study employs a fusion of three datasets that have the same picture structures. The classification performance of this fusion is then compared to both pre-trained and fine-tuned approaches. Table 1 compares the state-of-the-art methods with the methodology and dataset used.
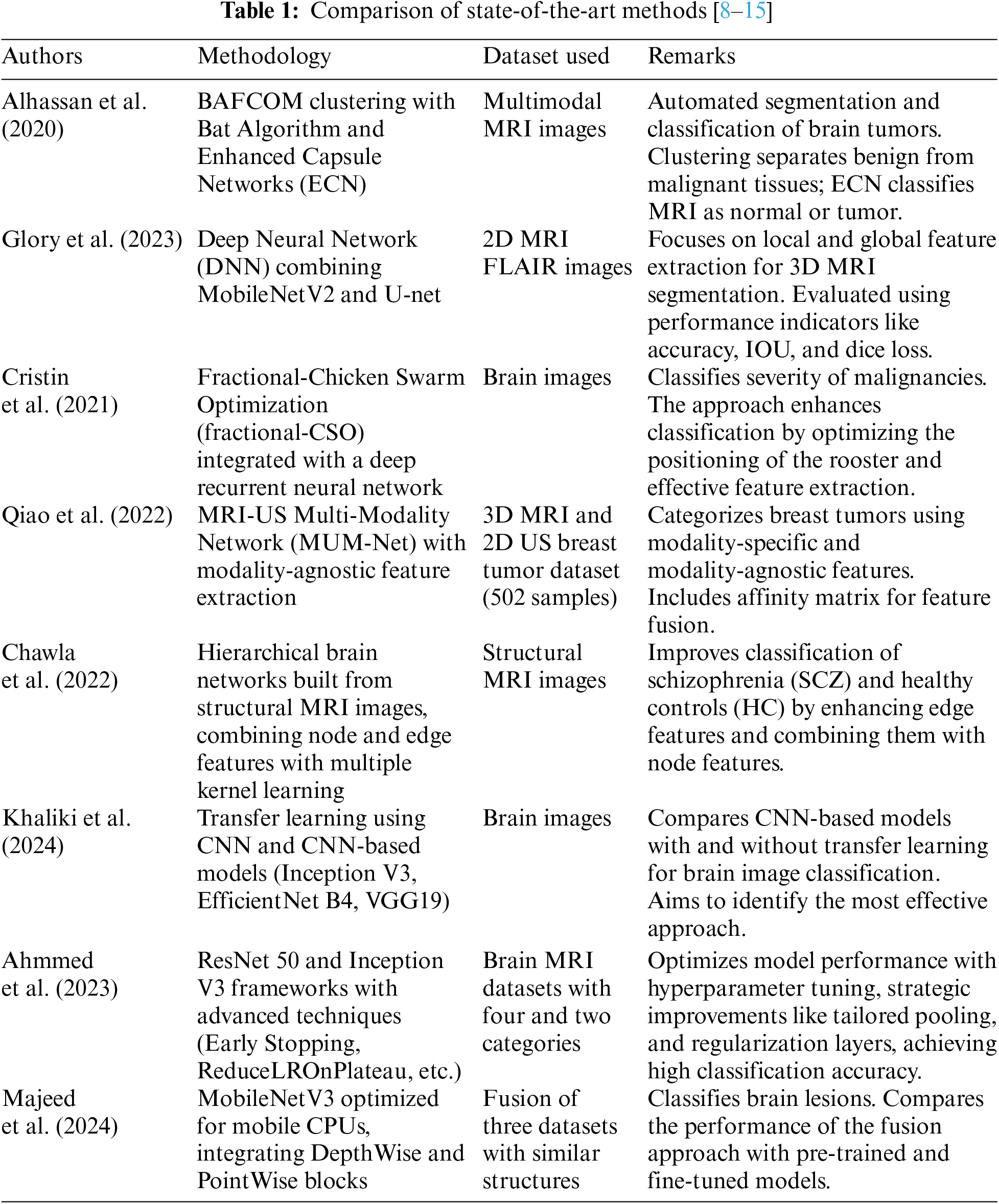
1.3 Challenges and Contributions
MRI data analysis to find tumors is generally the responsibility of the neurology field. However, the process faces a huge obstacle and necessitates a high level of topic knowledge through intensive formal skill acquisition. As a prelude to classification, researchers have suggested feature selection and reduction techniques as distinct approaches to this classification task. The main contributions are:
1. AlexNet 50 is trained with three different datasets and three different learning models: AlexNet 50 with dropped layers and AleNet 50 with augmented source domain.
2. To classify the small target domain, the knowledge that was gained in training the source domain is used. Thus, the knowledge gained in the three architectures is used to classify the images.
3. The performance of the proposed model has been analyzed with different hyperparameter optimizers.
Since this is a supervised learning technique, the initial stage involves loading the image samples together with their corresponding labels or class names. Next in the sequence is batch normalization, which is followed by cross-validation. Cross-validation involves dividing the dataset into separate training and validation sets. To address the issue of overfitting, we have employed diverse data augmentation approaches to generate virtual reproductions of brain MRI scans. This document outlines many approaches for increasing the size of datasets, including zooming, flipping, rotating, mirroring, and other methods.
Transfer learning is the application of prior experience knowledge to new cases. By utilizing their previously acquired model parameters in training data, this method makes the application of deep learning networks that have been pre-trained for novel tasks. Data-driven neural networks restrict access to its secrets to the concealed layers, which function inverted from the output layer. This makes the layers to be dropped selectively. So, this impedes any modifications to their weights and biases during the training process. Transfer learning is advantageous when enormous datasets are used to construct pre-trained models. A decrease in computation time may ensue when a subset of the model is trained as opposed to the entire model.
Transfer learning has three key components. Domain (D), task (T), and marginal characteristics (P) are their names. According to Eqs.(1) and (2), two values are defined, namely the input space (X) and the prior probability (P(X)).
So, in the previous illustration,
where
A transfer learning setting is defined as follows: where the source domain
With A is the space of input and B is the space of labels, consider the classification job T. Assuming two collections of examples from the source domain and the target domain as shown here.
Transfer learning aims to develop a mapping
Let’s say we have two tasks, X and Y. Let A represent the input space,
Fig. 1 gives an overview of the proposed transfer learning method. Initially, it is required to prepare the dataset, which involves loading the brain MRI image data along with any associated class labels. After that, batch normalization and cross-validation are required. Next, batch normalization and cross-validation, separated into training sets and validation sets, must be performed. We have created virtual replicas of brain MRI scans using a variety of data augmentation techniques to combat overfitting. This contains techniques to up-sample the datasets, such as image zoom, image flip, rotation, and image mirroring. The transfer learning with three stages is shown in Fig. 2.

Figure 1: Proposed framework

Figure 2: Transfer learning with AlexNet50
The AlexNET50 model has a zero padding layer followed by five convolution stages and a pooling layer with a fully connected layer. The first stage has a convolution layer, batch normalization, ReLu activation layer, and a max pooling layer. Stage 2 and Stage 3 have a convolution block and an identity block with different dimensions. The knowledge obtained through these features will be transferred for classification.
Hyper-parameter adjustment is essential for improving model performance but can be time-consuming. We employed the learning rate optimizer technique to improve model generalization performance to discover a stable set of learning rates [16]. The learning rate represents the step size utilized to estimate the training weight of the model. This impacts the pace of convergence. If the learning rate is too low, convergence to the error surface’s optimum takes a long time, and only minor adjustments to the model weights are made. The optimization method shoots over the minimum when the learning rate is very high, causing divergence and reducing model performance. The process used to determine the step size critically influences performance in out-of-sample generalization.
In the proposed work, hyperparameter optimizers like Adam, Stochastic Gradient Descent (SGD), Root Mean Square (RMS) prop, Adamax and AdamW are used. By default, the Adam optimizer is used, and others are compared with it. Adam is a technique used for efficient stochastic optimization that relies solely on first-order gradients and has minimal memory requirements. Stochastic optimization refers to the act of maximizing an objective function while taking into account the existence of random elements. The SGD optimizer is highly effective when dealing with large amounts of data and a high number of parameters. During each step, SGD computes an approximation of the gradient by using a randomly selected subset of the data, known as a mini-batch, unlike Gradient Descent, which takes into account the complete dataset at every iteration.
RMSProp adjusts the learning rate by considering the average of the recent gradient magnitudes. RMSProp calculates and stores the average of the squared gradients. Therefore assigning greater significance to more recent changes in gradients. AdamW optimization is a variant of stochastic gradient descent that incorporates an adaptive estimate of first-order and second-order moments, together with a weight decay methodology. Adamax is a first-order optimization approach that is derived from Adam and is based on the infinite norm. Because it can adapt the learning rate according to the features of the data, it is well-suited for learning processes that change over time.
This research uses three datasets to analyze the performance of transfer learning. The first dataset is the Harvard whole brain atlas (HWBA-dataset) [17] with five classes of T2-weighted contrast-enhanced images. The classes are normal, neoplastic, degenerative, inflammatory, infectious, and cerebrovascular diseases, with 65, 277, 223, 189, and 376 slices, respectively. Brain MRI dataset from the School of Biomedical Engineering of Southern Medical University (SMU-dataset) [18], which includes 3064 samples of contrast-enhanced T1-weighted images of 233 research participants. There are three classes in this dataset. They are Meningioma, Glioma, and Pituitary tumors with 708, 1426, and 930 slices, respectively. The National Institute of Neuroscience and Hospitals brain MRI dataset (NINS-dataset) [18], and the Computer Science and Engineering Department, University of Bangladesh, collaborated to curate the third dataset. There are 37 categories and 5285 T1-weighted, contrast-enhanced brain MRI pictures in total.
4 Result Analysis and Discussion
The Anaconda runtime environment was used to test the suggested strategy. It is suitable for massively parallel machine learning (ML) and deep learning (DL) model training since it has faster GPUs, more RAM, and more disc space. We used Python 3.6 and other Python libraries like TensorFlow and Keras to implement the suggested models. Our dataset was loaded using the OpenCV library, and its division and results were computed using the scikit-learn package. Also utilized was Matplotlib to display the plots. The primary component specifications of the computer also include the following: Intel Core i5 processor clocked at 2.40 GHz, NVIDIA Quadro RTX 3000 graphics card, 32 GB of RAM, and more parts. x64-based CPU operating system type and 64-bit operating system. The classification process aims to find the maximum log-likelihood, associating a larger probability mass with the correct class and a smaller probability mass with the incorrect class. As a result, the loss and cost functions are defined as follows:
where s is the probabilities of the class in unnormalized logarithmic form.
4.1 Analysis of Dataset 1: Harvard Whole Brain Atlas (HWBA-Dataset)
The HWBA-dataset has five classes of MRI brain images. Table 2 summarises the classes and their subcategories. The transfer learning model has three stages. Retraining of AlexNet50, training with augmented dataset, and retraining AlexNet50 with augmented dataset. The accuracy of the proposed framework is evaluated with a learning rate of 0.5, and the confusion matrix in Stage 1 is shown in Fig. 3.
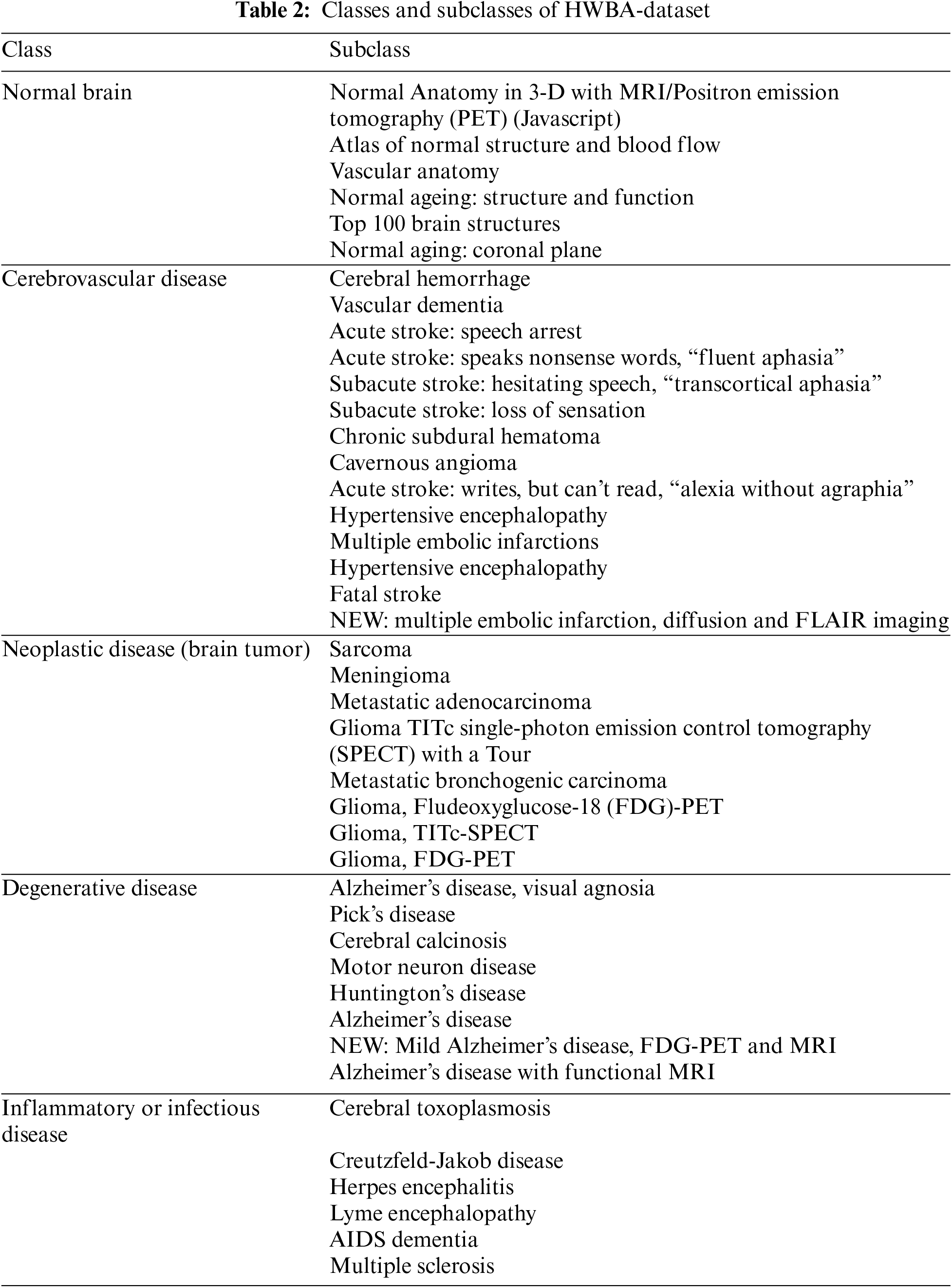

Figure 3: Confusion matrix for the HWBA-dataset in Stage 1
The results show that the overall accuracy is 84.5% with a precision of 82.2%, recall of 84.4%, and F1-score of 82.6%. The top 5 misclassified samples in Stage 1 are shown in Fig. 4. The Receiver Operating Characteristic (ROC) curve is an important performance metric to analyze the classwise classification performance [19,20]. Obtained with the default optimizer Adam has been shown in Fig. 5 for Stage 1. The class normal (Class 0) registers an area under curve (AUC) with 1 and the classes degenerative disease (Class 1), neoplastic disease (Class 2), inflammatory infectious disease (Class 3), and cerebrovascular disease (Class 4) with an AUC of 0.96, 0.96, 0.97 and 0.98, respectively.

Figure 4: Top 5 misclassified samples of the HWBA-dataset in Stage 1

Figure 5: ROC curve for HWBA-dataset in Stage 1
The accuracy of the HWBA-dataset in Stage 2 is 87.43%. The precision of the same is 84.8%, recall is 87.2%, and F1-score is 85.6%. The confusion matrix of Stage 2 classification is shown in Fig. 6. The top misclassified samples in Stage 2 are shown in Fig. 7. The normal class (Class 0) has an AUC of 1, while the degenerative disease class (Class 1), neoplastic disease class (Class 2), inflammatory infectious disease class (Class 3), and cerebrovascular disease class (Class 4) have AUC values of 0.96, 0.95, 0.96, and 0.98 correspondingly as shown in Fig. 8.

Figure 6: Confusion matrix for the HWBA-dataset in Stage 2

Figure 7: Top 5 misclassified samples of HWBA-dataset in Stage 2

Figure 8: ROC curve for HWBA-dataset in Stage 2
The confusion matrix of Stage 3 is shown in Fig. 9. The accuracy of the HWBA-dataset in Stage 3 is 97.96%. The precision is 96.6%, recall is 97.4%, and the F1-score is 97.4%. The top misclassified samples are shown in Fig. 10. The normal class (Class 0) has an AUC of 1, while the degenerative disease class (Class 1), neoplastic disease class (Class 2), inflammatory infectious disease class (Class 3), and cerebrovascular disease class (Class 4) have AUC values of 0.96, 0.95, 0.97, and 0.98 correspondingly as shown in Fig. 11. The training accuracy of transfer learning with AlexNet50, ResNet 50, VGG16 and ResNet32 are shown in Fig. 12. Sixty epochs were run in total, and each stage of transfer learning will be run in each of the 20 epochs.

Figure 9: Confusion matrix for the HWBA-dataset in Stage 3

Figure 10: Top 5 misclassified samples of HWBA-dataset in Stage 3

Figure 11: ROC curve for HWBA-dataset in Stage 3

Figure 12: Training accuracy for the HWBA-dataset through three stages
4.2 Analysis of Dataset 2: School of Biomedical Engineering of Southern Medical University Brain MRI Dataset (SMU-Dataset)
SMU-dataset has three classes with 5285 slices of MRI images. This dataset is also used to train the model with 60 epochs, 20 epochs for each stage of transfer learning. The confusion matrix for Stage 1 is shown in Fig. 13. Stage 1 ensures the classification with 96% of precision with 95.6%, recall of 96% and F1-score of 95.6% The overall accuracy is 98.07% with a precision of 97%, recall of 98%, and F1-score of 98.8%. The top misclassified samples are shown in Stage 2 and are listed in Fig. 14. The Meningioma class (Class 0) has an AUC of 0.96, while the Glioma class (Class 1) registers an AUC of 0.98 and Pitutary (Class 2) with 0.99 as shown in Fig. 15. The overall accuracy of the Stage 2 classification of the SMU dataset is 97.2%. The confusion matrix of Stage 2 is shown in Fig. 16. Precision, recall, and F1-score achieved are 96.6%, 97.33% and 97%, respectively. The top misclassified samples in Stage 2 are shown in Fig. 17. The Meningioma class (Class 0) has an AUC of 0.97, while the glioma class (Class 1) registers an AUC of 0.98 and Pituitary with 0.99, as shown in Fig. 18.

Figure 13: Confusion matrix of SMU-dataset in Stage 1

Figure 14: Top 5 misclassified samples of SMU-dataset in Stage 1

Figure 15: ROC curve for SMU dataset in Stage 1

Figure 16: Confusion matrix of SMU-dataset in Stage 2

Figure 17: Top 5 misclassified samples of SMU-dataset in Stage 2

Figure 18: ROC curve for SMU dataset in Stage 2
In Stage 3, the SMU dataset achieves 98.08% accuracy. The other metrics are 97% of precision, recall of 98.8%, and F1-score of 98.9%. The confusion matrix of Stage 3 classification is shown in Fig. 19. The top 5 misclassified samples in Stage 3 are shown in Fig. 20. The Meningioma class (Class 0) has an AUC of 0.97, while the glioma class (Class 1) registers an AUC of 0.98 and Pituitary with 0.98, as shown in Fig. 21. The accuracy obtained in the three stages using the state-of-the-art CNN models is shown in Fig. 22.

Figure 19: Confusion matrix of SMU-dataset in Stage 3

Figure 20: Top 5 misclassified samples of SMU dataset in Stage 3

Figure 21: ROC curve for SMU dataset in Stage 3

Figure 22: Training accuracy for SMU-dataset
4.3 Analysis of Dataset 3: NINS Brain MRI Dataset (NINS-Dataset)
The NINS dataset contains 5285 T1-weighted MRI images divided into 37 categories. In total, 60 epochs are run, with 20 epochs for each stage.
Stage 1 achieved 82% of accuracy with 81% of precision, 82% of recall, and 81% of F1-score. The confusion matrix of the Stage 1 classification with the NINS dataset is shown in Fig. 23. The misclassified samples in Stage 1 are shown in Fig. 24. Fig. 25 shows the classwise ROC in Stage 1, the AUC values are listed in labels.

Figure 23: Confusion matrix of NINS-dataset in Stage 1

Figure 24: Top 5 misclassified samples of NINS-dataset in Stage 1

Figure 25: ROC curve for NINS dataset in Stage 1
Stage 2 achieved 87% of accuracy with 85% of precision, 86% of recall, and 84% of F1-score. The confusion matrix of the Stage 1 classification with the NINS dataset is shown in Fig. 26. The misclassified samples in Stage 2 are shown in Fig. 27. The classification of the NINS dataset in Stage 3 has a 92% accuracy with 90% precision, 90% recall, and 92% of F1-score. Fig. 28 shows the classwise ROC in Stage 2, the AUC values are listed in labels.
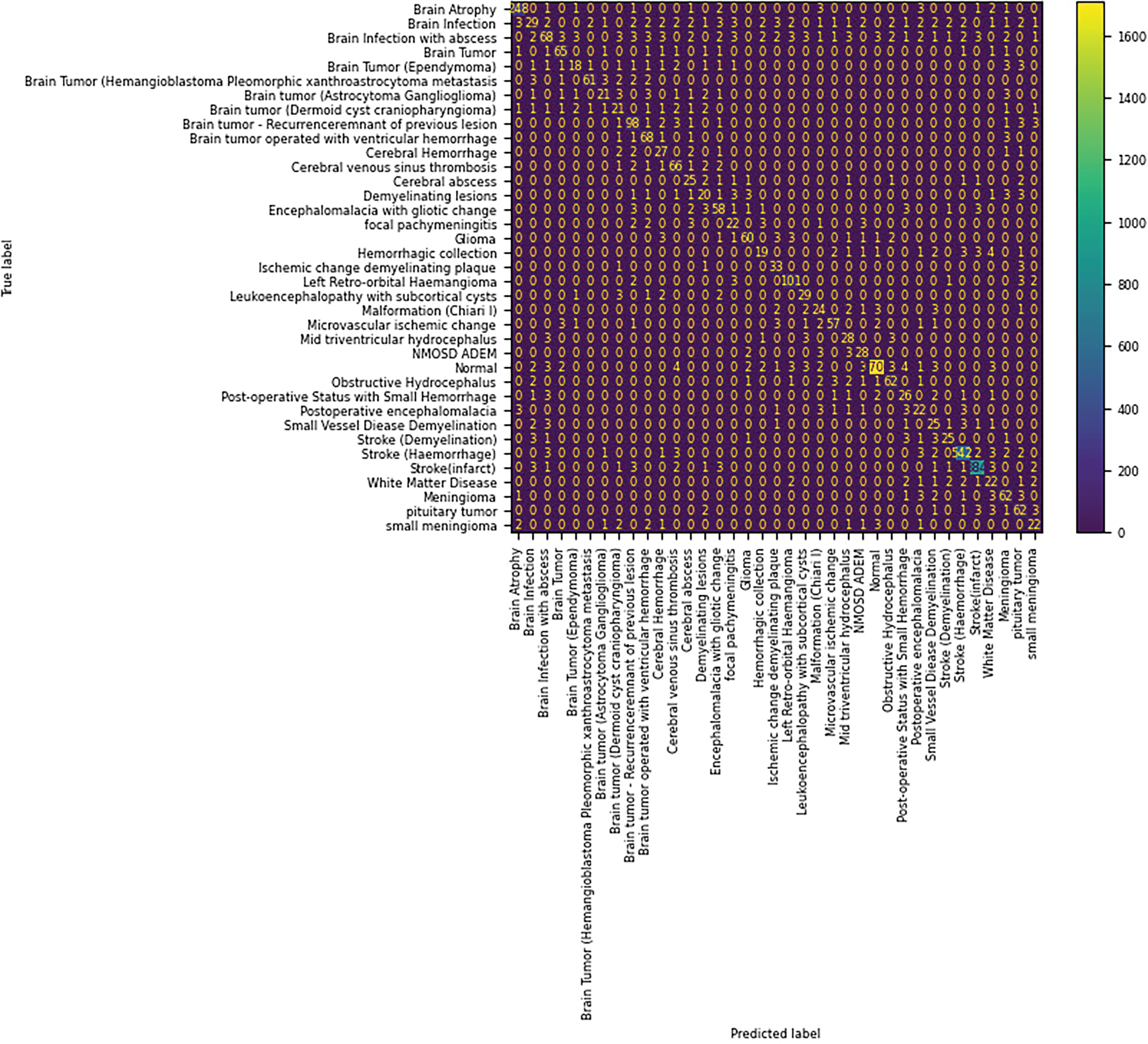
Figure 26: Confusion matrix of NINS-dataset in Stage 2

Figure 27: Top 5 misclassified samples of NINS-dataset in Stage 2

Figure 28: ROC curve for NINS dataset in Stage 2
The confusion matrix of the Stage 3 classification with the NINS dataset is shown in Fig. 29. The misclassified samples in Stage 3 are shown in Fig. 30. Fig. 31 shows the classwise ROC in Stage 3, the AUC values are listed in labels.

Figure 29: Confusion matrix of NINS-dataset in Stage 3

Figure 30: Top 5 misclassified samples of NINS-dataset in Stage 3
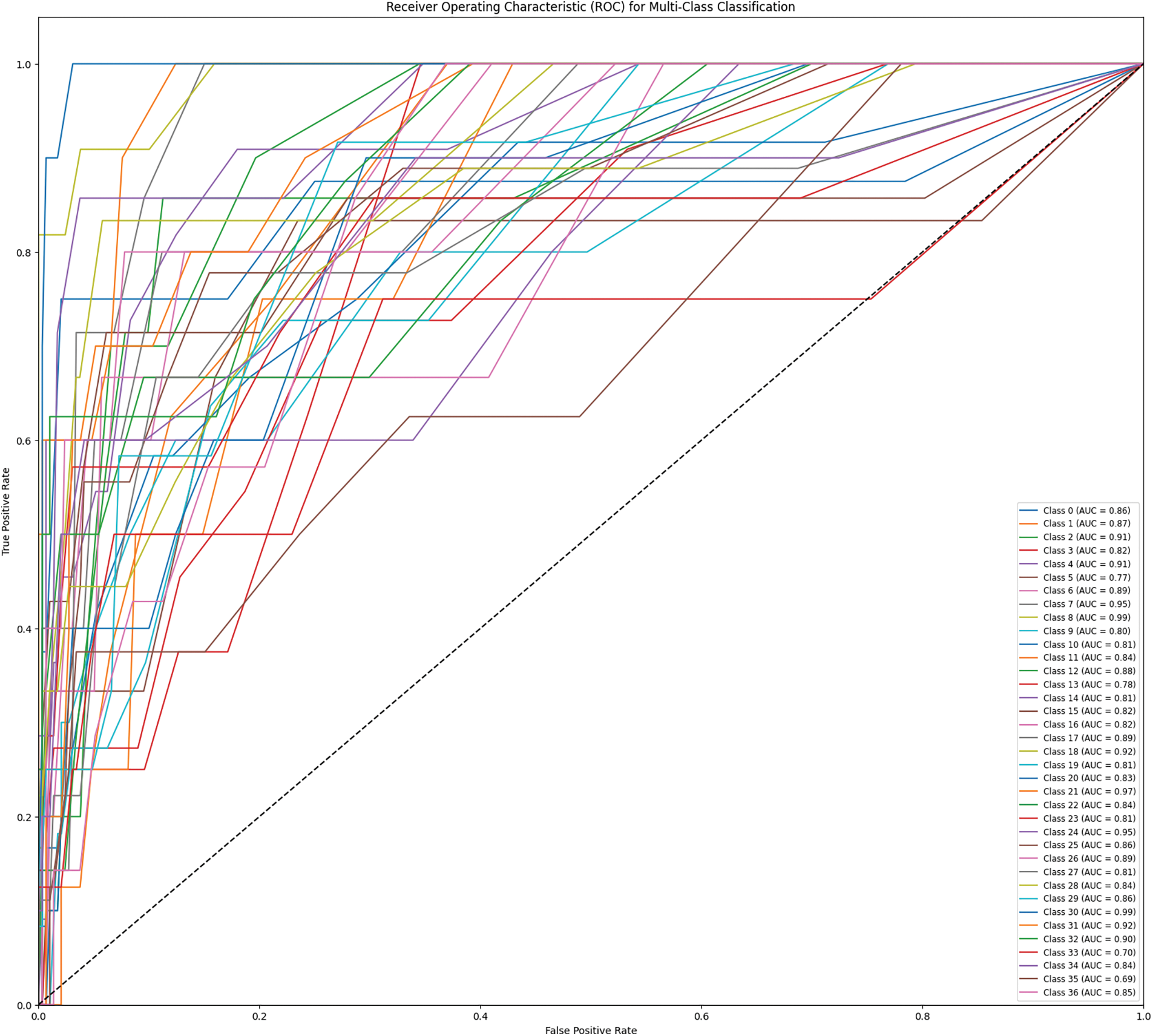
Figure 31: ROC curve for SMU dataset in Stage 3
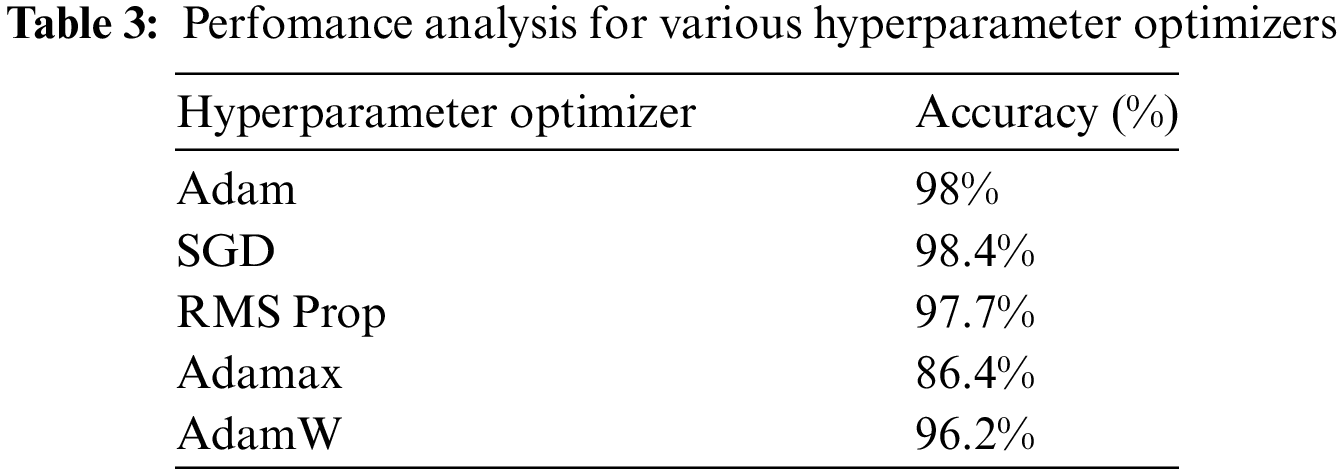
The performance of various hyperparameter optimizers has been discussed in Table 3. The AlexNet50 has 62,378,344 learnable parameters. The CNN model with a frozen convolution layer has 58,631,144 learnable parameters. The proposed model has been analyzed with the default optimizer Adam. Similarly, the other optimizers were also used, and the performance has been analyzed in terms of accuracy. It registers that the SGD optimizer registers a maximum average accuracy of 98.4%. Adamax registers the poor performance with 86% of accuracy as it suits well for the data that change over time.
To illustrate the performance of convolution layers, the Gradcam analysis is performed on the trained model. The super-imposed feature sets are shown in Fig. 32.

Figure 32: (a) Sample image from SMU-dataset, (b) Super-imposed heat map of (a), (c) Sample image from HWBA-dataset, (d) Super-imposed heat map of (c), (e) Sample image from NINS-dataset, (f) Super-imposed heat map of (e)
Besides the deep learning models, the proposed method was compared with the traditional image processing algorithms. It is compared with the MRI binary classification algorithm [21], which uses a multi-level wavelet transform-based feature extraction with a Support Vector Machine (SVM) classifier. The proposed method outperforms with 98% accuracy, whereas the existing one obtained 96%. On the other hand, the SVM-based MRI classifier [22] with feature extraction using Discrete Wavelet Transform (DWT), curvelet transform, and shearlet transform with particle swarm optimization obtained an accuracy of 97.3% with an MRI dataset with 612 samples using Shearlet Transform. In another diagnosis process [23] with Discrete Wavelet Packet Transform (DWPT), multi-level entropies for binary MRI classification achieved 99.33% accuracy using a dataset of 255 brain MRI images. The proposed method achieved 99.2% of accuracy on the same dataset.
Another MRI classifier [21] with DWT and principal component analysis (PCA) for dimensionality reduction was compared with the proposed method with 101 MRI samples. The proposed method achieves 99% of accuracy, whereas the other method achieved 98.6%. The MRI image classifier [24] with a pipeline of Stationary Wavelet Transform for feature extraction, PCA with Particle swarm optimization (PSO), and Artificial Vee Colony to classify MRI brain images obtained an accuracy of 99.45%. Where the proposed method achieved 99.5% of accuracy. The proposed method is compared with a traditional histogram equalization-based image enhancement-based diagnosis system [24], and the existing method reported 99.45% of accuracy on 255 MRI images. The proposed method reported 99.2% of accuracy. The recent transfer learning method [25] for binary classification Transfer Learning to perform MRI tumor classification used ResNet 50 on the three datasets that are used in this paper. The HWBA-dataset reported 84.4% accuracy, the NINS dataset 93.8% [26] and the Biomedical School of Engineering obtained 97.05% accuracy.
Deep Transfer Learning has demonstrated its efficacy in terms of performance metrics and exhibits the ability to quickly adapt an architecture for tackling a specific problem rather than building one from the ground up. Transfer learning, a significant area of deep learning research, emphasizes the ability to quickly adapt models to various challenges by leveraging transferable and invariant underlying information. We conducted three case studies using publicly accessible datasets. To address the limitations of the small-sized dataset in the multi-class classification problem, our research employed data augmentation techniques. The AlexNet 50 model has acquired knowledge from both original and augmented data, namely from the frozen convolution layers. This knowledge is now being transferred for further learning but without the frozen layers. The inadequate learning performance in certain classes of the NINS dataset limits the suggested study. The reason for this is the resemblance between those classes, which leads to an increase in both false positive and false negative instances. We have successfully showcased the viability of employing transfer learning as a potential solution for the multi-class classification challenge in brain MRI, regardless of the number of classes involved. The performance has been evaluated using different hyperparameter optimizers. In the future, Progressive Learning will facilitate the continual enhancement of MRI classification models by transferring information from easier classification tasks to more intricate MRI datasets, hence improving diagnosis accuracy without necessitating complete retraining. This methodology can likewise be expanded to adaptively assimilate MRI data adaptively, facilitating real-time model changes when fresh patient information is incorporated, resulting in enhanced accuracy and prompt diagnosis in a dynamic clinical setting.
Acknowledgement: We would like to thank our institutions to permit us to carry out this research. We acknowledge the patients who participated in the data collection process of the datasets used.
Funding Statement: The authors received no specific funds for this study.
Author Contributions: Conceptualization, A. Robert Singh; Data curation, A. Robert Singh and Suganya Athisayamani; Formal analysis, A. Robert Singh; Funding acquisition, Woong Cho; Methodology, A. Robert Singh and Suganya Athisayamani; Project administration, Woong Cho and Gyanendra Prasad Joshi; Resources, Woong Cho and Gyanendra Prasad Joshi; Software, Suganya Athisayamani; Supervision, Woong Cho and Gyanendra Prasad Joshi; Validation, Woong Cho and Gyanendra Prasad Joshi; Visualization, Suganya Athisayamani and Woong Cho; Writing–original draft, A. Robert Singh; Writing–review & editing, Gyanendra Prasad Joshi. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: 1. Harvard Whole Brain Atlas, Available online: https://www.med.harvard.edu/aanlib/home.html (accessed on 10 October 2024); 2. NINS, Available online: https://figsharhe.com/articles/dataset/Brain_MRI_Dataset/14778750/1?file=28399149 (accessed on 10 October 2024); 3. SMU dataset: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427 (accessed on 10 October 2024).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Mohan G, Subashini MM. MRI based medical image analysis: survey on brain tumor grade classification. Biomed Signal Proc and Ctrl. 2018;39:139–61. doi:10.1016/j.bspc.2017.07.007. [Google Scholar] [CrossRef]
2. Panayides AS, Amini AA, Filipovic N, Sharma A, Tsaftaris SA, Young AA, et al. AI in medical imaging informatics: current challenges and future directions. IEEE J Biomed Health Inform. 2020;24(7):1837–57. doi:10.1109/JBHI.2020.2991043. [Google Scholar] [PubMed] [CrossRef]
3. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA: 2016; p. 770–8. [Google Scholar]
4. Qi Y, Guo Y, Wang Y. Image quality enhancement using a deep neural network for plane wave medical ultrasound imaging. IEEE Trans on Ultrasonics Ferroelectrics and Freq Contl. 2021;68(4):926–34. doi:10.1109/TUFFC.2020.3023154. [Google Scholar] [PubMed] [CrossRef]
5. Valverde JM, Imani V, Abdollahzadeh A, de Feo R, Prakash M, Ciszek R, et al. Transfer learning in magnetic resonance brain imaging: a systematic review. J Imaging. 2021;7(4):66. doi:10.3390/jimaging7040066. [Google Scholar] [PubMed] [CrossRef]
6. Franke J, Heinen U, Lehr H, Weber AE, Jaspard F, Ruhm W, et al. System characterization of a highly integrated preclinical hybrid MPI-MRI scanner. IEEE Trans Med Imaging. 2016;35(9):1993–2004. doi:10.1109/TMI.2016.2542041. [Google Scholar] [PubMed] [CrossRef]
7. Singh R, Athisayamani S. Survival prediction based on brain tumor classification using convolutional neural network with channel preference. In: Data Engineering and Intelligent Computing. Singapore: Springer Nature; 2022. [Google Scholar]
8. Alhassan AM, Zainon WMNW. BAT Algorithm with Fuzzy C-Ordered Means (BAFCOM) clustering segmentation and Enhanced Capsule Networks (ECN) for brain cancer MRI images classification. IEEE Access. 2020;8:201741–51. doi:10.1109/ACCESS.2020.3035803. [Google Scholar] [CrossRef]
9. Glory J, Angeline Kirubha SP, Premkumar R, Evangeline IK. Automatic 2D and 3D segmentation of glioblastoma brain tumor. Biomed Engg: Applns, Basis Commun. 2023;35(2):2250055. doi:10.4015/S1016237222500557. [Google Scholar] [CrossRef]
10. Cristin R, Kumar KS, Anbhazhagan P. Severity level classification of brain tumor based on MRI images using fractional-chicken swarm optimization algorithm. Comp J. 2021;64(10):1514–30. doi:10.1093/comjnl/bxab057. [Google Scholar] [CrossRef]
11. Qiao MY, Liu CC, Li Z, Zhou J, Xiao Q, Zhou SC, et al. Breast tumor classification based on MRI-US images by disentangling modality features. IEEE J Biomedi Health Informati. 2022 Jul;26(7):3059–67. doi:10.1109/JBHI.2022.3140236. [Google Scholar] [PubMed] [CrossRef]
12. Chawla R, Beram SM, Murthy CR, Thiruvenkadam T, Bhavani NPG, Saravanakumar R, et al. Brain tumor recognition using an integrated bat algorithm with a convolutional neural network approach. Meas: Sens. 2022;24:100426. doi:10.1016/j.measen.2022.100426. [Google Scholar] [CrossRef]
13. Khaliki MZ, Başarslan MS. Brain tumor detection from images and comparison with transfer learning methods and 3-layer CNN. Sci Rep. 2024 Jan;14(1):2664. doi:10.1038/s41598-024-52823-9. [Google Scholar] [PubMed] [CrossRef]
14. Ahmmed S, Podder P, Mondal MRH, Rahman SMA, Kannan S, Hasan MJ, et al. Enhancing brain tumor classification with transfer learning across multiple classes: an in-depth analysis. Bio Med Inform. 2023;3(4):1124–44. doi:10.3390/biomedinformatics3040068. [Google Scholar] [CrossRef]
15. Majeed AF, Salehpour P, Farzinvash L, Pashazadeh S. Multi-class brain lesion classification using deep transfer learning with MobileNetV3. IEEE Access. Early Access. 2024;12:155295–308. doi:10.1109/ACCESS.2024.3413008. [Google Scholar] [CrossRef]
16. Sommer C, Straehle C, Köthe U, Hamprecht FA. Ilastik: interactive learning and segmentation toolkit. In: IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2011; Chicago, IL, USA. p. 230–3. [Google Scholar]
17. Johnson KA and Becker JA. Harvard whole brain atlas. Available from: www.med.harvard.edu/AANLIB/home.html. [Accessed 2023]. [Google Scholar]
18. Cheng J. Brain tumor dataset. Available from: https://doi.org/10.6084/m9.figshare.1512427.v5. [Accessed: 2023]. [Google Scholar]
19. Hajian-Tilaki K. Receiver Operating Characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 2013;4(2):627–35. [Google Scholar] [PubMed]
20. Nahm F. Receiver operating characteristic curve: overview and practical use for clinicians. Korean J Anesthesiol. 2022;75(1):25–36. doi:10.4097/kja.21209. [Google Scholar] [PubMed] [CrossRef]
21. Chaplot S, Patnaik LM, Jagannathan NR. Classification of magnetic resonance brain images using wavelets as input to support vector machine and neural network. Biomed Sig Proc and Ctrl. 2006;1(1):86–92. doi:10.1016/j.bspc.2006.05.002. [Google Scholar] [CrossRef]
22. Zhang Y, Dong Z, Wang S, Ji G, Yang J. Preclinical diagnosis of Magnetic Resonance (MR) brain images via discrete wavelet packet transform with tsallis entropy and Generalized Eigenvalue Proximal Support Vector Machine (GEPSVM). Entropy. 2015;17(4):1795–813. doi:10.3390/e17041795. [Google Scholar] [CrossRef]
23. Wang S, Zhang Y, Dong Z, Du S, Ji G, Yan J, et al. Feed-forward neural network optimized by hybridization of PSO and ABC for abnormal brain detection. Img Sys and Tech. 2015;25(2):1795–813. doi:10.1002/ima.22132. [Google Scholar] [CrossRef]
24. Nayak DR, Dash R, Majhi B. Brain MR image classification using two-dimensional discrete wavelet transform and AdaBoost with random forests. Neuro Comp. 2016;177:188–97. doi:10.1016/j.neucom.2015.11.034. [Google Scholar] [CrossRef]
25. Shen C, Gonzalez Y, Chen L, Jiang SB, Jia X. Intelligent parameter tuning in optimization-based iterative CT reconstruction via deep reinforcement learning. IEEE Trans Med Imaging. 2018;37(6):1430–9. doi:10.1109/TMI.2018.2823679. [Google Scholar] [PubMed] [CrossRef]
26. Brima Y, Tushar MHK, Kabir U, and Islam T. Brain MRI Dataset. Available from: https://doi.org/10.6084/m9.figshare.14778750.v2. [Accessed 2023]. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools