 Open Access
Open Access
ARTICLE
Segmentation of Head and Neck Tumors Using Dual PET/CT Imaging: Comparative Analysis of 2D, 2.5D, and 3D Approaches Using UNet Transformer
1 Information and Computer Science Department, College of Computer Science and Engineering, University of Ha’il, Ha’il, 55476, Saudi Arabia
2 Software Engineering Department, College of Computer Science and Engineering, University of Ha’il, Ha’il, 55476, Saudi Arabia
3 Computer Engineering Department, College of Computer Science and Engineering, University of Ha’il, Ha’il, 55476, Saudi Arabia
4 Department of Computer and Information Sciences, Universiti Teknologi Petronas, Seri Iskandar, 32610, Malaysia
5 Center for Research in Computer Vision (CRCV), University of Central Florida, Orlando, FL 32816, USA
* Corresponding Author: Rizwan Qureshi. Email:
(This article belongs to the Special Issue: Artificial Intelligence Emerging Trends and Sustainable Applications in Image Processing and Computer Vision)
Computer Modeling in Engineering & Sciences 2024, 141(3), 2351-2373. https://doi.org/10.32604/cmes.2024.055723
Received 05 July 2024; Accepted 11 September 2024; Issue published 31 October 2024
Abstract
The segmentation of head and neck (H&N) tumors in dual Positron Emission Tomography/Computed Tomography (PET/CT) imaging is a critical task in medical imaging, providing essential information for diagnosis, treatment planning, and outcome prediction. Motivated by the need for more accurate and robust segmentation methods, this study addresses key research gaps in the application of deep learning techniques to multimodal medical images. Specifically, it investigates the limitations of existing 2D and 3D models in capturing complex tumor structures and proposes an innovative 2.5D UNet Transformer model as a solution. The primary research questions guiding this study are: (1) How can the integration of convolutional neural networks (CNNs) and transformer networks enhance segmentation accuracy in dual PET/CT imaging? (2) What are the comparative advantages of 2D, 2.5D, and 3D model configurations in this context? To answer these questions, we aimed to develop and evaluate advanced deep-learning models that leverage the strengths of both CNNs and transformers. Our proposed methodology involved a comprehensive preprocessing pipeline, including normalization, contrast enhancement, and resampling, followed by segmentation using 2D, 2.5D, and 3D UNet Transformer models. The models were trained and tested on three diverse datasets: HeckTor2022, AutoPET2023, and SegRap2023. Performance was assessed using metrics such as Dice Similarity Coefficient, Jaccard Index, Average Surface Distance (ASD), and Relative Absolute Volume Difference (RAVD). The findings demonstrate that the 2.5D UNet Transformer model consistently outperformed the 2D and 3D models across most metrics, achieving the highest Dice and Jaccard values, indicating superior segmentation accuracy. For instance, on the HeckTor2022 dataset, the 2.5D model achieved a Dice score of 81.777 and a Jaccard index of 0.705, surpassing other model configurations. The 3D model showed strong boundary delineation performance but exhibited variability across datasets, while the 2D model, although effective, generally underperformed compared to its 2.5D and 3D counterparts. Compared to related literature, our study confirms the advantages of incorporating additional spatial context, as seen in the improved performance of the 2.5D model. This research fills a significant gap by providing a detailed comparative analysis of different model dimensions and their impact on H&N segmentation accuracy in dual PET/CT imaging.Keywords
Head and neck (H&N) cancers represent a significant health challenge worldwide, necessitating advanced diagnostic and treatment approaches to improve patient outcomes [1–5]. Positron Emission Tomography (PET) combined with Computed Tomography (CT) imaging offers a powerful modality for the accurate detection and characterization of tumors in this region [5,6]. The integration of metabolic and anatomical information through PET/CT imaging facilitates enhanced tumor localization, staging, and treatment planning [7].
Despite the advantages, accurate and automated segmentation of tumors in PET/CT images remains a complex task due to the variability in tumor shapes, sizes, and intensities, as well as the presence of artifacts and noise in the imaging data [8,9]. Traditional segmentation methods often rely on manual delineation by experts, which is time-consuming and subject to inter-observer variability [10]. Consequently, there is a critical need for robust and efficient automated segmentation techniques [11,12].
Recent advancements in deep learning, particularly Convolutional Neural Networks (CNNs) and transformer-based architectures, have demonstrated promising results in medical image analysis [13–16]. The Vision Transformer (ViT) and its variants have shown superior performance in capturing long-range dependencies and contextual information in images. UNetR (UNet with Transformer) leverages the strengths of both CNNs and transformers, providing a powerful framework for medical image segmentation [17].
In the literature, most studies have focused on either 2D or 3D approaches for tumor segmentation [18–20], each with its own set of challenges and benefits. 2D approaches, while computationally efficient, often fail to capture the full spatial context of the tumors [21,22]. On the other hand, 3D approaches provide comprehensive spatial information but are computationally intensive and require large, annotated datasets [23]. The intermediate 2.5D approach, which combines multiple 2D slices, offers a compromise but still lacks the full context provided by 3D models. Given these gaps, our research aims to systematically investigate the performance of 2D, 2.5D, and 3D approaches using the ViTUnet (UNetR) model for the segmentation of head and neck tumors in dual PET/CT images. Our contributions can be summarized as follows:
• We propose a unified ViTUnet (UNetR) framework that innovatively combines the strengths of convolutional neural networks and transformer networks for 2D, 2.5D, and 3D segmentation of head and neck tumors in dual PET/CT imaging.
• We introduce a novel preprocessing pipeline that includes resampling and prior attention mechanisms to enhance the quality of input PET/CT images.
• Our study pioneers the application of a 2.5D segmentation approach within a transformer-based architecture, presenting a new way to capture the intricate spatial relationships in medical imaging data.
The motivation for our work stems from the pressing need for accurate and automated tumor segmentation methods that can be readily integrated into clinical workflows. By leveraging the dual modality of PET/CT imaging and advanced deep learning techniques, we aim to enhance the accuracy and efficiency of head and neck tumor segmentation, ultimately contributing to better patient care and outcomes.
The structure of this paper is systematically divided into five main sections to effectively present our research. Section 2, related work, provides an overview of the existing studies and developments in the field, setting the stage for our research. In Section 3, methods, and materials, we detail the techniques and resources utilized in our study, emphasizing the methodologies specific to PET/CT tumor segmentation. Section 4, results, and discussion is dedicated to showcasing the outcomes of our research, along with a comprehensive analysis and interpretation of these results in the context of dual PET/CT imaging. The paper culminates in Section 5, conclusion, and future work, where we summarize our findings, underscore their significance in medical imaging, and suggest potential avenues for future research.
The rapid advancements in deep learning techniques have significantly impacted the field of oncology, particularly in the automatic analysis of multimodal images. Automation in tumor and lymph node delineation is crucial for diagnostic tasks, staging, quantitative assessment, radiotherapy planning, and outcome prediction, offering substantial benefits in terms of speed, robustness, and reproducibility compared to manual contouring [24–28].
Multimodal image analysis combined with machine learning for patient-level segmentation and outcome prediction enables predictive and prognostic modeling. This approach includes therapy response prediction, recurrence, and overall survival, utilizing image-derived data alongside clinical information to develop decision-support tools that enhance personalized patient management [29,30]. Myronenko et al. [31] developed SegRes-Net, a 3D U-Net-like architecture enhanced with an auto-encoder and deep supervision, based on the MONAI platform. This system is tailored for tasks such as PET/CT analysis and employs the Auto3DSeg system for parameter selection. It integrates various steps, including image normalization, tumor region detection, and isotropic resampling, and employs 5-fold cross-validation along with model ensembling. Their approach leverages anatomical positions for tumor region detection and employs random 3D cropping during training, focusing on foreground classes with designated probabilities for tumors, lymph nodes, and background. Zhu et al. [32] introduced the SDV-TUNet (Sparse Dynamic Volume TransUNet), a 3D magnetic resonance imaging (MRI) brain tumor segmentation network designed to enhance clinical diagnosis and treatment. Unlike traditional methods, which often neglect voxel details and inter-layer connections, SDV-TUNet employs an encoder-decoder architecture combining voxel information and multi-axis features. The Sparse Dynamic (SD) encoder-decoder module extracts global spatial features, while the Multi-Level Edge Feature Fusion (MEFF) module enhances edge information. Zhu et al. [33] proposed a 3D brain tumor segmentation model incorporating multimodal spatial information enhancement and boundary shape correction via three modules: Modality Information Extraction (MIE), Spatial Information Enhancement (SIE), and Boundary Shape Correction (BSC). These modules improve the input, backbone, and loss functions of deep convolutional networks, effectively addressing challenges in modality information utilization, spatial information acquisition, and boundary segmentation. The method, was validated on BraTS2017, 2018, and 2019 datasets.
Sun et al. [34] adopted a multi-stage, coarse-to-fine strategy utilizing a series of neural networks for precise tumor segmentation. Initially, a 3D U-Net identifies the head region in CT scans, followed by nnU-Net performing an initial rough segmentation of the primary and nodal tumors in PET/CT images. The final detailed segmentation within the refined bounding box is achieved through an ensemble of nnU-Nets and nnFormers, further enhanced by a 3D SE-norm U-Net. Jiang et al. [35] employed a standard nnU-Net with simple pre- and post-processing techniques, focusing on cropping images around the primary tumor. Their post-processing includes outlier removal based on criteria such as minimum volume and spatial proximity between predicted tumor volumes. They also integrated their segmentation results into a web-based platform for visualizing segmented regions, including Organs at Risk (OAR). Rebaud et al. implemented a straightforward nnU-Net-based method, adapting it with specific image resampling and training techniques, followed by median filtering to smooth the masks [36]. Salahuddin et al. developed a 3D U-Net with channel-wise attention, grid-attention gates, and specialized residual connections, complemented by outlier removal in post-processing and non-isotropic resampling for the input images [37]. Wang et al. introduced an enhanced nnU-Net with a Transformer model to refine segmentation, particularly focusing on tumor boundaries, using octree decomposition for patch selection [38]. Another study by Wang et al. utilized a simple segmentation approach with nnU-Net, employing a dense patch-based approach and post-processing based on the spatial relation between primary and nodal tumor volumes [39]. Jain et al. compared several deep learning models, including 2D/3D nnU-Net, MNet, and SwinU-Net, using resampled images registered to a common reference and cropped based on skull center location. They achieved promising results with average Dice Similarity Coefficients (DSC) of 0.77 for primary tumors and 0.70 for nodes in the HECKTOR2022 challenge [40]. Chen et al. created an ensemble of three 3D nnU-Nets with different loss functions, utilizing CT images for initial input and PET images in post-processing to refine predictions [41]. Meng et al. employed a multi-step approach with an organ localizer and 3D U-Net for organ segmentation, followed by a 3D ResU-Net for tumor segmentation, using a weighted combination of registered PET and CT images. Meng et al. proposed a novel approach combining a U-Net-based segmentation network with a cascaded survival network built on a DenseNet architecture, enabling simultaneous prediction of patient survival risk scores and tumor region segmentation [42]. Despite these advancements, significant challenges remain, such as handling the high variability in tumor shapes and sizes, the need for large, annotated datasets, and the difficulty in generalizing models across different medical imaging protocols and equipment. Addressing these gaps is essential to enhance the robustness and adaptability of segmentation models. Our study aims to contribute to this ongoing research by systematically investigating 2D, 2.5D, and 3D approaches using ViTUnet (UNetR) for dual PET/CT head and neck tumor segmentation, ultimately improving precision and effectiveness in cancer treatment planning.
The proposed methodology for head and neck tumor segmentation using dual PET/CT imaging involves several key steps, as illustrated in Fig. 1. The dataset consists of PET and CT images of the head and neck region. PET images provide metabolic information, while CT images offer detailed anatomical structures. These complementary modalities are crucial for accurate tumor localization and segmentation. The acquired PET and CT images are subjected to a resampling process to ensure uniform spatial resolution. This step is essential because PET and CT images often have different voxel sizes and resolutions. By resampling, we standardize the voxel dimensions, facilitating the subsequent integration of the two modalities. After resampling, the images are processed using a prior attention mechanism. This step involves highlighting the ROIs that are most likely to contain tumors. The prior attention mechanism leverages anatomical knowledge and intensity-based criteria to focus on potential tumor areas, reducing the search space for the segmentation algorithm. The core of our methodology is the UNetR (UNet with Transformer) model. The resampled and attention-enhanced images are fed into the UNetR architecture, which combines CNNs with transformer blocks. This hybrid model captures both local and global context, enhancing the segmentation accuracy. The UNetR processes the images through a series of convolutional layers, attention mechanisms, and upsampling operations to generate precise segmentation maps. The output of the UNetR model is a detailed segmentation map of the head and neck tumors. To further analyze the segmented regions, we perform shape analysis, which involves assessing the geometric properties of the segmented tumors. This step provides insights into tumor morphology, which can be valuable for clinical decision-making and treatment planning. This methodology outlines the comprehensive process of acquiring, preprocessing, and segmenting PET/CT images to accurately delineate head and neck tumors. The integration of advanced deep learning techniques with dual-modality imaging aims to improve the precision and effectiveness of tumor segmentation in clinical practice.
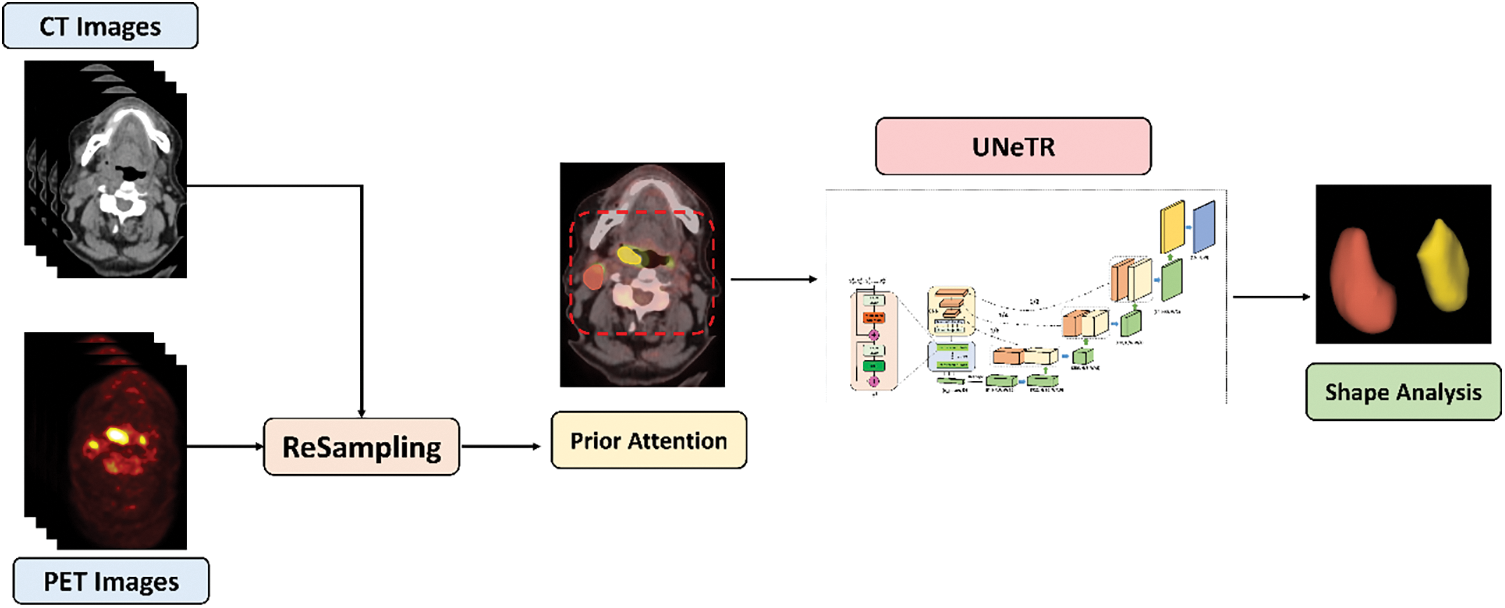
Figure 1: Overview of the proposed methodology for head and neck tumor segmentation using dual PET/CT imaging
In this study, the proposed method was validated on HECKTOR MICCAI 2023 Challenge dataset [43], SegRap2023 dataset [44] and AutoPET2023 dataset [45], (Fig. 2). The HECKTOR Challenge 2023 dataset acquired from nine different centers (Fig. 2). It comprises FDG-PET/CT images from patients diagnosed with H&N cancer, specifically located in the oropharynx region. This diverse and multi-center dataset is crucial for ensuring the robustness and generalizability of the developed models for tumor segmentation. The original annotations for the training and test sets varied across different medical centers.
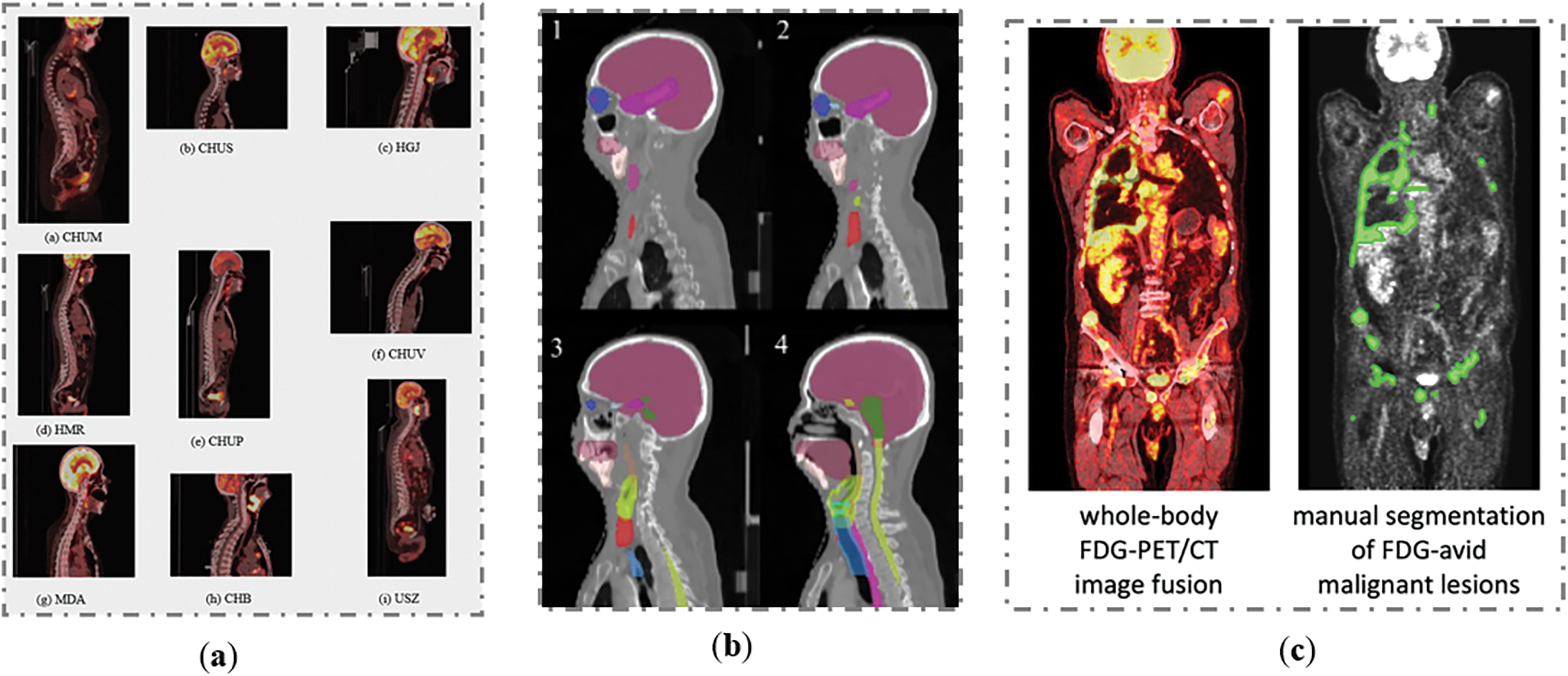
Figure 2: Samples from the H&N dataset: (a) HeckTor2022 dataset, (b) SegRap2023 dataset and (c) AutoPET2023 dataset
In the SegRap2023 dataset [44], a comprehensive collection of CT scans from patients diagnosed with nasopharyngeal carcinoma (NPC) was provided. The segmentation targets in this dataset include Organs at Risk (OARs), Gross Target Volume of the nasopharynx (GTVnx), and Gross Target Volume of the lymph nodes (GTVnd). The dataset comprises CT scans from 200 patients, divided into 120 for training, 20 for validation, and 60 for testing. Each patient has two CT scans: a no-contrast CT and a contrast-enhanced CT, both with pixel-level annotations for GTVnx, GTVnd, and 45 OARs. This extensive and detailed dataset provides a robust foundation for developing and evaluating advanced segmentation models.
The AutoPET2023 dataset [45] comprises PET/CT scans from patients with histologically confirmed malignant melanoma, lymphoma, or lung cancer, as well as negative control patients. These scans were collected from two large medical centers: University Hospital Tübingen and University Hospital of the LMU in Munich, Germany. The PET/CT data were acquired using state-of-the-art PET/CT scanners (Siemens Biograph mCT, mCT Flow, biograph 64, GE Discovery 690) following standardized protocols and international guidelines. The dataset includes 3D volumes of whole-body examinations, typically extending from the skull base to the mid-thigh, with some scans covering the entire body if clinically relevant. Patients fasted for at least 6 h before the injection of 18F-FDG, and whole-body PET/CT images were obtained approximately 60 min post-injection. Diagnostic CT scans were performed with contrast agents, with PET images reconstructed iteratively and smoothed with Gaussian post-reconstruction. The slice thickness for contrast-enhanced CT was 2–3 mm. This dataset provides a robust foundation for developing and evaluating advanced segmentation models in oncological imaging.
In our study, we applied the following preprocessing processes. Normalization is an essential preprocessing procedure that seeks to normalize the intensity distribution among various patients and imaging modalities. The goal is to synchronize the dynamic range of the photos, creating a more consistent dataset that can be easily analyzed by computational models. Mathematically, this might entail the process of z-score normalization, where each voxel intensity Ixyz in a two-dimensional (2D) and three-dimensional (3D) picture is subjected to a transformation:
The term
Contrast enhancement methods [41] are used on each modality to increase the visibility of important elements. We applied a technique designed to improve the visibility of important features within the PET/CT images. The objective of PET scans is to enhance regions with significant radiotracer uptake, which frequently indicates the presence of cancer. CT enhancement methods are designed to improve the visibility of anatomical features. The contrast enhancement transformation function can be expressed as:
where
Cropping narrows down the analysis to the specific Region of Interest (ROI) by eliminating extraneous backdrop and decreasing the computing burden. The technique entails choosing a sub-volume that encompasses the tumor and other anatomical markers that are crucial for diagnosis and planning of therapy. The cropped picture, denoted as Icrop, is determined by spatial limits inside the original volume, referred to as Ioriginal:
where
Voxel spacing homogeneity was used to address the varying resolutions between PET and CT scans. The utilization of voxel spacing homogenization helps to standardize the voxel spacing. This procedure entails adjusting the pictures to have uniform voxel dimensions, which enables precise image fusion and comparison. The process of homogenization may be mathematically expressed by:
where
The function RandomCrop randomly chooses a subvolume and does the RandomFlip operation. RandomFlip utilizes a conditional mirror transformation on a randomly chosen axis. These preprocessing processes are necessary for the precise, replicable, and strong segmentation of tumors. They tackle the natural diversity in multi-modal imaging datasets and improve the quality of the input data, which is crucial for the effectiveness of subsequent deep learning models employed for the segmentation tasks.
In Fig. 3, we present our advanced deep learning framework designed for the segmentation of head and neck tumors within dual PET/CT images, leveraging the synergistic benefits of multimodal imaging. This framework innovatively integrates transformer networks with CNNs, optimizing the pipeline for the complex task of tumor delineation. The process begins with the acquisition of multimodal inputs, where PET and CT images are fused to combine high-resolution anatomical information from CT scans with the functional metabolic data from PET scans. This fusion is critical for accurately identifying regions of neoplastic activity. By using bilinear interpolation for the image data and nearest neighbor interpolation for the masks, we maintain both the detailed structural integrity and the exact categorical distinctions necessary for effective multimodal analysis. These choices facilitate more accurate tumor localization and delineation in the fused images, enhancing the overall quality and reliability of the segmentation results. The multimodality input in Fig. 3 highlights the separate channels for the CT and PET data, as well as their combined representation. The individual CT and PET images are processed through separate pathways to retain their unique information before integration. The core of our methodology is the UNetR (UNet with Transformer) model. The input begins with a series of patches extracted from the PET/CT fusion volume. Each input patch undergoes normalization followed by a multi-head attention mechanism within the transformer block.
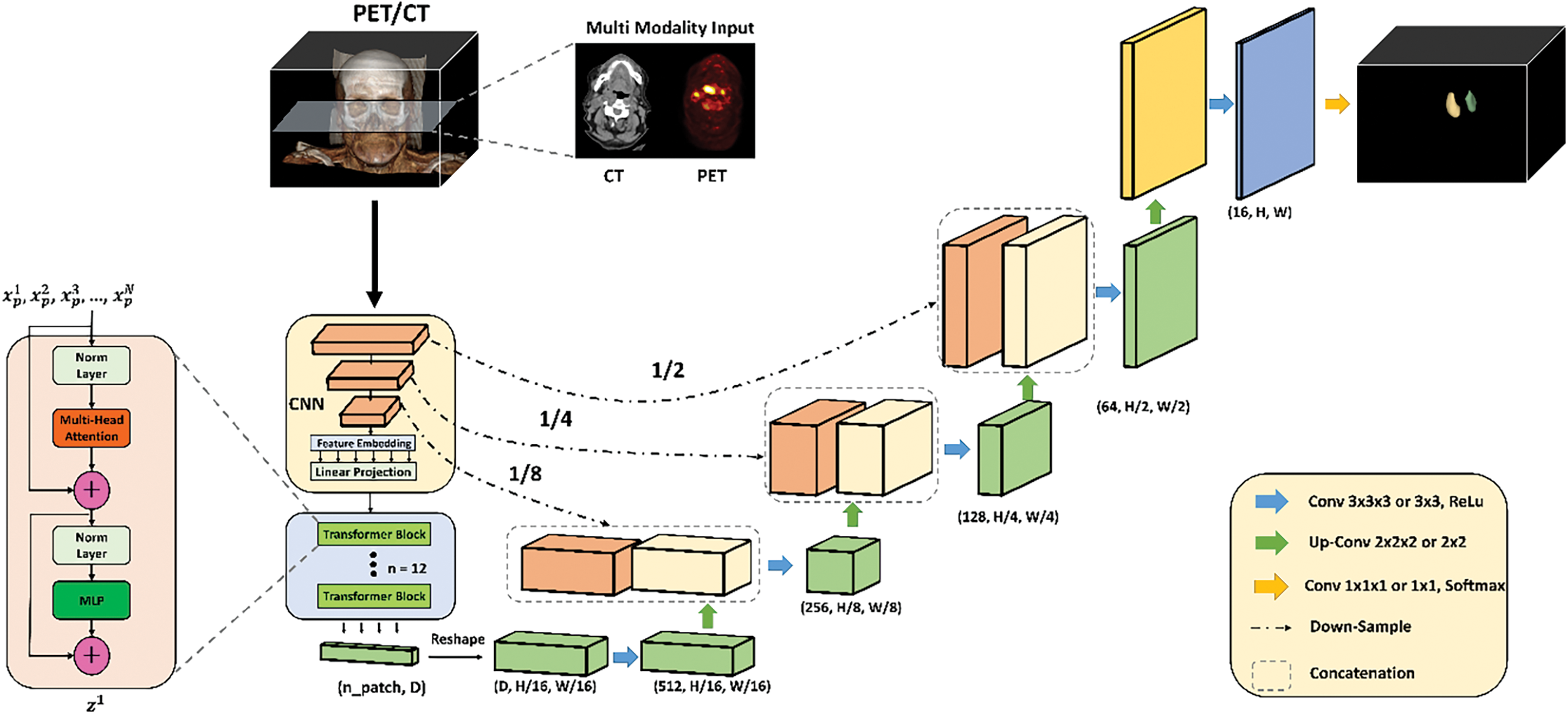
Figure 3: Detailed architecture of the proposed deep learning framework for head and neck tumor segmentation using dual PET/CT imaging. The framework integrates multimodal input data, applying a combination of CNNs and transformer blocks within the UNetR model. The process includes normalization, multi-head attention mechanisms, MLP blocks, and residual connections. The network is further refined through convolutional layers, up-sampling layers, and concatenation steps to produce a precise segmentation map, which is then analyzed for tumor shape and morphology
Additionally, the attention mechanism allows the model to focus on the most relevant parts of the input data for tumor segmentation. Following this, the data passes through another normalization layer and a Multi-Layer Perceptron (MLP) block for further processing. Residual connections within these blocks help preserve information across layers, aiding in the training of deep networks by mitigating the vanishing gradient problem. The repeated application of transformer blocks ensures the networks depth is adequate to capture complex features necessary for accurate segmentation. The processed data is then reshaped and passed through a series of convolutional and up-sampling layers. Convolutional layers (Conv 3 × 3 × 3) with ReLU activation function are used to extract spatial features, while up-convolutional layers (Up-Conv 2 × 2 × 2) progressively restore the spatial resolution. A 1 × 1 × 1 convolutional layer with Softmax activation finalizes the segmentation output. Throughout this process, concatenation steps combine features from different layers to enhance the model’s ability to delineate tumor boundaries accurately. The final output of our framework is a detailed segmentation map of the tumors, which is then subjected to shape analysis to evaluate their geometric properties. This comprehensive methodology aims to improve the precision and effectiveness of tumor segmentation in clinical practice, ultimately contributing to enhanced diagnostic and therapeutic planning for patients with head and neck cancer.
Table 1 provides a detailed overview of the hyperparameters used in our proposed 3D UNet Transformer segmentation model for the three different H&N datasets: HeckTor2022, AutoPET2023, and SegRap2023. Each of these models employs the 3D UNet Transformer architecture, which integrates the capabilities of CNNs with transformer networks, optimizing them for complex tumor segmentation tasks. All three models utilize a combination of Dice loss and Binary Cross Entropy (BCE) loss as their objective function. This combination is beneficial as it balances the need for accurate pixel-wise classification with the necessity of achieving a high degree of overlap between the predicted segmentation and the ground truth. To handle large volumetric data efficiently, these models use a 3D sliding window inference approach with a batch size of 4 and an overlap of 0.25, enabling the processing of smaller, overlapping sub-volumes. The AdamW optimizer is employed across all models, providing adaptive learning rates and incorporating weight decay to prevent overfitting. Additionally, data augmentation is enabled for all models, introducing variability into the training data and enhancing the model’s ability to generalize to unseen data. The training data for these models is formatted into patches of size (96, 96, 96), and voxel spacing is standardized to (2, 2, 1.5) to ensure consistent spatial resolution between the PET and CT images. The base layer of each network comprises 16 feature maps, forming the foundation for initial feature extraction. Within the transformer blocks, each model uses 12 attention heads, allowing them to focus on different aspects of the input data concurrently. The Multi-Layer Perceptron (MLP) dimension is set to 768, enabling the models to learn complex representations and capture intricate patterns within the data. Training is conducted over 1000 epochs, allowing ample opportunity for the models to learn and refine their parameters. A batch size of 32 is used during training, meaning that 32 samples are processed before the model parameters are updated. The initial learning rate is set to 0.001, carefully chosen to ensure stable and efficient convergence of the training process.
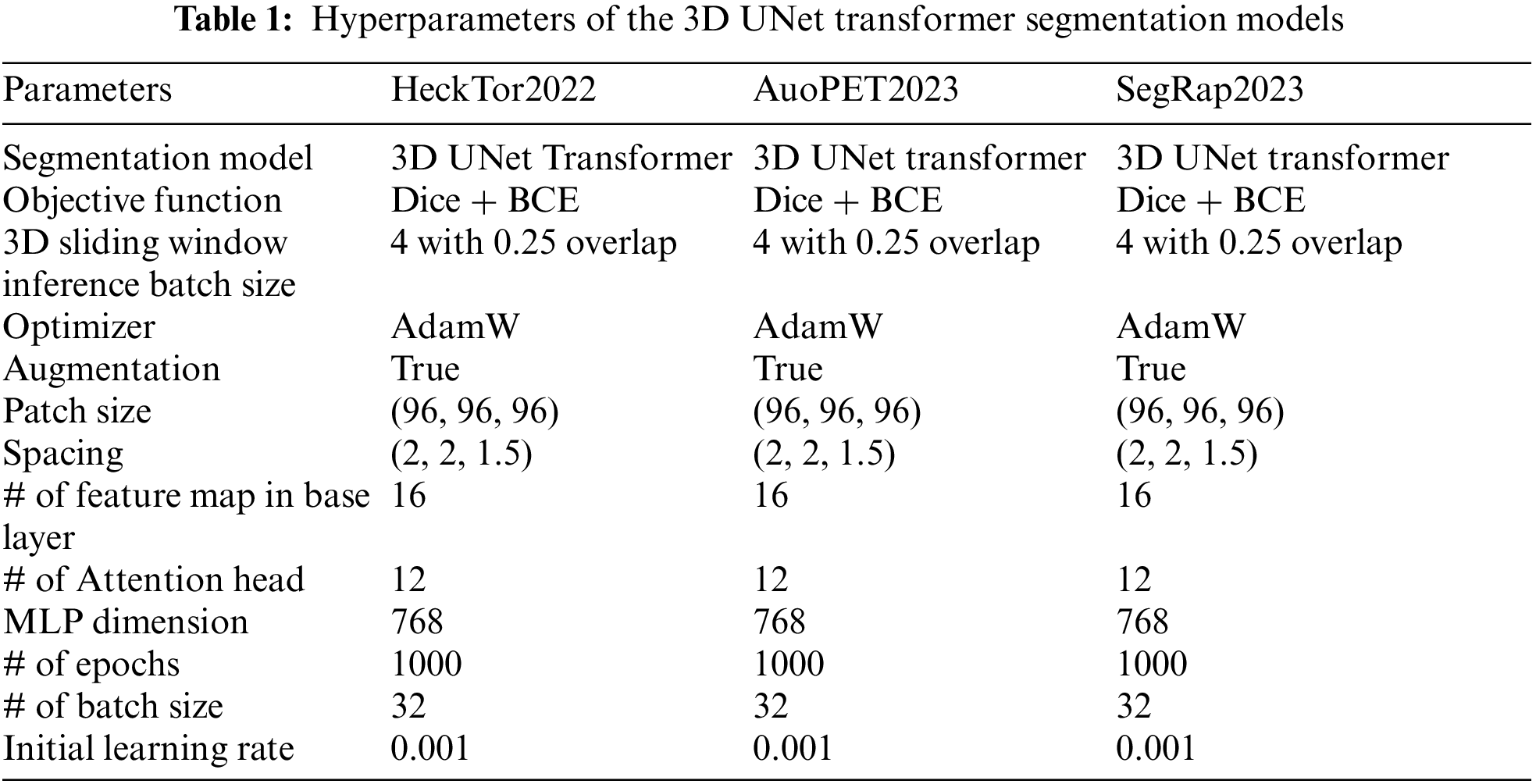
Table 2 outlines the hyperparameters used in our 2D and 2.5D segmentation models: HeckTor2022, AutoPET2023, and SegRap2023. Each model utilizes the 2D UNet Transformer architecture. The objective function for all models is a combination of Dice loss and BCE loss, balancing pixel-wise accuracy with overlap between predicted and ground truth segmentations. A 3D sliding window inference approach with a batch size of 4 and 0.25 overlap is employed, allowing efficient processing of larger data volumes. Training is optimized using the stochastic gradient descent (SGD) optimizer. Data augmentation is enabled to enhance generalization. The input size is (256, 256), with 3 channels for 2D and 7 channels for 2.5D models. The base layer includes 16 feature maps, and the transformer blocks have 12 attention heads with an MLP dimension of 768, ensuring the network captures detailed representations. Each model is trained for 300 epochs with a batch size of 16. The initial learning rate is set at 0.001, ensuring controlled learning. This configuration ensures robust and accurate segmentation of head and neck tumors in PET/CT images, enhancing diagnostic and therapeutic planning.
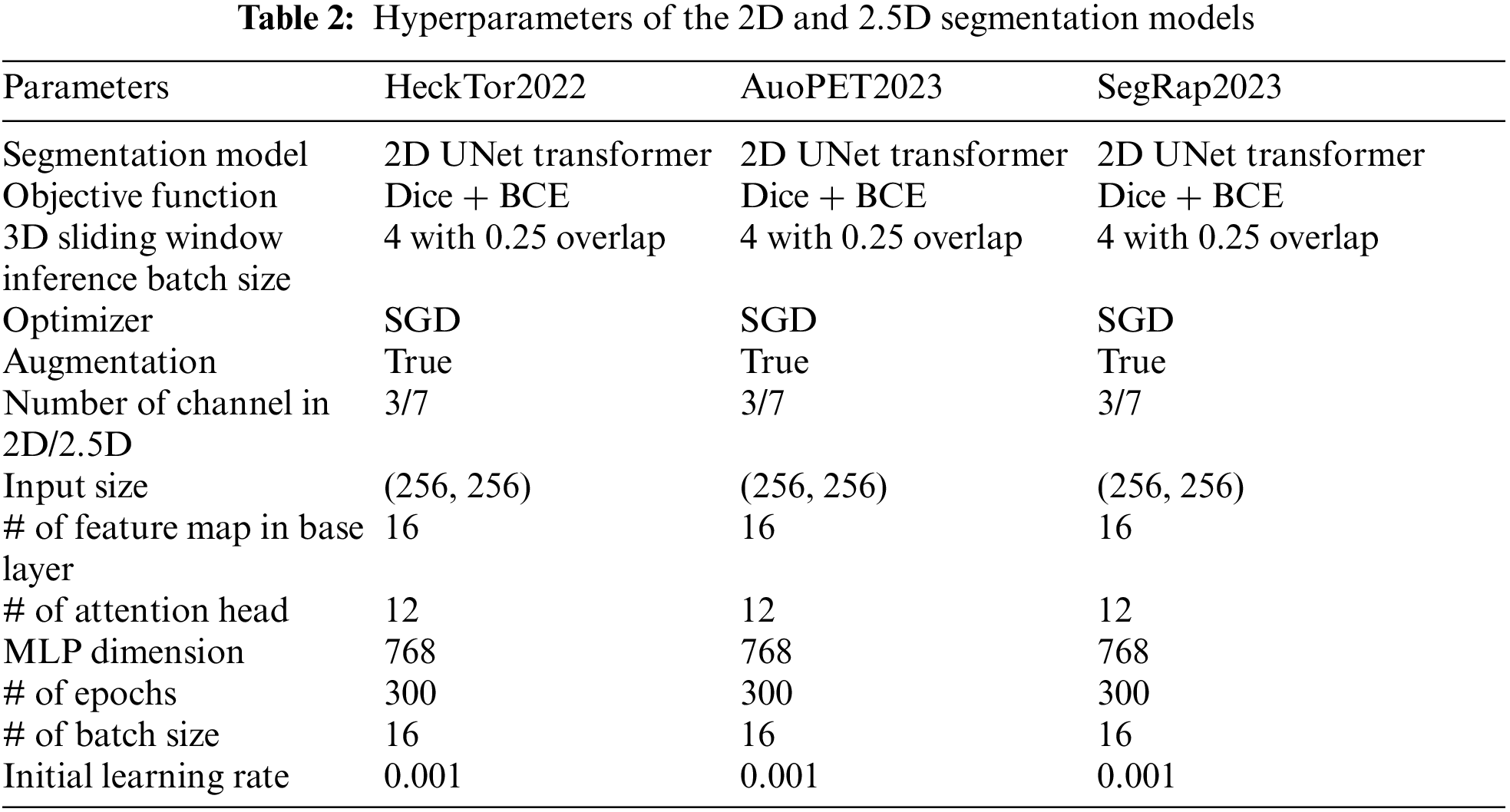
The assessment of segmentation performance utilizes the aggregated Dice Similarity Coefficient (DSCagg), a measure of volumetric overlap between the algorithm’s predictions and expert annotations. DSCagg is advantageous for evaluating the segmentation of small regions within large images. While effective for ranking algorithms, particularly in tumor segmentation, its limitation is apparent when no ground truth volume is present, potentially resulting in a DSC of 0. The metric is carefully chosen for its suitability in assessing segmentation accuracy for both primary tumors (GTVp) and nodal tumors (GTVn), despite the inability to measure standard deviation across patient data.
where it calculates the ratio of twice the shared information between the predicted segmentation (A) and the ground truth (B), over the total size of both individual segmentations. The higher the DSC, the more accurate the prediction is in relation to the ground truth. This aggregated version of the coefficient implies a summation over multiple comparisons, providing an overall effectiveness measure for segmentation across a dataset, rather than for a singular instance.
• The Relative Absolute Volume Difference (RAVD) is a metric used to quantify the difference in volume between the segmented region and the ground truth region. It measures the relative difference in the volume of the predicted segmentation compared to the actual ground truth volume. RAVD is particularly useful in medical imaging for assessing how accurately a segmentation algorithm predicts the volume of a tumor or other anatomical structure. RAVD is expressed as a percentage, indicating how much the predicted volume deviates from the ground truth volume. A lower RAVD value indicates better segmentation performance, with a value of zero representing a perfect match between the predicted and ground truth volumes. The RAVD is calculated using the following formula:
where
• Jaccard Index (JI): Also known as the Intersection over Union (IoU), the Jaccard index is another common metric for evaluating the similarity between the predicted and actual values. It is defined as:
where P represents the predicted segmentation and GT is the ground truth.
• Average Hausdorff Distance (AHD): The AHD measures the distance between the surfaces of the predicted and ground truth segmentations, offering a surface distance metric:
where
In this section, we present the results of our comprehensive evaluation of the proposed 2D, 2.5D, and 3D UNet Transformer models using the HeckTor2022, AutoPET2023, and SegRap2023 datasets. The primary goal of this analysis is to assess the performance, accuracy, and robustness of our models in segmenting head and neck tumors from dual PET/CT imaging data. We conducted extensive experiments to compare the effectiveness of different model configurations and hyperparameters across the three datasets. The results are systematically presented to highlight the strengths and weaknesses of each approach. We analyze the impact of varying dimensionality (2D, 2.5D, and 3D) approaches on the segmentation performance, providing insights into the suitability of each method for clinical applications. Furthermore, we discuss the influence of different hyperparameters and training strategies on the models’ outcomes. Our analysis also includes visual comparisons of segmentation outputs to qualitatively assess the models’ performance. By examining both quantitative metrics and qualitative results, we aim to provide a holistic view of the models’ capabilities and their potential for clinical deployment.
4.1 Performance Evaluation on HeckTor2022
Fig. 4 presents a comprehensive analysis of the performance of the 2D, 2.5D, and 3D UNet Transformer models on the HeckTor2022 dataset across various metrics. This section synthesizes the models’ effectiveness in segmenting Primary Gross Tumor Volume (GTV), Lymph Node GTV (LN GTV), and Aggregate GTV.
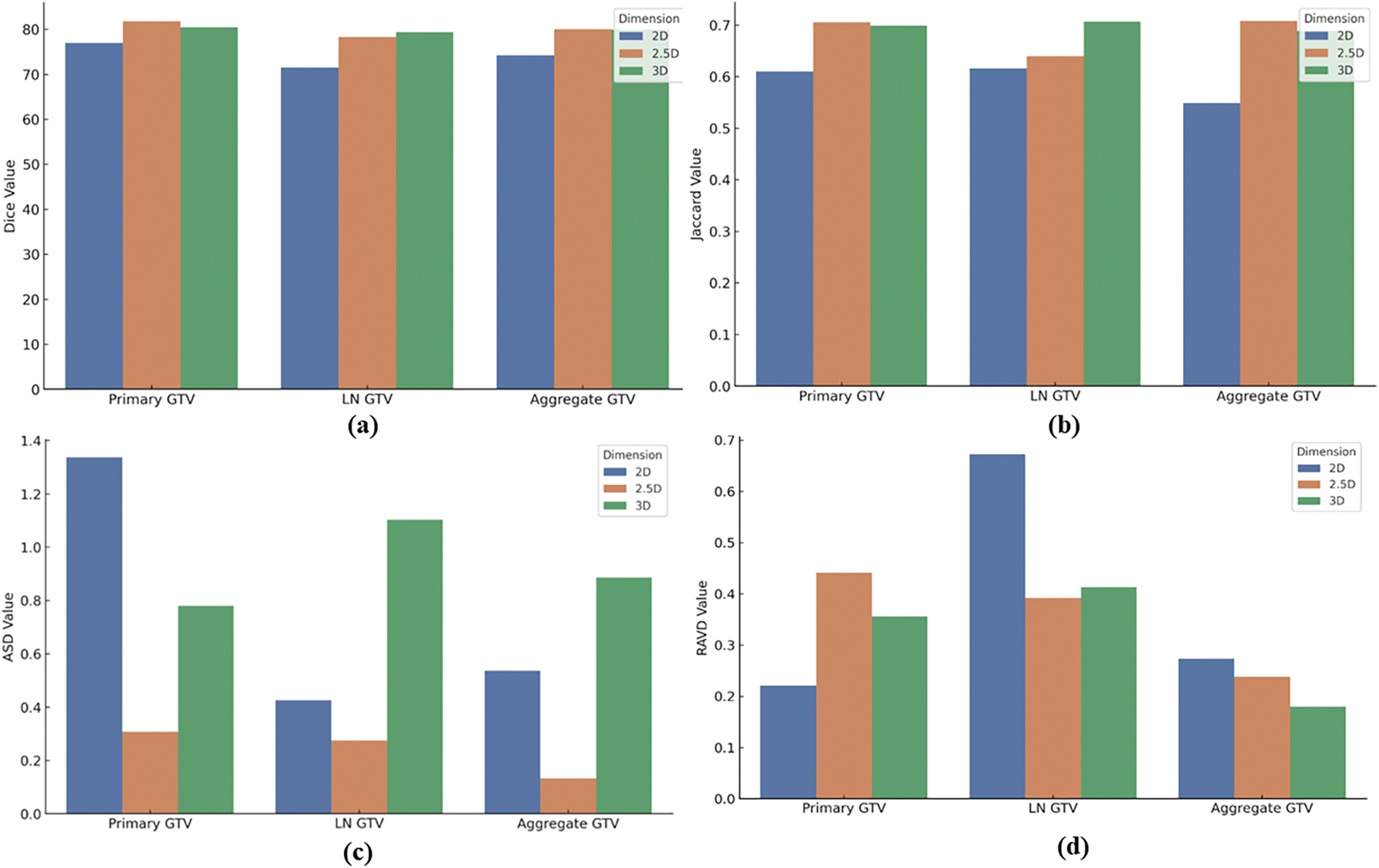
Figure 4: 2D, 2.5D, and 3D UNet Transformer models performance on HeckTor2022 Dataset: (a) Dice similarity coefficient scores, (b) Jaccard index scores, (c) Average surface distance scores, (d) Relative absolute volume difference scores for primary GTV, LN GTV, and aggregate GTV
Fig. 4a shows the dice similarity coefficient scores, where both the 2.5D and 3D models outperform the 2D model, demonstrating superior segmentation accuracy. This trend continues in Fig. 4b, with the Jaccard Index results, particularly noting the 2.5D model’s strength in primary GTV and the 3D model’s proficiency in LN GTV segmentation. Fig. 4c illustrates the average surface distance scores. The 2.5D model consistently achieves the lowest ASD values across all categories, indicating its precision in boundary delineation. Although the 3D model offers enhanced spatial context, it does not consistently translate to improved surface accuracy, showing mixed results across tumor types. Lastly, Fig. 4d evaluates the RAVD scores. The 2D model excels in volume accuracy for Primary GTV, while the 2.5D model demonstrates balanced performance across all tumor types. The 3D model, despite its detailed volumetric analysis, exhibits higher RAVD in LN GTV, highlighting some challenges in accurate volume estimation.
Table 3 presents a comparative analysis of the performance metrics for the 2D, 2.5D, and 3D UNet Transformer models across three categories: Primary Gross Tumor Volume (GTVp), Lymph Node GTV (GTVn), and Aggregate GTV (GTVT). The evaluation metrics include Dice Similarity Coefficient (Dice), Jaccard Index, ASD, and RAVD. The 2.5D model consistently achieves the highest Dice scores across all categories, with 81.777 for GTVp, 78.297 for GTVn, and 80.037 for GTVT. This indicates a superior overlap between the predicted and ground truth segmentations compared to the 2D and 3D models. The 3D model also performs well, especially for GTVn and GTVT, showing slightly lower Dice scores than the 2.5D model but outperforming the 2D model. The Jaccard Index results follow a similar trend to the Dice scores. The 2.5D model achieves the highest values for GTVp (0.705), GTVn (0.64), and GTVT (0.707), indicating better segmentation accuracy. The 3D model closely follows, particularly for GTVn and GTVT, while the 2D model shows the lowest Jaccard values across all categories. In terms of ASD, the 2.5D model demonstrates the lowest values across all tumor volumes, with 0.308 for GTVp, 0.275 for GTVn, and 0.133 for GTVT. This suggests that the 2.5D model provides the most precise boundary delineation. The 2D model has higher ASD values, particularly for GTVp (1.335), while the 3D model shows mixed performance with the lowest ASD for GTVn but higher values for GTVp and GTVT. The 2.5D model also performs well in terms of RAVD, with the lowest values for GTVp (0.441) and GTVT (0.238), although the 3D model achieves a lower RAVD for GTVn (0.3921). The 2D model exhibits the highest RAVD for GTVn (0.672), indicating less accurate volume predictions compared to the 2.5D and 3D models.
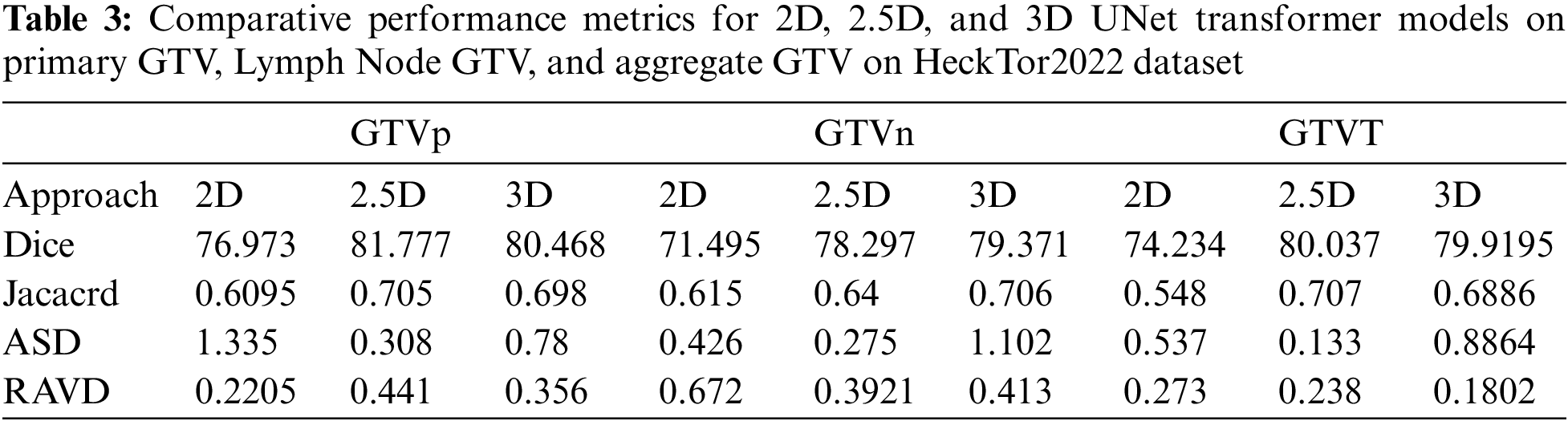
Overall, the 2.5D model demonstrates superior performance across most metrics, providing a balanced approach with high segmentation accuracy and precise boundary delineation. The 3D model also performs well, particularly for lymph node GTV, but shows some variability in boundary accuracy. The 2D model, while still effective, generally underperforms compared to the 2.5D and 3D models, highlighting the benefits of incorporating additional spatial context in the 2.5D and 3D approaches.
Fig. 5 illustrates the segmentation accuracy of the UNetR model on the HeckTor2022 dataset, showcasing results from three patients (P1, P9, P29) across axial, sagittal, and coronal views. The model delineates head and neck tumors with high precision, evident from the segmentation contours overlaid on dual PET/CT images. Each view highlights the model’s effectiveness in capturing tumor boundaries, confirming its robustness in handling complex anatomical variations. This visualization supports the UNetR model’s potential in enhancing diagnostic and therapeutic planning for head and neck cancers.
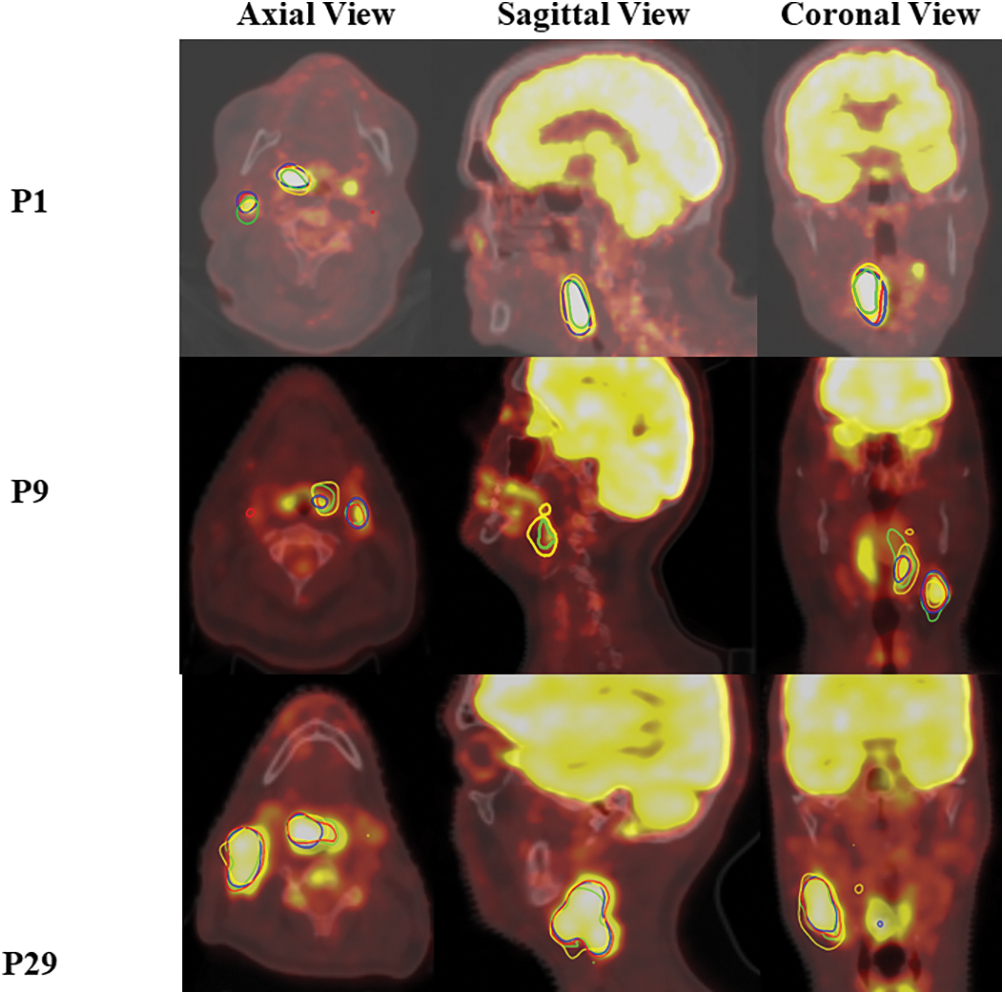
Figure 5: Segmentation performance of the UNetR model on the HeckTor2022 dataset
4.2 Performance Benchmarking on AutoPET2023 and SegRap2023
In this section, we extend the evaluation of our proposed 2D, 2.5D, and 3D UNet Transformer models by testing them on two additional datasets: AutoPET2023 and SegRap2023. These datasets have been selected to further validate the robustness and generalizability of our models in segmenting head and neck tumors from PET/CT images The AutoPET2023 dataset, as described in the paper available at AutoPET2023 [45], provides high-quality annotations and diverse clinical cases, making it an excellent resource for testing the efficacy of our models. This dataset includes a wide range of PET/CT images, ensuring that the models are evaluated under varied conditions. The SegRap2023 dataset, detailed in the publication SegRap2023 [44], offers a comprehensive set of annotated images from multiple centers and vendors. This dataset is particularly valuable for assessing the models’ performance in a real-world, multi-institutional context, ensuring that the segmentation algorithms are robust and adaptable to different imaging protocols and equipment. The performance metrics in Table 4 provide a detailed comparative analysis of the 2D, 2.5D, and 3D UNet Transformer models on the AutoPET2023 and SegRap2023 datasets. These metrics include Dice, ASD, and RAVD.
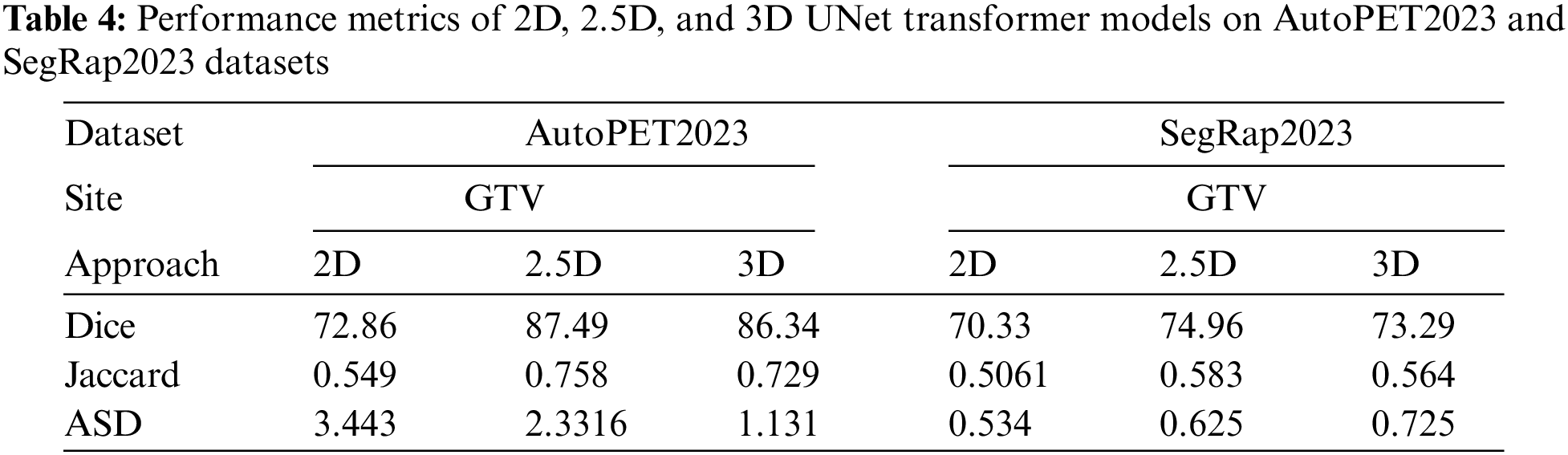
For the AutoPET2023 dataset, the 2.5D model demonstrates superior performance with the highest Dice score of 87.49, indicating strong overlap between the predicted and ground truth segmentations. The 3D model follows closely with a Dice score of 86.34, while the 2D model lags behind at 72.86. The Jaccard Index mirrors these results, with the 2.5D model achieving the highest value of 0.758, further supporting its higher segmentation accuracy, followed by the 3D and 2D models at 0.729 and 0.549, respectively. However, when considering boundary delineation, as reflected by the ASD, the 3D model performs best with the lowest value of 1.131, suggesting more precise boundary identification. The 2.5D model also performs well, albeit with a slightly higher ASD of 2.331, whereas the 2D model shows the highest ASD of 3.443, indicating less precision in boundary demarcation. The RAVD results emphasize the 2.5D model’s capability to maintain volumetric accuracy, as it achieves the lowest RAVD of 0.2946. This is closely followed by the 3D model at 0.343, while the 2D model shows a higher RAVD of 0.4888. Overall, the 2.5D model offers the best balance between segmentation accuracy and boundary precision, with the 3D model also exhibiting strong performance, particularly in boundary precision.
On the SegRap2023 dataset, the 2.5D model again leads with a Dice score of 74.96, outperforming both the 3D model (73.29) and the 2D model (70.33). The Jaccard Index results are consistent with these findings, with the 2.5D model achieving the highest value of 0.583, followed by the 3D model at 0.564 and the 2D model at 0.5061. Interestingly, the ASD values reveal that the 2D model performs best in boundary delineation on this dataset, with the lowest value of 0.534. The 2.5D and 3D models show slightly higher ASD values of 0.625 and 0.725, respectively. This could be due to the additional spatial context considered by the 2.5D and 3D models, which may introduce complexity in boundary handling. The RAVD analysis for the SegRap2023 dataset indicates that the 2.5D and 3D models are nearly identical in their volumetric accuracy, with RAVD values of 0.255 and 0.256, respectively, whereas the 2D model shows a higher RAVD of 0.585. This underscores the consistent volumetric accuracy of the 2.5D model across different datasets.
Finally, the 2.5D model demonstrates a consistent advantage in segmentation accuracy and volume prediction across all datasets, making it the most robust and reliable model among the three. The 3D model also shows strong performance but exhibits variability in boundary precision. The 2D model, while effective, generally underperforms compared to the 2.5D and 3D models, highlighting the benefits of incorporating additional spatial context in the 2.5D and 3D approaches. This conclusion is further supported by our statistical analysis, which reveals statistically significant p-values across multiple datasets (AutoPET2023, SegRap2023, and HeckTor2022), confirming that the observed differences in performance metrics are not only meaningful but also statistically robust. Fig. 6a–d illustrates the comparative performance of the 2D, 2.5D, and 3D UNet Transformer models across the AutoPET2023 and SegRap2023 datasets. The 2.5D model consistently outperforms the others, achieving the highest Dice and Jaccard values, indicating superior overlap and segmentation accuracy. Specifically, the 2.5D model achieves Dice scores of 87.49 on AutoPET2023 and 74.96 on SegRap2023. The 3D model also performs well, particularly on AutoPET2023, though it shows a slight decline on SegRap2023. The 2D model lags behind, reflecting its limitations in capturing complex tumor structures.
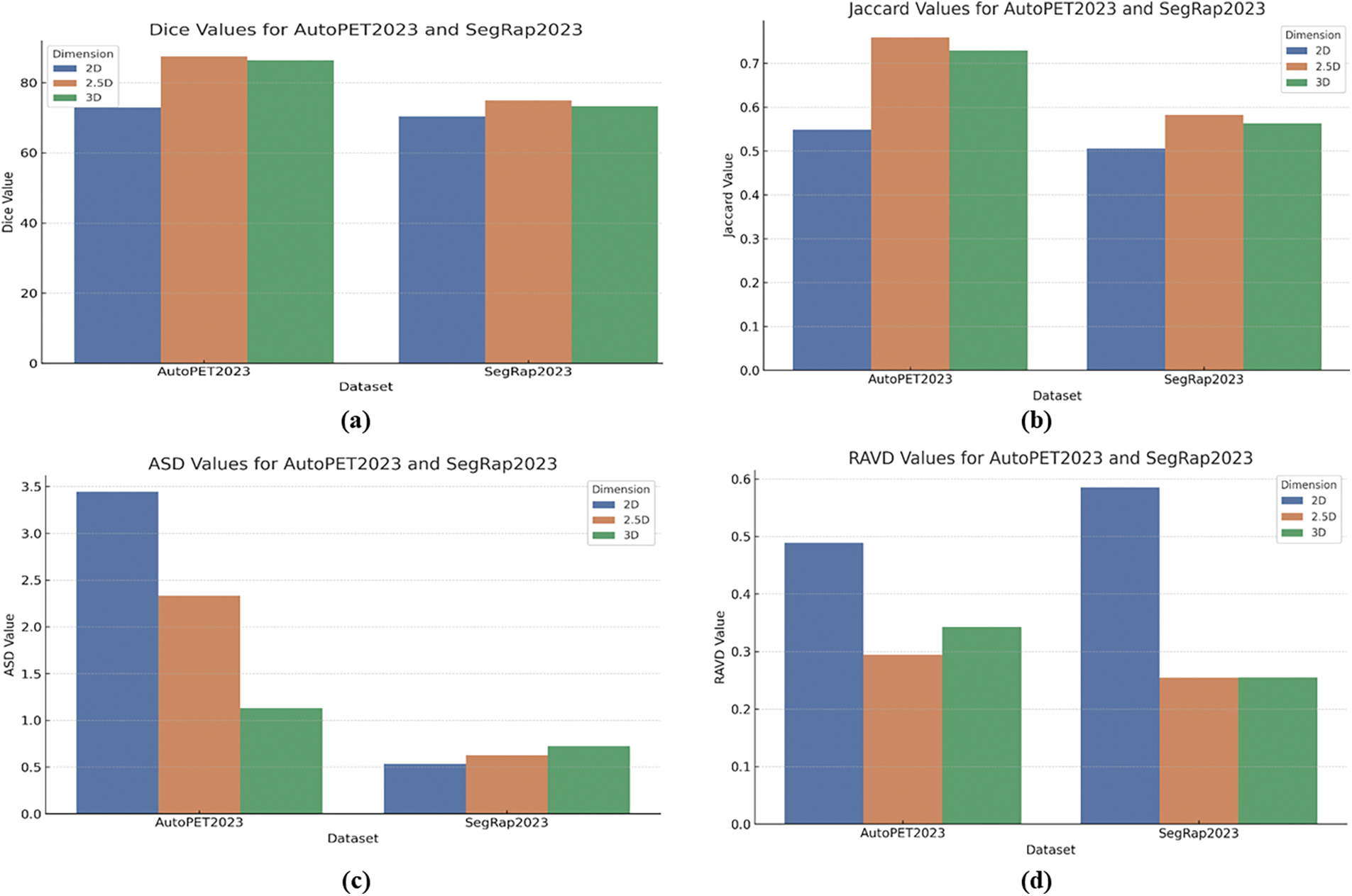
Figure 6: (a) Dice values, (b) Jaccard values, (c) ASD values, and (d) RAVD values for AutoPET2023 and SegRap2023 datasets across 2D, 2.5D, and 3D UNet transformer model
Additionally, ASD analysis highlights that the 3D model excels in boundary precision on AutoPET2023, while the 2.5D model remains competitive across both datasets. Interestingly, the 2D model achieves the lowest ASD on SegRap2023, suggesting dataset-specific strengths. In terms of RAVD, the 2.5D model again demonstrates the best volume prediction accuracy, closely followed by the 3D model, with the 2D model showing less accuracy in this regard. Overall, these results reinforce the effectiveness of incorporating additional spatial context in the 2.5D and 3D models for more accurate and reliable tumor segmentation.
4.3 Statistical Analysis of Performance Metrics
To rigorously assess the significance of the observed differences in performance among the 2D, 2.5D, and 3D UNet Transformer models, we conducted a statistical analysis using p-values calculated for each pairwise comparison across the three datasets: AutoPET2023, SegRap2023, and HeckTor2022. This analysis aimed to determine whether the differences in metrics such as Dice, Jaccard Index, ASD, and RAVD are statistically significant or merely attributable to random variation. The results of the statistical analysis are summarized in Fig. 7. A p-value of less than 0.05 was used as the threshold for statistical significance.
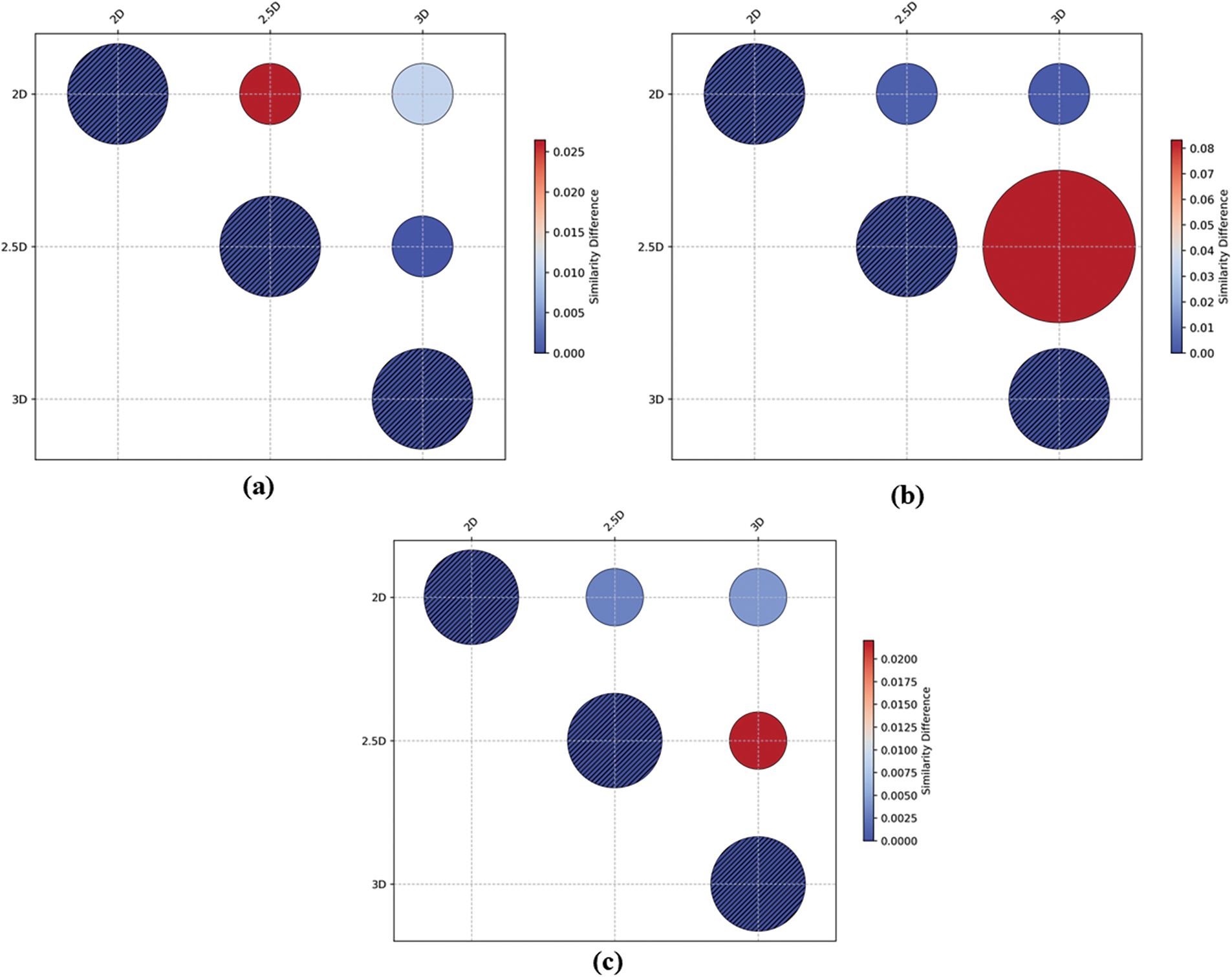
Figure 7: Comparative analysis of segmentation performance p-value across different approaches in (a) AutoPET2023 Datasets; (b) SegRap2023 datasets and (c) HeckTor2022 dataset
• For the AutoPET2023 dataset (Fig. 7a), the 2D model showed significantly lower performance compared to both the 2.5D and 3D models, with p-values of 0.026438 and 0.010532, respectively, for Dice scores, indicating that the observed differences are statistically significant.
• The analysis of the SegRap2023 dataset revealed similar trends (Fig. 7b), with the 2.5D and 3D models significantly outperforming the 2D model (p-values of 0.003085 and 0.004225, respectively). However, the comparison between the 2.5D and 3D models yielded a higher p-value, suggesting that the difference in performance between these two models might not be statistically significant in this dataset.
• In the HeckTor2022 dataset (Fig. 7c), the 2.5D model once again demonstrated superior performance, with p-values as low as 0.000163 when compared to the 2D and 3D models, confirming the statistical significance of these results.
Lastly, the statistical analysis substantiates that the performance enhancements observed with the 2.5D and 3D models over the 2D model are statistically significant across all evaluated datasets. These findings underscore the benefits of incorporating additional spatial context in medical image segmentation tasks and provide a robust basis for the conclusions drawn in this study. The results further emphasize the importance of selecting an appropriate model based on both statistical significance and practical clinical applicability.
4.4 Comparison with Literature
In this study, we compared our proposed 2D, 2.5D, and 3D UNet Transformer models for tumor and lymph node segmentation on the HECKTOR2022 dataset with several approaches reported in the literature. Table 5 presents the Dice coefficients for the tumor and lymph node segmentation tasks. The results show that our 2.5D model achieves a Dice score of 0.81777 for tumor segmentation, which surpasses the scores of other methods like the 3D nnU-Net [40] (0.82) and SwinUNet [40] (0.8047). Similarly, for lymph node segmentation, our 3D model achieves a Dice score of 0.79371, outperforming the nnM-Net [40], SwinUNet [40] and 3D ResU-Net [42]. These comparisons highlight the effectiveness of our proposed approach, particularly in the 2.5D configuration, which consistently demonstrates superior performance across both tumor and lymph node segmentation tasks.
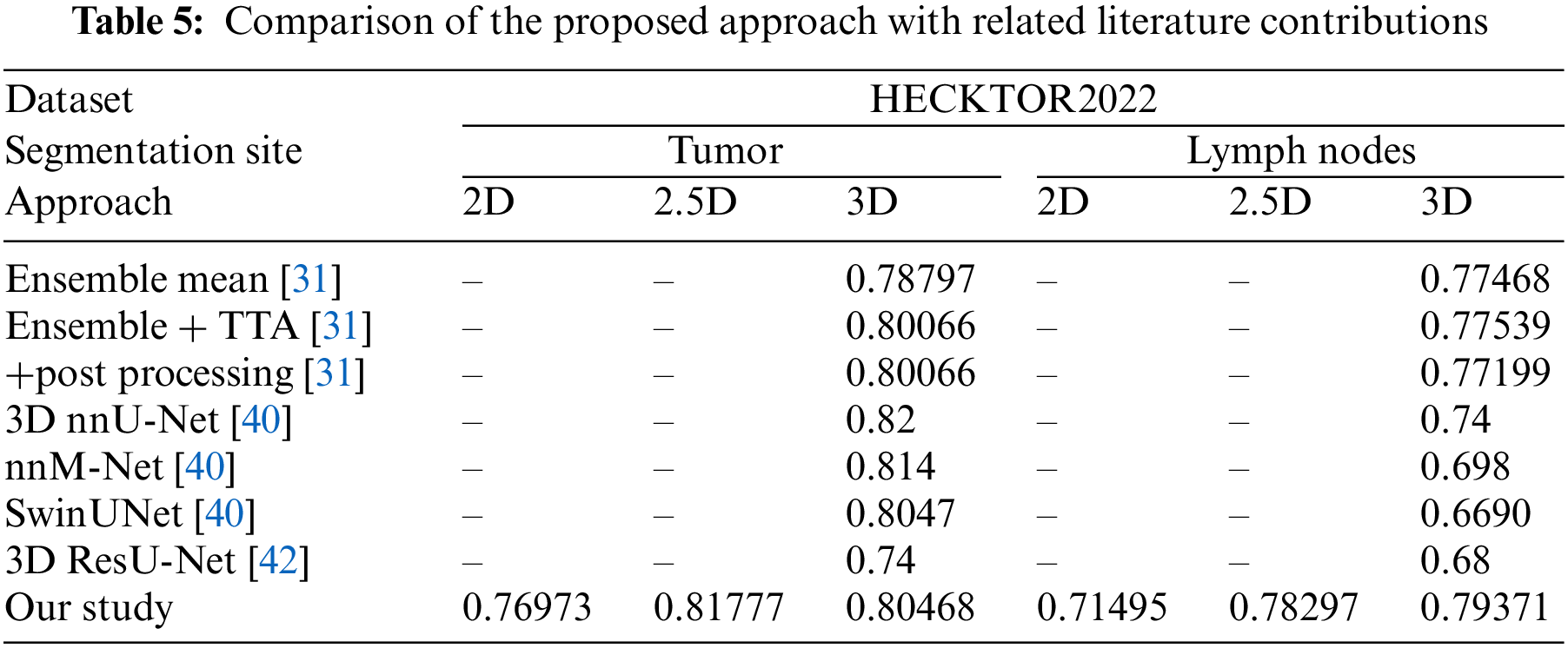
As can be seen from Table 5, the proposed 2.5D UNet Transformer model achieves superior segmentation performance on the HECKTOR2022 dataset without requiring additional complex post-processing steps. This effectiveness can be attributed to the model’s ability to capture and leverage spatial context, allowing it to learn detailed and accurate representations of tumor and lymph node structures automatically.
4.5 Clinical Implications and Potential Impact
This study significantly advances the field of medical imaging by demonstrating the effectiveness of 2D, 2.5D, and 3D UNet Transformer models in accurately segmenting H&N tumors using dual PET/CT imaging. The clinical implications of these findings are profound, offering several key benefits that can enhance diagnostic accuracy, improve treatment planning, and potentially transform patient outcomes in oncology. The improved segmentation accuracy provided by our models, particularly the 2.5D UNet Transformer, enables more precise delineation of tumor boundaries. This precision is critical for oncologists and radiologists as it directly influences the assessment of tumor stage, size, and potential metastasis, thereby facilitating more accurate diagnoses and tailored treatment strategies. Accurate tumor segmentation is essential for effective treatment planning, especially in radiation therapy and surgery. The detailed segmentation results from our models allow for the precise calculation of tumor volume and configuration, which can be used to tailor radiation doses and surgical margins more effectively. This not only ensures that the tumor receives sufficient treatment but also helps in sparing healthy tissues, thus minimizing side effects and enhancing overall treatment efficacy. The computational efficiency of our 2.5D model supports its use in real-time clinical scenarios, where quick processing of imaging data is essential. This capability makes it an invaluable tool in dynamic clinical environments, aiding medical professionals in making faster and more informed decisions during diagnostic and therapeutic procedures. While our study focuses on PET/CT imaging, the methodologies developed are adaptable to other imaging modalities such as MRI and ultrasound. This versatility enhances the utility of our models across different branches of medicine, potentially benefiting a broader range of patients with various conditions.
In this study, we aimed to enhance the segmentation of head and neck tumors using dual PET/CT imaging by developing and evaluating advanced 2D, 2.5D, and 3D UNet Transformer models. The integration of CNNs with transformer networks was intended to leverage the strengths of both architectures for accurate and robust tumor delineation. Our methodology involved preprocessing steps such as normalization, contrast enhancement, and resampling, followed by segmentation using the UNet Transformer models. We evaluated the performance of these models on three datasets: HeckTor2022, AutoPET2023, and SegRap2023. The findings from our evaluation indicate that the 2.5D UNet Transformer model generally outperformed the 2D and 3D models across several key metrics, including Dice and Jaccard values, which demonstrate superior overlap and segmentation accuracy. For instance, the 2.5D model achieved a Dice similarity coefficient of up to 0.81777, significantly higher than the 2D model, with p-values as low as 0.000163 across the datasets, indicating that these differences are statistically significant. However, it is important to acknowledge that these conclusions are based on a specific set of metrics and datasets, and the 2.5D model’s performance, while strong, should not be interpreted as definitively superior in all scenarios. Specifically, the 3D model showed notable strengths in boundary accuracy, particularly on the AutoPET2023 dataset, though it also exhibited variability across different datasets. For example, while the 3D model performed well with a Jaccard Index score of 0.706 on the HeckTor2022 dataset, it showed less consistency on SegRap2023. Additionally, the 2D model, while effective, generally underperformed compared to the 2.5D and 3D models, underscoring the benefits of incorporating additional spatial context. Despite these promising results, the study’s limitations, such as variability in model performance across different datasets, suggest that further work is needed to improve generalizability. Furthermore, the computational complexity of the 3D model poses challenges for practical deployment, especially in resource-constrained clinical settings. Future research should explore optimizing these models for real-time applications and reducing computational demands without compromising accuracy. We plan to investigate the integration of more advanced attention mechanisms and multi-scale feature extraction techniques to further enhance segmentation performance. Expanding our dataset to include more diverse clinical cases and testing our models on additional imaging modalities will also be key areas of focus. Additionally, developing lightweight versions of our models could facilitate their adoption in routine clinical practice.
Acknowledgement: This work was supported by Scientific Research Deanship at University of Ha’il, Saudi Arabia.
Funding Statement: This work was supported by Scientific Research Deanship at University of Ha’il, Saudi Arabia through project number RG-23 137.
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, Mohammed A. Mahdi, Sawsan A. Saad and Alawi Alqushaibi; Data curation, Shahanawaj Ahamad and Sawsan A. Saad; Methodology, Mohammed A. Mahdi and Rizwan Qureshi; Project administration, Mohammed A. Mahdi and Rizwan Qureshi; Resources, Sawsan A. Saad, Alawi Alqushaibi; Software, Mohammed A. Mahdi and Rizwan Qureshi; Validation, Mohammed A. Mahdi; Visualization, Alaa Dafhalla; Writing—original draft, Mohammed A. Mahdi, Shahanawaj Ahamad, Sawsan A. Saad and Alawi Alqushaibi; Writing—review & editing, Alaa Dafhalla and Rizwan Qureshi. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets used in this study are publicly available and can be accessed as follows: HeckTor2022: https://hecktor.grand-challenge.org/ (accessed on 02 February 2024); AutoPET2023 dataset: https://autopet-ii.grand-challenge.org/ (accessed on 25 March 2024); SegRap2023 dataset: https://segrap2023.grand-challenge.org (accessed on 25 March 2024).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Gatta G, Capocaccia R, Botta L. Descriptive epidemiology of the head and neck cancers in old patients. Front Oncol. 2023;13:1102236. doi:10.3389/fonc.2023.1102236. [Google Scholar] [CrossRef]
2. Rocke J, Mclaren O, Hardman J, Garas G, Smith ME, Ishii H, et al. The role of allied healthcare professionals in head and neck cancer surveillance: a systematic review. Clin Otolaryngol. 2020;45(1):83–98. doi:10.1111/coa.v45.1 [Google Scholar] [PubMed] [CrossRef]
3. Pulumati A, Pulumati A, Dwarakanath BS, Verma A, Papineni RV. Technological advancements in cancer diagnostics: improvements and limitations. Cancer Rep. 2023;6(2):e1764. doi:10.1002/cnr2.v6.2. [Google Scholar] [CrossRef]
4. Chung CH, Dietz A, Gregoire V, Guzzo M, Hamoir M, Leemans CR, et al. Head and neck cancer. In: Oxford textbook of oncology. UK: Oxford University Press; 2016. p. 329. [Google Scholar]
5. Abdalla AS, Sheybani ND, Khan SA. Clinical role of positron emission tomography/computed tomography imaging in head and neck squamous cell carcinoma. PET Clinics. 2022;17(2):213–22. doi:10.1016/j.cpet.2021.12.008 [Google Scholar] [PubMed] [CrossRef]
6. Zhou Q-P, Zhao Y-H, Gao L. Positron emission tomography and magnetic resonance imaging combined with computed tomography in tumor volume delineation: a case report. World J Clin Cases. 2022;10(1):249. doi:10.12998/wjcc.v10.i1.249 [Google Scholar] [PubMed] [CrossRef]
7. Ghidini M, Vuozzo M, Galassi B, Mapelli P, Ceccarossi V, Caccamo L, et al. The role of positron emission tomography/computed tomography (PET/CT) for staging and disease response assessment in localized and locally advanced pancreatic cancer. Cancers. 2021;13(16):4155. doi:10.3390/cancers13164155 [Google Scholar] [PubMed] [CrossRef]
8. Li L, Zhao X, Lu W, Tan S. Deep learning for variational multimodality tumor segmentation in PET/CT. Neurocomputing. 2020;392:277–95. doi:10.1016/j.neucom.2018.10.099 [Google Scholar] [PubMed] [CrossRef]
9. Andrearczyk V, Oreiller V, Boughdad S, Rest CCL, Elhalawani H, Jreige M, et al. Overview of the HECKTOR challenge at MICCAI 2021: automatic head and neck tumor segmentation and outcome prediction in PET/CT images. In: 3D head and neck tumor segmentation in PET/CT challenge, 2021; Strasbourg, France: Springer; p. 1–37. [Google Scholar]
10. Nikolov S, Blackwell S, Zverovitch A, Mendes R, Livne M, De Fauw J, et al. Clinically applicable segmentation of head and neck anatomy for radiotherapy: deep learning algorithm development and validation study. J Med Internet Res. 2021;23(7):e26151. doi:10.2196/26151 [Google Scholar] [PubMed] [CrossRef]
11. Groendahl AR, Knudtsen IS, Huynh BN, Mulstad M, Moe YM, Knuth F, et al. A comparison of methods for fully automatic segmentation of tumors and involved nodes in PET/CT of head and neck cancers. Phys Med Biol. 2021;66(6):65012. doi:10.1088/1361-6560/abe553. [Google Scholar] [CrossRef]
12. Ye X, Guo D, Ge J, Yan S, Xin Y, Song Y, et al. Comprehensive and clinically accurate head and neck cancer organs-at-risk delineation on a multi-institutional study. Nat Commun. 2022;13(1):6137. doi:10.1038/s41467-022-33178-z [Google Scholar] [PubMed] [CrossRef]
13. Shamshad F, Khan S, Zamir SW, Khan MH, Hayat M, Khan FS, et al. Transformers in medical imaging: a survey. Med Image Anal. 2023;88:102802. doi:10.1016/j.media.2023.102802 [Google Scholar] [PubMed] [CrossRef]
14. Li J, Chen J, Tang Y, Wang C, Landman BA, Zhou SK. Transforming medical imaging with transformers? A comparative review of key properties, current progresses, and future perspectives. Med Image Anal. 2023;85:102762. doi:10.1016/j.media.2023.102762 [Google Scholar] [PubMed] [CrossRef]
15. Xia K, Wang J. Recent advances of transformers in medical image analysis: a comprehensive review. MedComm-Futur Med. 2023;2(1):e38. doi:10.1002/mef2.v2.1. [Google Scholar] [CrossRef]
16. Mohammadi R, Shokatian I, Salehi M, Arabi H, Shiri I, Zaidi H. Deep learning-based auto-segmentation of organs at risk in high-dose rate brachytherapy of cervical cancer. Radiot Oncol. 2021;159:231–40. doi:10.1016/j.radonc.2021.03.030. [Google Scholar] [CrossRef]
17. Yao W, Bai J, Liao W, Chen Y, Liu M, Xie Y. From CNN to transformer: a review of medical image segmentation models. J Imaging Inform Med. 2024;37:1–19. [Google Scholar]
18. Andrearczyk V, Oreiller V, Vallières M, Castelli J, Elhalawani H, Jreige M, et al. Automatic segmentation of head and neck tumors and nodal metastases in PET-CT scans. In: Medical imaging with deep learning. PMLR; 2020. vol. 121, p. 33–43. [Google Scholar]
19. Chen Z, Li C, He J, Ye J, Song D. A novel hybrid convolutional neural network for accurate organ segmentation in 3D head and neck CT images. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, 2021 Sep 27–Oct 1; Strasbourg, France: Springer. [Google Scholar]
20. Naser MA, van Dijk LV, He R, Wahid KA, Fuller CD. Tumor segmentation in patients with head and neck cancers using deep learning based-on multi-modality PET/CT images. In: 3D head and neck tumor segmentation in PET/CT challenge, 2020; Lima, Peru: Springer; p. 85–98. [Google Scholar]
21. Kumar A, Jiang H, Imran M, Valdes C, Leon G, Kang D, et al. A flexible 2.5 D medical image segmentation approach with in-slice and cross-slice attention. arXiv preprint arXiv:240500130. 2024. [Google Scholar]
22. Chakrabarty S, LaMontagne P, Shimony J, Marcus DS, Sotiras A. MRI-based classification of IDH mutation and 1p/19q codeletion status of gliomas using a 2.5D hybrid multi-task convolutional neural network. Neuro-Oncol Adv. 2023;5(1):1–13. [Google Scholar]
23. Singh SP, Wang L, Gupta S, Goli H, Padmanabhan P, Gulyás B. 3D deep learning on medical images: a review. Sensors. 2020;20(18):5097. doi:10.3390/s20185097 [Google Scholar] [PubMed] [CrossRef]
24. Suh Y, Amelio I, Guerrero Urbano T, Tavassoli M. Clinical update on cancer: molecular oncology of head and neck cancer. Cell Death Dis. 2014;5(1):e1018. doi:10.1038/cddis.2013.548 [Google Scholar] [PubMed] [CrossRef]
25. Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA: A Cancer J Clin. 2019;69(2):127–57. [Google Scholar]
26. Vaz SC, Adam JA, Bolton RCD, Vera P, van Elmpt W, Herrmann K, et al. Joint EANM/SNMMI/ESTRO practice recommendations for the use of 2-[18F] FDG PET/CT external beam radiation treatment planning in lung cancer V1.0. Eur J Nucl Med Mol Imaging. 2022;49:1–21. [Google Scholar]
27. Forghani R, Savadjiev P, Chatterjee A, Muthukrishnan N, Reinhold C, Forghani B. Radiomics and artificial intelligence for biomarker and prediction model development in oncology. Comput Struct Biotechnol J. 2019;17(3):995–1008. doi:10.1016/j.csbj.2019.07.001. [Google Scholar] [CrossRef]
28. Nikulin P, Zschaeck S, Maus J, Cegla P, Lombardo E, Furth C, et al. A convolutional neural network with self-attention for fully automated metabolic tumor volume delineation of head and neck cancer in [18 F] FDG PET/CT. Eur J Nucl Med Mol Imaging. 2023;50(9):2751–66. doi:10.1007/s00259-023-06197-1. doi:10.1007/s00259-023-06197-1. [Google Scholar] [CrossRef]
29. Lambin P, Zindler J, Vanneste BG, Van De Voorde L, Eekers D, Compter I, et al. Decision support systems for personalized and participative radiation oncology. Adv Drug Deliv Rev. 2017;109(93):131–53. doi:10.1016/j.addr.2016.01.006. [Google Scholar] [CrossRef]
30. Marcu LG, Boyd C, Bezak E. Feeding the data monster: data science in head and neck cancer for personalized therapy. J Am Coll Radiol. 2019;16(12):1695–701. doi:10.1016/j.jacr.2019.05.045. doi:10.1016/j.jacr.2019.05.045. [Google Scholar] [CrossRef]
31. Myronenko A, Siddiquee MM, Yang D, He Y, Xu D. Automated head and neck tumor segmentation from 3D PET/CT HECKTOR 2022 challenge report. In: 3D head and neck tumor segmentation in PET/CT challenge, 2022 Sep 22; Cham: Springer Nature Switzerland; p. 31–7. [Google Scholar]
32. Zhu Z, Sun M, Qi G, Li Y, Gao X, Liu Y. Sparse dynamic volume TransUNet with multi-level edge fusion for brain tumor segmentation. Comput Biol Med. 2024;172(8):108284. doi:10.1016/j.compbiomed.2024.108284. [Google Scholar] [CrossRef]
33. Zhu Z, Wang Z, Qi G, Mazur N, Yang P, Liu Y. Brain tumor segmentation in MRI with multi-modality spatial information enhancement and boundary shape correction. Pattern Recognit. 2024;153(1):110553. doi:10.1016/j.patcog.2024.110553. [Google Scholar] [CrossRef]
34. Sun X, An C, Wang L. A coarse-to-fine ensembling framework for head and neck tumor and lymph segmentation in CT and PET images. In: 3D head and neck tumor segmentation in PET/CT challenge, 2022; Singapore: Springer; p. 38–46. [Google Scholar]
35. Jiang H, Haimerl J, Gu X, Lu W. A general web-based platform for automatic delineation of head and neck gross tumor volumes in PET/CT images. In: 3D head and neck tumor segmentation in PET/CT challenge, 2022; Singapore: Springer; p. 47–53. [Google Scholar]
36. Rebaud L, Escobar T, Khalid F, Girum K, Buvat I. Simplicity is all you need: out-of-the-box nnUNet followed by binary-weighted radiomic model for segmentation and outcome prediction in head and neck PET/CT. In: 3D head and neck tumor segmentation in PET/CT challenge, 2022; Singapore: Springer; p. 121–34. [Google Scholar]
37. Salahuddin Z, Chen Y, Zhong X, Rad NM, Woodruff HC, Lambin P. HNT-AI: an automatic segmentation framework for head and neck primary tumors and lymph nodes in FDG-PET/CT images. In: 3D head and neck tumor segmentation in PET/CT challenge, 2022; Singapore: Springer; p. 212–20. [Google Scholar]
38. Wang A, Bai T, Nguyen D, Jiang S. Octree boundary transfiner: efficient transformers for tumor segmentation refinement. In: 3D head and neck tumor segmentation in PET/CT challenge, 2022; Singapore: Springer; p. 54–60. [Google Scholar]
39. Wang K, Li Y, Dohopolski M, Peng T, Lu W, Zhang Y, et al. Recurrence-free survival prediction under the guidance of automatic gross tumor volume segmentation for head and neck cancers. In: 3D head and neck tumor segmentation in PET/CT challenge, 2022; Singapore: Springer; p. 144–53. [Google Scholar]
40. Jain A, Huang J, Ravipati Y, Cain G, Boyd A, Ye Z, et al. Head and neck primary tumor and lymph node auto-segmentation for PET/CT scans. In: 3D head and neck tumor segmentation in PET/CT challenge, 2022; Singapore: Springer; p. 61–9. [Google Scholar]
41. Chen J, Martel AL. Head and neck tumor segmentation with 3D UNet and survival prediction with multiple instance neural network. In: 3D head and neck tumor segmentation in PET/CT challenge, 2022; Singapore: Springer; p. 221–9. [Google Scholar]
42. Meng M, Bi L, Feng D, Kim J. Radiomics-enhanced deep multi-task learning for outcome prediction in head and neck cancer. In: 3D head and neck tumor segmentation in PET/CT challenge, 2022; Singapore: Springer; p. 135–43. [Google Scholar]
43. Andrearczyk V, Oreiller V, Hatt M, Depeursinge A. Head and neck tumor segmentation and outcome prediction. UK: Springer; 2022. [Google Scholar]
44. Challenge G. Segmentation of organs-at-risk and gross tumor volume of NPC for radiotherapy planning (SegRap2023). 2023. Available from: https://segrap2023.grand-challenge.org/. [Accessed 2024]. [Google Scholar]
45. Gatidis SKT. A whole-body FDG-PET/CT dataset with manually annotated tumor lesions (FDG-PET-CT-Lesions) [Dataset]. The Cancer Imaging Archive, 2022. Available from: https://autopet-ii.grand-challenge.org/dataset/. [Accessed 2024]. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools