 Open Access
Open Access
ARTICLE
Transfer Learning Empowered Skin Diseases Detection in Children
1 Department of Computer Science, College of Computer Science and Information Technology, Imam Abdulrahman Bin Faisal University, P.O. Box 1982, Dammam, 31441, Saudi Arabia
2 Department of Computer Information Systems, College of Computer Science and Information Technology, Imam Abdulrahman Bin Faisal University, P.O. Box 1982, Dammam, 31441, Saudi Arabia
3 Department of Computer Engineering, College of Computer Science and Information Technology, Imam Abdulrahman Bin Faisal University, P.O. Box 1982, Dammam, 31441, Saudi Arabia
4 ICS Department, King Fahd University of Petroleum and Minerals, Dhahran, 31261, Saudi Arabia
* Corresponding Author: Atta Rahman. Email:
(This article belongs to the Special Issue: Intelligent Medical Decision Support Systems: Methods and Applications)
Computer Modeling in Engineering & Sciences 2024, 141(3), 2609-2623. https://doi.org/10.32604/cmes.2024.055303
Received 22 June 2024; Accepted 12 September 2024; Issue published 31 October 2024
Abstract
Human beings are often affected by a wide range of skin diseases, which can be attributed to genetic factors and environmental influences, such as exposure to sunshine with ultraviolet (UV) rays. If left untreated, these diseases can have severe consequences and spread, especially among children. Early detection is crucial to prevent their spread and improve a patient’s chances of recovery. Dermatology, the branch of medicine dealing with skin diseases, faces challenges in accurately diagnosing these conditions due to the difficulty in identifying and distinguishing between different diseases based on their appearance, type of skin, and others. This study presents a method for detecting skin diseases using Deep Learning (DL), focusing on the most common diseases affecting children in Saudi Arabia due to the high UV value in most of the year, especially in the summer. The method utilizes various Convolutional Neural Network (CNN) architectures to classify skin conditions such as eczema, psoriasis, and ringworm. The proposed method demonstrates high accuracy rates of 99.99% and 97% using famous and effective transfer learning models MobileNet and DenseNet121, respectively. This illustrates the potential of DL in automating the detection of skin diseases and offers a promising approach for early diagnosis and treatment.Keywords
As the largest organ in the human body, the skin is essential to our physical appearance. The skin is a barrier between a person’s body and environmental surroundings [1]. Almost all skin comprises 5–10% of melanocytes [2]. Recent research shows skin diseases are the fourth leading cause of the worldwide nonfatal disease burden [3]. There are several infections and skin illnesses that are contagious and can affect newborns, children, and teenagers [4]. Healthcare practitioners receive about 2.3 million medical appointments yearly from infants with skin problems. Children and their caretakers are significantly affected by persistent dermatologic disorders, and the apparent presence of such disorders significantly impacts a child’s self-image and interactions between the family and classmates from a young age [4].
Researchers have discovered that kids with skin diseases evaluated poor life quality as low as children with other disorders on issues such as disruption in educational and sports activities, poor sleep, taunting, and discrimination [5].
The research conducted [6] in Saudi Arabia commonly presented that skin infections appear during the first six months. Approximately 60% of pediatric patients have dermatitis symptoms during the first year of age and about 85% by the age of 5 [7]. Eczema was about 37%. On the other hand, psoriasis and ringworm were an effect of 6% [8]. Early detection and prevention of these malicious skin diseases play a vital role in the lives of children and their parents. Examining skin disease scans appropriately is critical in determining a child’s health [9]. Nonetheless, it heavily relies on a dermatologist’s medical competence, awareness of regional diseases, differences in levels of expertise, and the pattern of imaging scans add to the difficulty of diagnosis with the human vision system (HVS) [10].
Dermatologists usually quickly diagnose such disorders before moving on to the pathologic examinations and diagnostic procedures to confirm the diagnosis. However, fresher dermatologists and those with less expertise are more prone to mistakes in diagnosis. As a result, technologies are necessary to help dermatologists accurately diagnose these malicious skin diseases and preserve valuable children’s lives and self-image [11]. Three skin diseases that are selected based on previous studies are Eczema, Ringworm, and Psoriasis. These unhealthy infections cause different factors, such as inflamed skin, dryness, itchiness, skin-to-skin contact, or changes in skin color [12].
The proposed research investigates Artificial Intelligence (AI) techniques to diagnose skin diseases early. AI is a subsection of computer science that has emerged as a significant area of research in medical imaging. Although AI and machine learning (ML) have been around for a while and are employed in many healthcare sectors, their usage in dermatological operations is very recent and limited. AI techniques train specific samples to solve problems based on many layers and complex neurons. Deep learning (DL) techniques were employed to solve complex diagnostic problems that other methods can hardly solve [13]. In addition, a limited number of studies were observed in the literature conducted in the Kingdom of Saudi Arabia, particularly on the locally obtained datasets.
Based on a comprehensive review of the relevant literature, DL is a potential candidate in image-based disease detection and diagnostics, especially in skin disease detection, with enhanced accuracy and precision. For instance, Bibi et al. [9] achieved improved accuracies of 98.80% and 85.4% over two different datasets, namely ISIC2019 and ISIC2018. Similarly, Dillshad et al. [11] achieved the highest accuracy of 94.4% on HAM10000, a publicly available dataset.
Accordingly, this study aims to:
• Covering the lack of dermatologists in Saudi Arabia.
• Developing DL models to determine the type of condition.
• Improving the easier way to diagnose skin diseases without requiring medical intervention.
• Achieving Saudi Arabia’s 2030 vision of a high-quality health care system.
This paper is organized according to the following sections. The Section 2 provides a background of the selected skin diseases. Followed by the Section 3, which provides a review of related literature. Then, the Section 4 describes the proposed techniques. Materials and methodology, a description of the dataset, image preprocessing, and performance measurements are presented in the Section 5. Afterward, the Section 6 presents the results and discussion comprehensively compared to state-of-the-art studies in the literature. Lastly, the study is concluded with a conclusion and recommendations for future work.
This section provides an overview of skin diseases targeted for detection problems in the current study, including eczema, ringworm, and psoriasis.
Eczema belongs to a group of conditions causing inflammatory, itchy, dry, and flared skin [14]. Eczema is not contagious, usually diagnosed by skin lesions. Approximately 15–20% of people suffer from eczema or another dermatitis at some point in their age. Eczema equally affects infantile, childhood, and adulthood. The conditions of eczema can be managed by proper treatment, but there is no cure [15]. Genetic and environmental conditions are the major causes of eczema in children. Usually, dust and unhygienic conditions are the primary culprits behind severe eczema in children. In the case of genetic linkage, one of the patient’s parents or family members should be affected to pass on the mutant gene to the next generation. The most prevalent form of eczema in children is Atopic Dermatitis (AD), or atopic eczema [16]. It is the most chronic disease affecting 10–20% of children in developed countries [16].
The skin of atopic eczema-affected children is inflamed and flaky. The scaling and roughness of the skin usually characterize it. Children are susceptible to skin infections due to repeated scratching, including lesions, hyperpigmentation, red bumps, and lumps, that is, papules. Usually, eczema starts in the first three months of life in children. Firstly, it affects the children’s lips and scalp, and then after puberty, it eventually ends up on the skin itching and infections. The hands and fingers are the most affected areas because children mostly use them due to constant irritation. Clinical diagnosis is the primary diagnostic criterion for this disease because no laboratory test is available. If eczema becomes chronic, the treatment and management of the disease becomes difficult for parents and children [17].
Anti-inflammatory and immunosuppressive drugs, such as topical steroids, are the most effective for treating skin lesions and inflammation in children’s skin. Children with AD are usually treated with acyclovir and are referred to a specialized dermatologist for effective diagnosis and treatment. Eczema in children is a chronic condition, so educating the patient and guardian is necessary for a prolonged period of preventive therapy to treat skin flares, itching inflammation, and infection in children [18]. AD is a chronic skin condition affecting infants, characterized by itchy, dry, and flaked skin. Genetic and environmental factors are the primary culprits in this disease. There is no cure for the disease, but treatment and management can alleviate the chronic condition in children [19].
Ringworm is a fungus-based skin and nail disease that is quite common. It is called ringworm because it produces a circular, red, and itchy rash. Other names for ringworm include “tinea” and “dermatophytosis” [20]. Thus, it is common in tropical areas and during the summer. In warm, moist change rooms and indoor swimming pools, dermatophytes grow. Even cold outside, ringworms are quite contagious [21]. The following parts of the body are sensitive to ringworm infection, including feet, scalp, beards, hands, and nails, whether on the hands or feet, and other body parts, such as the arms or legs. Initial symptoms of tinea involve circular, flat, slightly elevated, scaly, crusty patches. The patches usually have a red or pink tint on fair skin, but on dark skin, they typically have a brown or grey hue, can gradually enlarge, spread to more areas of the body, and cause itching. A doctor must diagnose such a problem to administer treatment correctly. Body ringworm, known as tinea corporis, can be confused with other skin illnesses, including eczema or psoriasis. When the diagnosis is complex, a sample of the afflicted skin is removed and examined under a microscope to confirm tinea corporis. Because tinea corporis has distinct histological characteristics, the bacteria that cause it can be discovered on the skin’s surface [22].
Based on a medical examination and inspection made in the primary school children in Alexandria, 510 children were inspected. Approximately 54.1% of them carried a considerable percentage of tinea capitis (Ringworm of the Scalp). It is highly recommended that parents and elders know the effects and symptoms of such cases by closely monitoring their children to prevent them. In addition, children with ringworms can experience hair loss, red patches on the skin, swollen lymph glands in the neck, and a noticeable high fever. In conclusion, ringworms are a skin infection that parents and elders should teach since it affects children by a substantial percentage [23].
Inflammatory autoimmune diseases such as psoriasis have multifactorial and systemic manifestations and are complex and multifactorial. There is an interaction between genetics and the environment that contributes to the pathogenesis of the disease of psoriasis. In most cases, it occurs between the ages of 15 and 30. However, it can take place at any age. The disfiguring and debilitating effects of psoriasis, which is not contagious, result in significant social and psychological problems for those affected. The worst things are social rejection, a negative impact on their quality of life, and loss of productivity. In addition, psoriasis is widely regarded as a health concern that requires comprehensive treatment. Although there is no cure for this illness, a carefully supervised course of treatment can lessen morbidity and enhance patients’ quality of life. There is an estimated prevalence of 2% worldwide, but it can vary considerably from country to country. Most psoriasis varies across nations from 0.09% to 11.4% [24].
People from different cultures and ethnic groups have psoriasis to varying degrees of severity. Despite psoriasis’ immunogenic nature, its causative immunogen has yet to be identified. Psoriasis is considered an immune-mediated chronic disease with genetic components [24]. Psoriasis frequently appears in children. The diagnosis can be problematic if the condition is minor or the appearance is unusual. Children experience all forms recognized in adults (plaque, guttate, erythrodermic, and pustular).
In youngsters, flexural and guttate types are prevalent. It is necessary to educate the child’s parents on the disease’s progression and available treatments to successfully manage the disease. Environmental triggers should be identified and, whenever possible, removed. Providing childcare or inpatient care for individuals who do not respond is appropriate. Topical medications can be used in conjunction with Ultraviolet Radiation phototherapy. A small percentage of patients, typically those with pustular psoriasis, arthropathic psoriasis, and refractory or erythrodermic illness, require systemic therapy. The preferred systemic agent is probably retinoids [25].
3 Review of Related Literature
In recent years, the medical field has widely incorporated AI to detect skin diseases. AI methods have proven efficient and accurate in detecting and classifying skin diseases. This section aims to explore the related studies of AI in detecting skin diseases, focusing on specific aspects such as their aims, datasets, methods, and results.
According to the study presented in [26], a model has been developed to classify various skin diseases, such as acne, cherry angioma, melanoma, and psoriasis. This study’s dataset consists of 377 images representing different disease categories. The images were obtained from various open and publicly available sources, including DermNet NZ and Atlas Dermatologico. The proposed model involves five steps: image acquisition, preprocessing, segmentation, feature extraction, and classification. Machine learning algorithms and image processing techniques were employed to develop this model. This study utilized a Support Vector Machine (SVM), Random Forest (RF), and K-Nearest Neighbor (K-NN) as classifiers. Hence, with 90.7% accuracy, the SVM achieved the best results.
In addition, Bandyopadhyay et al. [27] developed a smartphone application for Android that uses a Convolutional Neural Network (CNN) to enable users to quickly diagnose skin diseases and overcome the shortcomings of the traditional approach. The TensorFlow library, integrated into the mobile application, was utilized to run the model. The MobileNetV2 model was employed for the project, with a dataset including 6318 images from online dermatology resources accessible to the public. The diseases to be detected were vitiligo, eczema, and acne. The smartphone app achieved an overall accuracy rate of 91%, and the inference took 317 milliseconds. The MobileNetV2 model is preferred for smartphone devices due to its higher performance, small size, and fast training.
In addition, Aijaz et al. [28] developed an application using DL to predict the appearance of normal skin and accurately categorize five forms of psoriasis, including plaques, guttate, inverse, pustules, and erythrodermic. They used a dataset of 301 psoriasis images and 172 healthy skin images. Long short-term memories (LSTM) and CNN are two DL algorithms used with classification models trained on 80% of the images. Compared to LSTM, CNN archived higher with 84.2% accuracy. The work presented in [29] used an automated psoriasis detection system that employs DL to distinguish between psoriasis and other skin diseases that look similar. A dataset of 815 images was used, including 330 images of psoriasis-affected body parts, 235 images of skin in good health, and 250 images of diseases that are Psoriasis-like conditions (Seborrheic Dermatitis, Pityriasis, Tinea Corporis, Lichen Planus, Dandruff, and Eczema). In addition, they used the CNN model, which is ResNeXt101, to identify skin diseases. The model achieved 94% accuracy.
In addition, Setiawan et al. [30] developed classifiers to detect psoriasis, ringworm, and eczema using the Deep Convolutional Neural Network (DCNN). Experts in medicine supported researchers in gathering the dataset, which comprised 8000 images of the selected skin conditions. This research applied several models, such as DCNN and Neural Network (NN). As a result, the model achieved 93% accuracy. Hameed et al. [31] proposed a new model for classifying the most common skin lesions. They indicated that a categorization framework can classify an input skin image into one of six categories: eczema, benign, acne, psoriasis, malignant melanoma, and healthy skin. The following four steps are preprocessing, segmentation, feature extraction, and classification. Datasets from various sources were collected, and over 1800 images were tested for the experiments. Numerous alternative algorithms were employed to train the classification model. Therefore, the SVM had the highest accuracy of 94.74% compared to the other algorithms. Rashed and Popescu [32] proposed a highly automated approach to identify dermatological diseases from collected lesion images, such as melanoma, herpes, eczema, and psoriasis. The dataset consisted of 10,000 skin disease images and was trained using SVM and CNN models. After combining both methods, it attained the highest level of accuracy at 95.3%.
In addition, Alam et al. [33] utilized various image processing techniques to develop an automated method for detecting eczema and evaluating its severity. The dataset included 85 images, 31 healthy skin, 24 mild eczemas, and 30 severe eczemas. Images were acquired from various sources. They employed two different segmentation algorithms for skin and eczema segmentation from identified skin. The segmentation procedure utilized morphological image processing, image dilation, K-means clustering, and erosion techniques. Two steps were involved in the classifying process. First, images of eczema and healthy skin were classified. Then, categorized eczema images were utilized to determine whether the severity measurement was mild or severe. The classification achieved 90% accuracy.
Although the review of studies demonstrated high levels of accuracy, the proposed study showed more elevated levels than existing ones. In addition, none of the studies reviewed concentrated on the most prevalent skin diseases affecting children in Saudi Arabia; therefore, this is the first study in the Kingdom of Saudi Arabia.
4 Description of Proposed Techniques
Fig. 1 represents the methodological steps followed in the proposed study.
Figure 1: Methodological steps
4.1 Convolutional Neural Network (CNN)
CNNs are DL algorithms that recognize and process images. Multiple layers are included in this algorithm, including convolutional layers, pooling layers, and fully connected layers. CNNs utilize convolutional layers that employ filters to extract features such as edges, textures, and shapes from input images. A pooling layer is then applied to the output of the convolutional layer, which reduces the spatial dimensions of the feature maps but retains the essential information. A fully connected layer takes the outcome of the pooling layers to make a prediction or classify the image [34]. Size of kernels, number of kernels, length of strides, and pooling size are among the most common and widely used hyperparameters [34].
MobileNet is a type of CNN that is open source by Google. It employs depth-separable convolutions. The separable convolutions are derived from two operations, namely, depth-wise and pointwise convolutions. Depth-wise convolution, as the name reveals, is derived from the idea that a filter’s depth and spatial dimensions can be separated. On the other hand, pointwise convolution consists of a depth-wise convolution with a kernel size of 1 × 1, combining the features produced by the depth-wise convolution. Compared to networks with regular convolutions of the same depth, it significantly reduces the number of parameters. Thus, the result is a relatively lightweight deep NN (DNN) [35]. Optimizer, type of cost function, classifier type, batch size, number of epochs, and dropout function are among the popular hyperparameters that are considered during the optimization process in the proposed study.
The DenseNet architecture is a type of CNN in which every layer is connected to every other layer in a feed-forward manner. Similarly, densely connected convolutional networks are referred to as densely connected networks. The primary objective of DenseNet is to ensure feature reuse and reduce the number of parameters in the model. This is accomplished by connecting each layer to all subsequent layers, allowing for the reuse of features from previous layers. The model becomes more accurate due to reducing the number of parameters needed for training [36].
The DenseNet architecture was first introduced by Huang et al. in 2016 [37]. They proposed a pair of architectures, DenseNet121 and DenseNet169, which vary in depth and width. They also proposed that each layer be connected to all subsequent layers using a new type of connection called a “dense block”, which allows for reusing features from previous layers and works as a knowledge accumulator. Optimizer, type of cost function, classifier type, batch size, number of epochs, and initial learning rate are among the popular hyperparameters considered during the optimization process in the proposed study. These models were chosen after carefully reviewing the literature. These models exhibit promising results for similar problems.
The study obtained skin disease images from various sources, including the Kaggle Dermnet dataset [38], the Google Images search engine, and Atlas [39]. A total of 285 images were obtained for all the selected skin diseases. Afterward, the images were annotated manually to identify the type of skin disease shown in each image. In addition, images from the university hospital were collected and used for validation. The dataset was divided into two sets: training and testing. The training set comprised 76 skin disease images (80%). In contrast, the testing set comprised 19 images for each type of skin disease (20%).
Image preprocessing removes undesirable features from an image while improving its significant features before they are fed into a classification model. The dataset has unwanted regions in terms of size and color. This study utilized image augmentation to improve the detection accuracy. Various techniques can be used, such as rotation, shifting, and flipping an image. The images can be rotated at any angle, generating new images. An image’s pixels are shifted from one position to another by changing their position. That includes the augmentation of horizontal shifts and vertical shifts, respectively. Lastly, image flipping refers to rotating an image horizontally or vertically [40].
This study evaluated the performance of CNN models based on the following standards: accuracy, precision, recall, and F-score. The metrics were shortlisted because of their applications in similar studies in the literature [41,42]. The model with the highest level of accuracy was selected to classify new skin disease images. The equations below show the formulas for each performance metric. Based on the formulas, TP indicates a True Positive, TN indicates a True Negative, FP indicates a False Positive, and FN indicates a False Negative.
This section describes dataset collection and image preprocessing. MobileNet and DenseNet121 are employed to classify the type of skin disease. Then, a confusion matrix is utilized to evaluate the model’s performance.
The size of the feature vector in skin cancer detection using images can vary depending on the specific method and model used for feature extraction and classification. Some methods can use smaller, fixed-sized areas (patches) of an image and reduce the pixel values within each patch to a single vector, while others can use different parameters such as lesion diameter, color change, border structure, and asymmetry. The optimization of these vectors is crucial for enhancing the model’s ability to discern patterns in the image data for accurate diagnosis [33]. This study utilized the same feature vectors to make the study comparable to the state-of-the-art studies in the literature.
The proposed experimental setup comprised dataset preprocessing and training with evaluation phases. These phases were performed in two experiments.
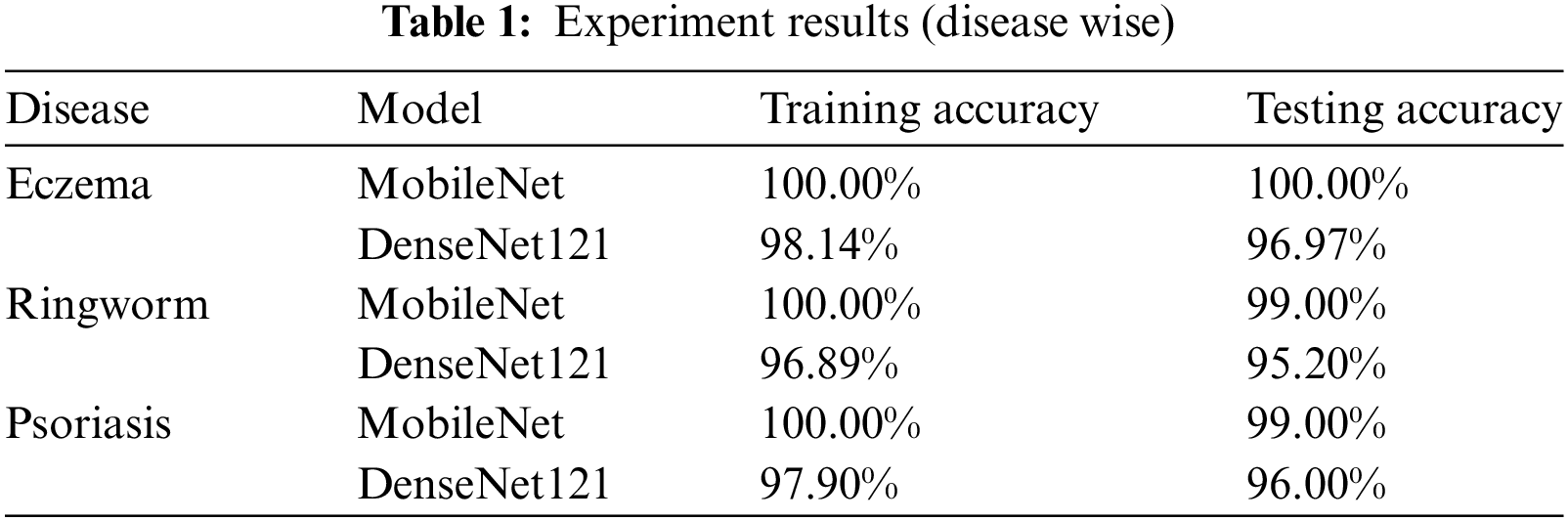
Firstly, MobileNet was trained on the dataset in this experiment. The preprocessing phase divides the dataset into 80% training and 20% testing images. The model was investigated for each disease separately, and training and testing accuracies were obtained. For all three diseases, the test accuracy was 100%, which shows the algorithm’s promising nature for the given dataset. Test accuracy was 99.7%, 99. 2% and 99.1%, respectively. The results are shown in Fig. 1 and Table 1.
Regarding hyperparameters, the following values were used: Adam as an optimizer, binary cross entropy as loss function, SoftMax as a classifier, 20 as maximum epochs with 64 batch sizes and a dropout rate of 0.3. These hyperparameters were obtained intuitively based on the well-known studies conducted in the literature for similar problems.
Secondly, DenseNet121 was trained on the dataset and investigated for the diseases individually. Regarding hyperparameters, the following values were used: Adam as an optimizer, binary cross entropy as a loss function, SoftMax as a classifier, 20 epochs with 64 batch sizes and a learning rate of 0.003. These hyperparameters were obtained intuitively based on the well-known studies conducted in the literature.
Fig. 2 and Table 1 indicate that the performance is slightly below the MobileNet model. Testing and training accuracies for eczema are 98.14% and 96.97%, respectively. For ringworm, testing and training accuracies are 96.89% and 95.20%, respectively. Testing and training accuracies are 97.90% and 96% for psoriasis, respectively. MobileNet outperforms DenseNet121 by 2–4% in the training and testing phases.
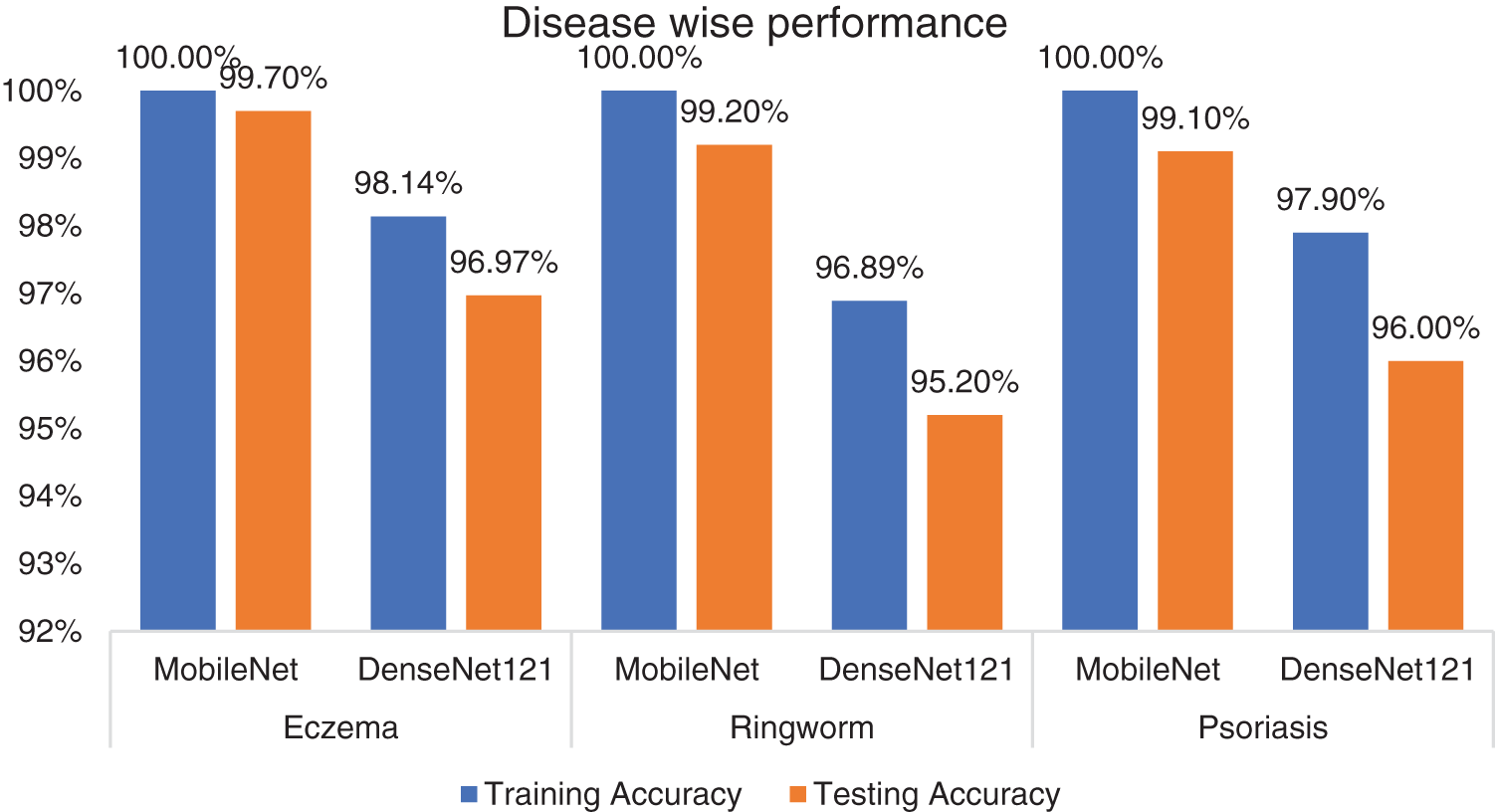
Figure 2: Disease-wise performance of the models
The obtained results were shared with, vetted, and validated by medical healthcare professionals and dermatologists at the College of Medicine. In addition, a local and unseen image set was employed to validate and verify the proposed techniques.
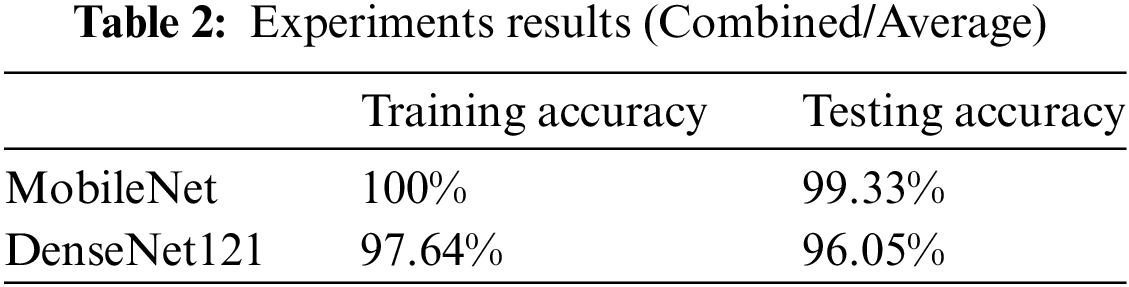
Thirdly, the performance of both models was aggregated for all the diseases and averaged. MobileNet achieved an average training accuracy of 100% and an average testing accuracy of 99.3%. DenseNet121 was investigated using the same dataset for the three skin conditions. In DenseNet121, the average training accuracy reached 97.64%, whereas the average testing accuracy reached 96%.
Fig. 3 and Table 2 indicate the experiment results. MobileNet outperforms DenseNet121 by 2.64% and 3.30% in the training and testing phases while comparing average accuracy for all three skin diseases in children.
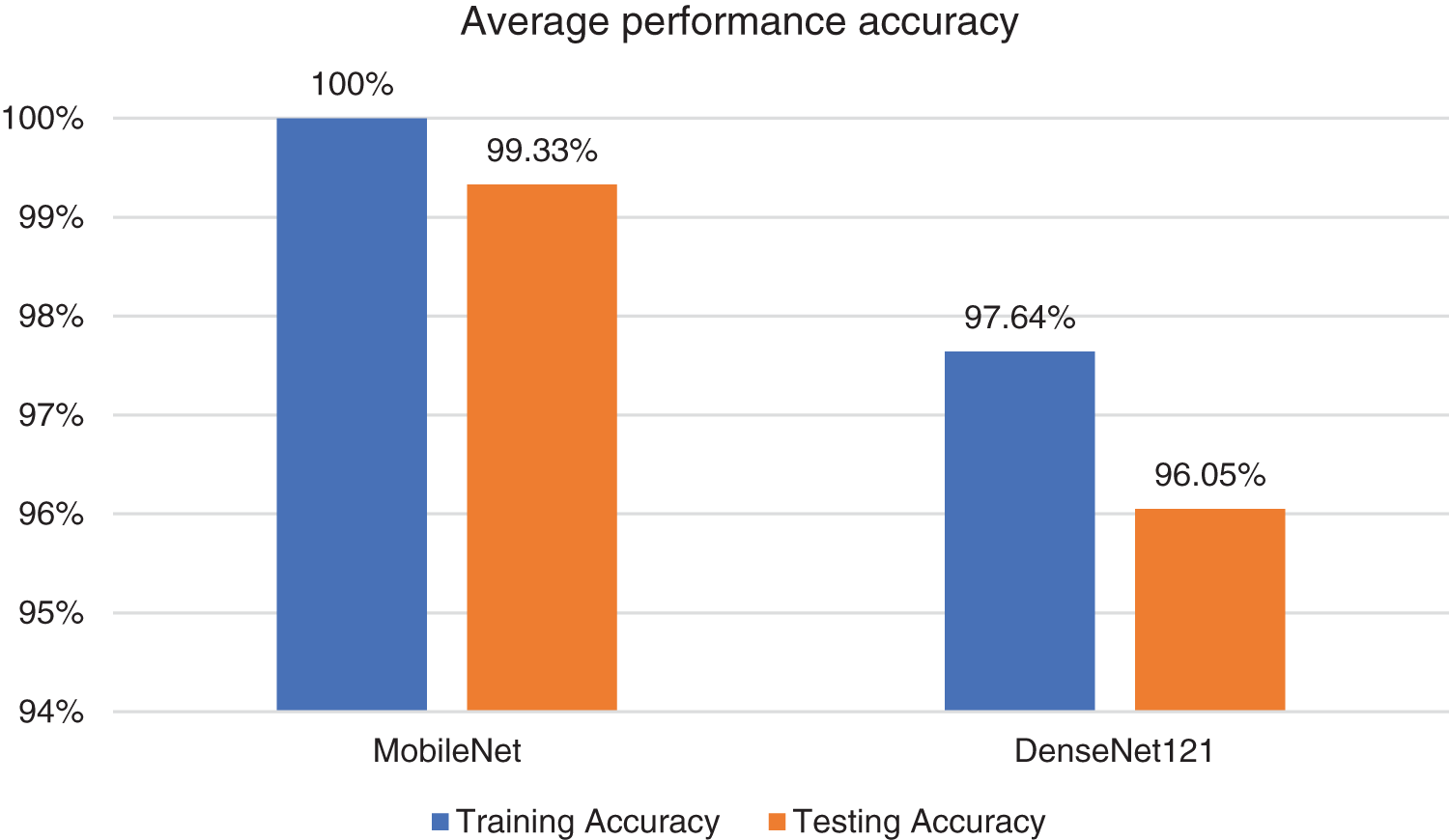
Figure 3: Average performance of the models
5.2 Comparison with State-of-the-Art
Finally, the classification performance of the proposed models was evaluated, and the results were compared to the performance of previous studies. First, the two models were trained using several sources, such as Kaggle, Google Images, and Atlas. Upon conducting several experiments, it was revealed that the results of the MobileNet architecture achieved the highest accuracy. Then, DenseNet121 was used with the same dataset. Both were imported as pre-trained models. The studies were carefully selected based on common grounds to provide a fair comparison.
Table 3 compares state-of-the-art approaches to similar datasets for skin disease in children. The schemes are shortlisted based on their suitability and alignment with the dataset. Training and test data accuracy continued to increase until the 1930s and gradually started to saturate for the later eras. Upon comparing the detection results of MobileNet and DenseNet121, the performance of the MobileNet model trained on images is generally superior to DenseNet121. The model was trained using only body images of eczema, psoriasis, and ringworm diseases. Hence, the average accuracy, precision, recall, and F1-score are listed in Table 3.
Upon comparison to state-of-the-art, the scheme in the literature [43] is close to the proposed models in terms of accuracy. However, other metrics are not mentioned in the study. In addition, the studies in [44] and [45] perform considerably below the proposed models. Numerically, that is around 6% and 15% below MobileNet while 4% and 12% below DenseNet121, respectively.
This study presents transfer learning approaches to skin disease classification among children. A diverse range of datasets are combined, and the pre-trained models are investigated. The proposed models are promising in accuracy and other merits, such as precision, recall, and F1-score. Afterward, the scheme is compared to state-of-the-art, which indicates that the proposed schemes outperform in classification accuracy. The proposed schemes incorporated three common skin diseases among children: eczema, ringworm, and psoriasis. In addition, the datasets are adequately preprocessed and annotated, and they contain high-resolution images. This is the main reason behind the overall good results.
As far as the study’s limitations are concerned, firstly, it is limited to only three diseases above, while studies in the literature [45] comprehend a greater number of diseases in their modeling and classification. Secondly, the current study only targets skin diseases among children, not adults. Thirdly, analyses are not made on gender bases. These limitations can be addressed in the future by incorporating large visual models (LVM) [46] with more diseases and a diverse range of age groups with gender segregation.
DL is successfully applied to detect skin diseases with high accuracy. This study achieved an accuracy of 99.99% utilizing the MobileNet model. Hence, patients suffering from skin conditions can receive faster, more accurate diagnoses and earlier treatment. Also, the results indicated that DL can potentially become a valuable tool for diagnosing skin diseases in the future. Although DL can be valid for future clinical applications, some challenges must be addressed before it can be implemented. There is a need to gather additional data for training and validating the models and develop more robust models that can demonstrate better generalization to unseen data. More sophisticated models can be developed to detect a broader array of skin diseases for future work. Additional data sources, such as a Saudi dataset, can be incorporated into these models to improve their accuracy. Research can be conducted on the methods to automatically interpret their results, allowing clinicians to receive actionable insights from DL models. Lastly, all these efforts will improve the accuracy and reliability of skin disease detection using DL. In the future, hybrid intelligence-based approaches must be investigated to improve the results of skin disease detection among different age groups by incorporating data augmentation techniques.
Acknowledgement: The authors would like to acknowledge the College of Computer Science and Information Technology (CCSIT) at Imam Abdulrahman Bin Faisal University (IAU), Dammam, for partially using the computing lab resources during the experiments phase for the current study.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, Mohammed Gollapalli, Atta Rahman; Data curation, Nourah S. Alqahtani and Aghiad Bakry; Formal analysis, Atta Rahman and Alaa Alahmadi; Investigation, Mohammed Gollapalli and Rashad Ahmed; Methodology, Meena N. Alnuaimi, Nourah S. Alqahtani, Dania AlKhulaifi and Alaa Alahmadi; Project administration, Alaa Alahmadi; Software, Meena Alnuaimi and Nourah S. Alqahtani; Supervision, Mohammed Gollapalli and Atta Rahman; Writing—original draft, Meena N. Alnuaimi; Writing—review & editing, Atta Rahman, Hesham Al-Musallam and Mustafa Youldash. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data are openly available in a public repository. The data that support the findings of this study are openly available in Kaggle (Dermnet) at https://www.kaggle.com/datasets/shubhamgoel27/dermnet and Atlas Dermatology https://atlasdermatology.com/ (accessed on 11 September 2024).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Lopez-Ojeda W, Pandey A, Alhajj M, Oakley AM, Anatomy, skin (Integument). In: StatPearls. Treasure Island (FLStatPearls Publishing; 2024 Jan. [Google Scholar]
2. Brodowski R, Pakla P, Dymek M, Migut M, Ambicki M, Stopyra W, et al. Clinical-pathological characteristics of patients treated for cancers of the eyelid skin and periocular areas. Adv Clin Exp Med. 2019;28(3):325–30. doi:10.17219/acem/78023 [Google Scholar] [PubMed] [CrossRef]
3. Hardalo C, Lodise TP, Bidell M, Flanagan S, De-Anda C, Anuskiewicz S, et al. Clinical safety and tolerability of tedizolid phosphate in the treatment of acute bacterial skin and skin structure infections. Expert Opin Drug Saf. 2018;17(4):359–67. doi:10.1080/14740338.2018.1446939 [Google Scholar] [PubMed] [CrossRef]
4. Bewley A. The neglected psychological aspects of skin disease. Br Med J. 2017;358:j3208. doi:10.1136/bmj.j3208 [Google Scholar] [PubMed] [CrossRef]
5. Goyal M, Yap MH, Hassanpour S. Multi-class semantic segmentation of skin lesions via fully convolutional networks. In: Bioinformatics 2020, 11th International Conference on Bioinformatics Models, Methods and Algorithms, Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies. (BIOSTEC 2020), 2020; Valletta, Malta; p. 290–4. doi:10.5220/0009380300002513. [Google Scholar] [CrossRef]
6. Alraey Y, Alhweti R, Almutairi H, Al-Qahtani AA, Alshahrani MA, Asiri ML, et al. Molecular characterization of leishmania species among patients with cutaneous leishmaniasis in Asir province, Saudi Arabia. Pathogens. 2022;11(12):1472. doi:10.3390/pathogens11121472 [Google Scholar] [PubMed] [CrossRef]
7. Elezbawy B, Fasseeh AN, Fouly E, AbuEsba LC, Abdulkarim HA, Al-Haddab M, et al. The humanistic and economic burden of atopic dermatitis among adults and adolescents in Saudi Arabia. J Med Econ. 2022;25(1):1231–9. doi:10.1080/13696998.2022.2152234 [Google Scholar] [PubMed] [CrossRef]
8. Ahmed AE, Jradi H, AlBuraikan DA, AlMuqbil BI, Albijan MA, Al-Shehri AM, et al. Rate and factors for scabies recurrence in children in Saudi Arabia: a retrospective study. BMC Pediatr. 2019;19(1):187. doi:10.1186/s12887-019-1565-9 [Google Scholar] [PubMed] [CrossRef]
9. Bibi S, Khan MA, Shah JH, Damaševičius R, Alasiry A, Marzougui M, et al. MSRNet: multiclass skin lesion recognition using additional residual block based fine-tuned deep models information fusion and best feature selection. Diagnostics. 2023;13(19):3063. doi:10.3390/diagnostics13193063 [Google Scholar] [PubMed] [CrossRef]
10. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017 Jun 28: 546(7660):686. doi:10.1038/nature21056. [Google Scholar] [CrossRef]
11. Dillshad V, Khan MA, Nazir M, Saidani O, Alturki N, Kardy S. D2LFS2Net: multi-class skin lesion diagnosis using deep learning and variance-controlled Marine Predator optimisation: an application for precision medicine. CAAI Trans Intell Technol. 2023;1(1):1–16. doi:10.1049/cit2.12267. [Google Scholar] [CrossRef]
12. Fasseeh AN, Elezbawy B, Korra N, Tannira M, Dalle H, Aderian S, et al. Burden of atopic dermatitis in adults and adolescents: a systematic literature review. Dermatol Ther. 2022;12(12):2653–68. doi:10.1007/s13555-022-00819-6 [Google Scholar] [PubMed] [CrossRef]
13. Schneider SL, Kohli I, Hamzavi IH, Council ML, Rossi AM, Ozog DM. Emerging imaging technologies in dermatology: part I: basic principles. J Am Acad Dermatol. 2019;80(4):1114–20. doi:10.1016/j.jaad.2018.11.042 [Google Scholar] [PubMed] [CrossRef]
14. Al-Adawiyah R, Putera AM, Astari L, Ariyanto FC. Determinant factors of recurrence atopic dermatitis symptoms in children: a cross-sectional study. Ann Med Surg. 2021;70:102847. doi:10.1016/j.amsu.2021.102847 [Google Scholar] [PubMed] [CrossRef]
15. Hernández CD, Casanello P, Harris PR, Castro-Rodríguez JA, Iturriaga C, Perez-Mateluna G, et al. Early origins of allergy and asthma (ARIESstudy protocol for a prospective prenatal birth cohort in Chile. BMC Pediatr. 2020;20(1):164. doi:10.1186/s12887-020-02077-x [Google Scholar] [PubMed] [CrossRef]
16. Lu CL, Liu XH, Stub T, Kristoffersen AE, Liang SB, Wang X, et al. Complementary and alternative medicine for treatment of atopic eczema in children under 14 years old: a systematic review and meta-analysis of randomized controlled trials. BMC Complem Altern Med. 2018 Sep 26;18(1):260. doi:10.1186/s12906-018-2306-6 [Google Scholar] [PubMed] [CrossRef]
17. Jackson-Browne MS, Henderson N, Patti M, Spanier A, Braun JM. The impact of early-life exposure to antimicrobials on asthma and eczema risk in children. Curr Environ Health Rep. 2019 Dec;6(4):214–24. doi:10.1007/s40572-019-00256-2 [Google Scholar] [PubMed] [CrossRef]
18. Szajewska H, Horvath A. Lactobacillus rhamnosus GG in the primary prevention of eczema in children: a systematic review and meta-analysis. Nutrients. 2018;10(9):1319. doi:10.3390/nu10091319 [Google Scholar] [PubMed] [CrossRef]
19. de-Lusignan S, Alexander H, Broderick C, Dennis J, McGovern A, Feeney C, et al. Atopic dermatitis and risk of autoimmune conditions: population-based cohort study. J Allergy Clin Immunol. 2022;150(3):709–13. doi:10.1016/j.jaci.2022.03.030 [Google Scholar] [PubMed] [CrossRef]
20. Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses. 2009 Jan;52(1):95. doi:10.3390/nu10091319. [Google Scholar] [CrossRef]
21. Gupta AK, Mays RR, Versteeg SG, Piraccini BM, Shear NH, Piguet V, et al. Tinea capitis in children: a systematic review of management. J Eur Acad Dermatol Venereol. 2018;32(12):2264–74. doi:10.1111/jdv.15088 [Google Scholar] [PubMed] [CrossRef]
22. Ringworm (for Parents)-Nemours KidsHealth, 2023. Available from: https://kidshealth.org/en/parents/fungal-ringworm.html [Accessed 2023]. [Google Scholar]
23. Gupta AK, Friedlander SF, Simkovich AJ. Tinea capitis: an update. Pediatr Dermatol. 2022;39(2):167–72. doi:10.1111/pde.14925 [Google Scholar] [PubMed] [CrossRef]
24. Alzeer F, AlOtair H, Aleisa A. Epidemiology and cutaneous manifestations of psoriasis in Saudi Arabia: a narrative review. Clin Cosmet Investig Dermatol. 2022;15:347–55. doi:10.2147/ccid.s352654 [Google Scholar] [PubMed] [CrossRef]
25. Hroch M, Chladek J, Simkova M, Vaneckova J, Grim J, Martinkova J. A pilot study of pharmacokinetically guided dosing of oral methotrexate in the initial phase of psoriasis treatment. J Eur Acad Dermatol Venereol. 2008;22(1):19–24. doi:10.1111/j.1468-3083.2007.02264.x [Google Scholar] [PubMed] [CrossRef]
26. Maduranga M, Nandasena D. Mobile-based skin disease diagnosis system using convolutional neural networks (CNN). Image Graph Signal Process. 2022;2022(1):47–57. doi:10.5815/ijigsp.2022.03.05. [Google Scholar] [CrossRef]
27. Bandyopadhyay SK, Bose P, Bhaumik A, Poddar S. Machine learning and deep learning integration for skin diseases prediction. Int J Eng Trends Technol. 2022;70(2):11–8. doi:10.14445/22315381/ijett-v70i2p202. [Google Scholar] [CrossRef]
28. Aijaz SF, Khan SJ, Azim F, Shakeel CS, Hassan U. Deep learning application for effective classification of different types of psoriasis. J Healthc Eng. 2022;2022(4):1–12. doi:10.1155/2022/7541583 [Google Scholar] [PubMed] [CrossRef]
29. Ahmed MS, Rahman A, AlGhamdi F, AlDakheel S, Hakami H, AlJumah A, et al. Joint diagnosis of pneumonia, COVID-19, and tuberculosis from chest X-ray images: a deep learning approach. Diagnostics. 2023;13(15):2562. doi:10.3390/diagnostics13152562 [Google Scholar] [PubMed] [CrossRef]
30. Setiawan NB, Natalia F, Ferdinand FV, Sudirman S, Ko CS. Classification of skin diseases and disorders using convolutional neural network on a mobile application. ICIC Express Letters: Part B: Appl. 2021;12(8):715–21. doi:10.24507/icicelb.12.08.715. [Google Scholar] [CrossRef]
31. Hameed N, Hameed F, Shabut A, Khan S, Cirstea S, Hossain A. An intelligent computer-aided scheme for classifying multiple skin lesions. Computers. 2019;8(3):62. doi:10.3390/computers8030062. [Google Scholar] [CrossRef]
32. Rashed BM, Popescu N. Critical analysis of the current medical image-based processing techniques for automatic disease evaluation. Syst Lit Rev Sens. 2022;22(18):7065. doi:10.3390/s22187065 [Google Scholar] [PubMed] [CrossRef]
33. Alam MN, Munia TTK, Tavakolian K, Vasefi F, Mackinnon N, Fazel-Rezai R. Automatic detection and severity measurement of eczema using image processing. In: Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 2016; Orlando, FL, USA; p. 1365–8. doi:10.1109/embc.2016.7590961 [Google Scholar] [PubMed] [CrossRef]
34. Rahman A, Youldhash M, Alshammari G, Sebiany A, Alzayat J, Alsayed M, et al. Diabetic retinopathy detection: a hybrid intelligent approach. Comput Mat Contin. 2024;80(3):1–16. doi:10.32604/cmc.2024.055106. [Google Scholar] [CrossRef]
35. Souid A, Sakli N, Sakli H. Classification and predictions of lung diseases from chest X-rays using mobilenet V2. Appl Sci. 2021;11(6):2751. doi:10.3390/app11062751. [Google Scholar] [CrossRef]
36. Chauhan T, Palivela H, Tiwari S. Optimization and fine-tuning of DenseNet model for classification of COVID-19 cases in medical imaging. Int J Inform Manage Data Insights. 2021 Nov;1(2):100020. doi:10.1016/j.jjimei.2021.100020. [Google Scholar] [CrossRef]
37. Srinivasu PN, SivaSai JG, Ijaz MF, Bhoi AK, Kim W, Kang JJ. Classification of skin disease using deep learning neural networks with MobileNet V2 and LSTM. Sensors. 2021 Apr 18;21(8):2852. doi:10.3390/s21082852 [Google Scholar] [PubMed] [CrossRef]
38. Goel S, Hall B. Dermnet | Kaggle. 2020. Available from: https://www.kaggle.com/datasets/shubhamgoel27/dermnet. [Accessed 2023]. [Google Scholar]
39. Rouge B. Baton Rouge, LA Dermatologist | Atlas Dermatology. 2024. Available from: https://atlasdermatology.com/. [Accessed 2023]. [Google Scholar]
40. Ibrahim NM, Gabr DG, Rahman A, Musleh D, AlKhulaifi D, AlKharraa M. Transfer learning approach to seed taxonomy: a wild plant case study. Big Data Cogn Comput. 2023;7(3):128. doi:10.3390/bdcc7030128. [Google Scholar] [CrossRef]
41. Gollapalli M, Rahman A, Kudos SA, Foula MS, Alkhalifa AM, Albisher HM, et al. Appendicitis diagnosis: ensemble machine learning and explainable artificial intelligence-based comprehensive approach. Big Data Cogn Comput. 2024;8(9):108. doi:10.3390/bdcc8090108. [Google Scholar] [CrossRef]
42. Arooj S, Khan MF, Shahzad T, Khan MA, Nasir MU, Zubair M, et al. Data fusion architecture empowered with deep learning for breast cancer classification. Comput Mat Contin. 2023;77(3):2813–31. doi:10.32604/cmc.2023.043013. [Google Scholar] [CrossRef]
43. Alghieth M. Skin disease detection for kids at school using deep learning techniques. Int J Onl Eng. 2022;18(10):114–28. doi:10.3991/ijoe.v18i10.31879. [Google Scholar] [CrossRef]
44. Lucius M, De All J, De All JA, Belvisi M, Radizza L, Lanfranconi M, et al. Deep neural frameworks improve the accuracy of general practitioners in the classification of pigmented skin lesions. Diagnostics. 2020 Nov 18;10(11):969. doi:10.3390/diagnostics10110969 [Google Scholar] [PubMed] [CrossRef]
45. Liu Y, Jain A, Eng C, Way DH, Lee K, Bui P, et al. A deep learning system for differential diagnosis of skin diseases. Nat Med. 2020;26(6):900–8. doi:10.1038/s41591-020-0842-3 [Google Scholar] [PubMed] [CrossRef]
46. Wang J, Liu Z, Zhao L, Wu Z, Ma C, Yu S, et al. Review of large vision models and visual prompt engineering. Meta-Radiology. 2023;1(3):100047. doi:10.1016/j.metrad.2023.100047. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF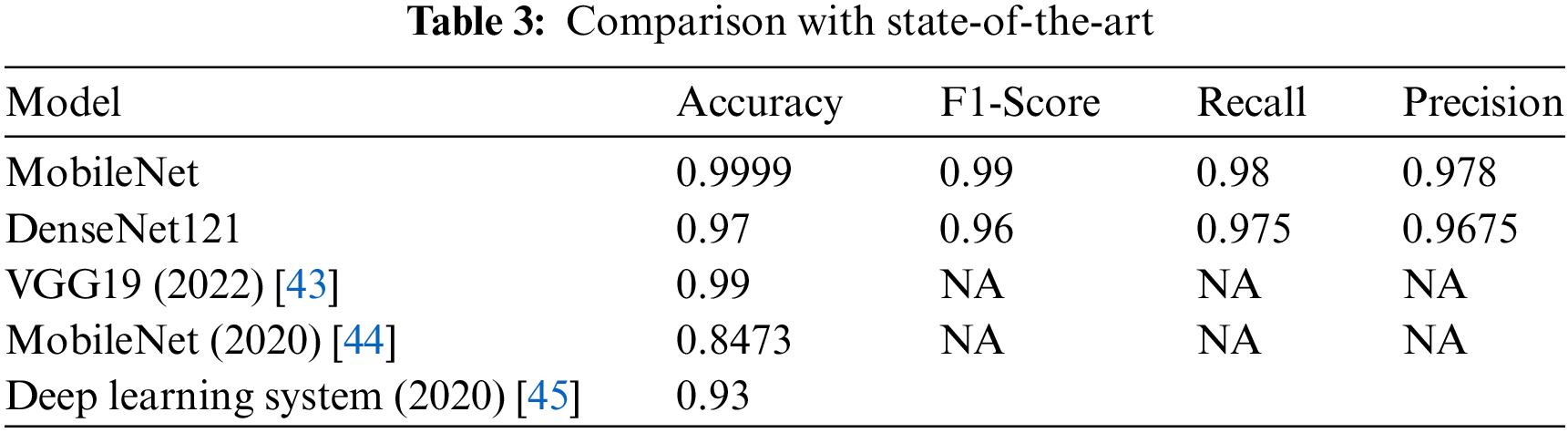
 Downloads
Downloads
 Citation Tools
Citation Tools