 Open Access
Open Access
ARTICLE
A Genetic Algorithm-Based Optimized Transfer Learning Approach for Breast Cancer Diagnosis
1 Department of Computer Science, College of Computer and Information Sciences, King Saud University, Riyadh, 11543, Saudi Arabia
2 Department of Information Systems, College of Computer and Information Sciences, King Saud University, Riyadh, 11543, Saudi Arabia
* Corresponding Author: Mohammad Mehedi Hassan. Email:
(This article belongs to the Special Issue: Intelligent Medical Decision Support Systems: Methods and Applications)
Computer Modeling in Engineering & Sciences 2024, 141(3), 2575-2608. https://doi.org/10.32604/cmes.2024.055011
Received 13 June 2024; Accepted 11 September 2024; Issue published 31 October 2024
Abstract
Breast cancer diagnosis through mammography is a pivotal application within medical image-based diagnostics, integral for early detection and effective treatment. While deep learning has significantly advanced the analysis of mammographic images, challenges such as low contrast, image noise, and the high dimensionality of features often degrade model performance. Addressing these challenges, our study introduces a novel method integrating Genetic Algorithms (GA) with pre-trained Convolutional Neural Network (CNN) models to enhance feature selection and classification accuracy. Our approach involves a systematic process: first, we employ widely-used CNN architectures (VGG16, VGG19, MobileNet, and DenseNet) to extract a broad range of features from the Medical Image Analysis Society (MIAS) mammography dataset. Subsequently, a GA optimizes these features by selecting the most relevant and least redundant, aiming to overcome the typical pitfalls of high dimensionality. The selected features are then utilized to train several classifiers, including Linear and Polynomial Support Vector Machines (SVMs), K-Nearest Neighbors, Decision Trees, and Random Forests, enabling a robust evaluation of the method’s effectiveness across varied learning algorithms. Our extensive experimental evaluation demonstrates that the integration of MobileNet and GA significantly improves classification accuracy, from 83.33% to 89.58%, underscoring the method’s efficacy. By detailing these steps, we highlight the innovation of our approach which not only addresses key issues in breast cancer imaging analysis but also offers a scalable solution potentially applicable to other domains within medical imaging.Keywords
The prevalence of breast cancer is significant in Saudi Arabia [1]. It was the ninth leading cause of death among women in 2010 [2]. In 2009, the number of newly reported breast cancer cases surpassed 1000. With an increasing population and aging demographic, this number is expected to rise significantly in the coming decades. The American Cancer Society recommends that every woman over the age of 40 should undergo breast cancer screening through mammograms. Patients with lesions visible in mammography typically undergo a follow-up ultrasound examination, during which ultrasound-guided biopsies are performed. Breast biopsies, which save many lives by identifying cancerous tumors, are performed annually in the United States, with 70%–90% of these biopsies being benign [3]. There is a need for a technique that can reliably distinguish between benign and malignant lesions without resorting to invasive methods, which would reduce unnecessary biopsies, lower healthcare costs, and decrease patient anxiety.
Since the advent of Deep Learning in 2012, these technologies have revolutionized medical diagnostics, showing superior performance over traditional machine learning methods. Notably, a team led by Tuggener et al. [4] employed deep learning for mitotic figure detection, demonstrating outstanding accuracy on the first publicly annotated breast cancer dataset at the ICPR 2020 (International Conference on Pattern Recognition). Another significant advancement in cancer diagnostics was made by [5], achieving a 99.48% accuracy rate when combined with human pathologists’ predictions. Such advancements suggest a promising future for deep learning in enhancing diagnostic precision and reducing reliance on human pathologists.
In this research, our goal is to assess various pre-trained deep learning methods, such as the models developed by the Visual Geometry Group (VGG) which called VGG16, VGG19, and other models such as MobileNet and DenseNet to extract features and detect whether lesions in ultrasound images are benign or malignant. Currently, ultrasound diagnosis relies on the expertise of human pathologists, which can be subjective. By utilizing the aforementioned techniques, we aim to provide compelling evidence for accurate diagnosis. The system will acquire knowledge from an extensive database of numerous patient cases, a task nearly impossible for a human pathologist to accomplish within a short period. Within a few hours, the system will be able to deliver a reliable verdict on the malignancy of lesions [6].
Wrapper feature selection methods create numerous models with different subsets of input features and select those features that result in the best-performing model according to a performance metric. The Wrapper methodology treats feature set selection as a search challenge, where various combinations are formed, assessed, and compared against others. To evaluate a set of features and allocate performance scores, a predictive model is employed [7]. Our approach involved utilizing a genetic algorithm in combination with the wrapper method for feature selection. This technique optimizes the learning algorithm’s performance by generating a population of candidate solutions and iteratively improving them using mutation and crossover operators. We applied this approach to each Convolutional Neural Network (CNN) model, and the resultant features were combined and further selected to obtain the most useful ones. The chosen features were then utilized by the Multiclass Support Vector Machine (MSVM) classifier. The Medical Image Analysis Society (MIAS) developed and supplied the digital mammography datasets, which are extensively used and available online for research purposes at no cost. The images are presented in Portable Gray Map (PGM) format and include both right- and left-oriented breasts. Each mammography in the Mini-Mammographic Image Analysis Society (MIAS) dataset is categorized as malignant, benign, or normal [8,9].
This paper seeks to create and implement automated artificial intelligence tools to serve as preliminary diagnostic aids in detecting breast cancer lesions. Radiologists have long used certain features in B-mode images of mammographic-visible breast lesions for preliminary determination of malignancy by reviewing related work and collecting a large data sample of breast cancer images. In addition, our intention is to enhance the clarity and distinction of acquired breast images using deep learning and image processing methods. In this study, we propose a hybrid optimized approach by integrating pre-trained CNN models with a genetic algorithm (GA) and various classifiers. The contributions of our work can be summarized as follows:
– Feature Extraction: Utilizing four wide-ranging pre-trained CNN models to speed up the classification process and meet the challenges posed by the limited size of breast image datasets.
– Feature Selection: Using Genetic Algorithm (GA) to select the most consistent, relevant, and non-redundant features extracted from the pre-trained CNN models.
– Comparative Analysis: Building an effective and accurate classification model through a comparative analysis using different classifiers, capable of handling complex extracted and selected features.
– Evaluation: Evaluating and comparing the results of both baseline and proposed approaches using the public dataset of MIAS Mammography Regions of Interest (ROIs) images.
– Accuracy Improvement: Achieving higher accuracy compared with the competition work on the public dataset.
This work presents an innovative approach that integrates GA with transfer learning to improve the accuracy and efficiency of breast cancer diagnosis. In the following parts, we break down the key features, advantages, and disadvantages of existing methods, and the motivation for this research. The first key feature of the proposed approach is the use of GA to optimize the selection of best features and enhancing the performance of breast cancer diagnosis. The second key feature is utilizing the transfer learning concept to leverage pre-trained models on large datasets, adapting them to the target domain (breast cancer diagnosis). This reduces the need for large annotated datasets in the medical field, where data can be scarce and expensive to label. The third key feature is the integration of GA with transfer learning aims to systematically and automatically optimize the number of features generated by transfer learning models, which can lead to better performance compared to using all features. The advantages of the proposed approach are enhancing the performance, reducing manual effort, adaptability, and effective use of limited data. The disadvantages of existing methods are the use of all features for classifying breast cancer, the large amounts of annotated data requirement, computational cost, and overfitting. The research motivation can be determined in improving diagnostic accuracy, efficiency, scalability, and addressing data scarcity. The research starting point begins from the perspective of combining the strengths of genetic algorithms and transfer learning to address the limitations of existing methods. By focusing on the optimization of model hyperparameters and structure through genetic algorithms, the research aims to develop a more effective and efficient diagnostic tool for breast cancer, addressing the challenges of data scarcity, manual tuning, and computational cost.
The rest of the paper is organized as follows: Section 2 gives an overall description of related work in breast cancer identification based on mammography images. Section 3 details the proposed approach and the used materials and methods. Section 4 presents the experimental results, and finally, Section 5 concludes our work.
Numerous studies have underscored the benefits of utilizing publicly accessible mammography images for the identification and categorization of breast cancer. Over the past decade, a variety of computer-aided diagnosis (CAD) models have been developed, focusing on three pivotal aspects: extracting features, reducing features, and classifying images. Many researchers have proposed diverse methods of feature extraction, leading to significant advancements in both detection and categorization stages [10,11]. A novel approach, known as MAR Row (Medical Active Learning and Retrieval), has been introduced to aid in breast cancer detection. This method emphasizes relevance feedback (RF) within the content-based image retrieval (CBIR) process, leveraging diversity and uncertainty levels to enhance outcomes [12].
Recent study [13] in breast cancer diagnosis emphasized the critical role of early detection in improving survival rates, leveraging advanced machine learning and genetic algorithms. This study focuses on classifying breast cancer mass pathology using radiologists’ annotations from the Breast Cancer Digital Repository screen-film mammograms. The research explores the effectiveness of precomputed features in the Breast Cancer Digital Repository (BCDR) combined with discrete wavelet transform and Radon transform. By employing four sequential feature selection methods and three genetic algorithms, the study enhances the classification accuracy. The fusion of features from craniocaudal and mediolateral oblique views demonstrated a significant performance boost for the classifier. For mass classification, the study utilized deep transfer learning models, incorporating ResNet50, NASNetLarge, and Xception networks. The implementation of an ensemble of Deep Transfer Learning (DTL) outperformed individual DTL models, resulting in superior classification performance. The Ensemble of Deep Transfer Learning (EDTL) approach achieved area under the receiver operating curve scores of 0.8843 and 0.9089 for the region of interest (ROI) and ROI union datasets, respectively. Notably, the proposed EDTL method achieved the highest breast cancer mass classification area under the curve (AUC) score on the BCDR dataset to date, indicating its potential for application to other datasets.
Another research [14] investigated the feature selection limitation in the field due to the complexity of breast cancer multifactorial nature. Authors proposed various hybrid models have been created to enhance the accuracy of breast cancer predictive models by selecting the best features. Achieving optimal parameters for these models can be challenging. Mainly, a hybrid teaching–learning optimization and GA-based approach, termed teaching–learning optimization (TLBOG), has been proposed to improve the reliability of evolutionary algorithms. The integration of GA aims to address the slow convergence rate and enhance the exploitation search capability observed in TLBOG. The primary objective of this approach is to optimize the parameters of support vector machines for higher accuracy compared to other machine learning models while simultaneously selecting the best feature subsets. Performance evaluation results indicate that the proposed method significantly outperforms traditional wrapper techniques in terms of accuracy, sensitivity, precision, and F-measure, as demonstrated on the Wisconsin Breast Cancer Database (WBCD) and Wisconsin Diagnostic Breast Cancer (WDBC) databases.
Recent work [15] investigated the application of GA to optimize the performance of a Multilayer Perceptron (MLP) model for breast cancer diagnosis. Various configurations of hidden layers in the MLP are explored, with accuracies ranging from 0.92 to 0.972. Robust evaluation is ensured through k-fold cross-validation and comprehensive dataset preprocessing, including normalization, scaling, and encoding. These methodologies contribute to consistent performance and enhanced generalization of the model. However, the incorporation of a genetic algorithm significantly improves the accuracy range, achieving values between 0.97 and 0.99 across different generations. The GA optimizes the MLP model by evolving a population of potential solutions (individuals) over multiple generations. Each individual represents a specific set of MLP parameters, such as the number of hidden layers, neurons per layer, and learning rate. The fitness of each individual is evaluated based on the MLP model’s accuracy on the breast cancer dataset. The fittest individuals are selected for reproduction, with genetic operators like crossover and mutation applied to generate new offspring. This iterative process of selection, crossover, and mutation gradually enhances the MLP model’s performance.
A new technique was introduced for rapidly and effectively identifying unclear regions in digital mammograms [16]. This method employs Electromagnetism-like Optimization (EML) for image segmentation, followed by 2D median noise filtering, and utilizes a support vector machines (SVM) classifier for feature extraction and classification, achieving an accuracy of 78.57% with just 56 images [17]. Another study presented a CAD system that combines deep convolutional neural networks (DCNN) with support vector machines (SVM) for breast mammography. This hybrid approach resulted in impressive detection accuracy, sensitivity, and specificity of 92.85%, 93.25%, and 90.56%, respectively [18].
In the same study [18], an automated algorithm for detecting breast cancer masses was introduced, relying on feature matching using Maximally Stable Extremal Regions (MSER). The system’s performance was evaluated with 85 images from the MIAS dataset, accurately identifying mass locations with a success rate of 96.47% [18]. Researchers also introduced a method that combines recurrent neural networks (RNN) with convolutional neural networks (CNN), using the Firefly updated chicken-based comprehensive learning particle swarm optimization (FC-CSO) technique to enhance segmentation accuracy and optimize integration of RNN and CNN, achieving accuracy rates of 90.6%, sensitivity of 90.42%, and specificity of 89.88% [19].
A breast cancer classification method called BDR-CNN-GCN was introduced, integrating dropout (DO), batch normalization (BN), and advanced neural networks (CNN and graph convolutional network (GCN)). Applied to the MIAS breast dataset, the BDR-CNN-GCN algorithm demonstrated specificity, sensitivity, and accuracy rates of 96.00%, 96.20%, and 96.10%, respectively [20,21]. Despite these advancements, a large proportion of existing techniques have struggled to achieve the necessary precision, particularly in differentiating between benign, malignant, and normal cases.
Consequently, the research demonstrated in study [22] aims to improve the automated classification of breast mammography patches by merging features extracted from three distinct pre-trained deep learning networks. Subsequently, the TV feature selection technique is employed to identify resilient and high-priority characteristics [22]. Another study proposed a deep learning approach combining feature extraction models with learning classifiers to diagnose breast cancer from mammograms, utilizing a pre-trained VGG16 model for feature extraction and an SVM for classification [22]. Additionally, another research seeks to enhance the precision of breast cancer classification using information gain-based feature selection and machine learning techniques on the Wisconsin Diagnostic Breast Cancer (WDBC) dataset [23].
The decision to omit radiotherapy was linked to a higher likelihood of local recurrence, but it did not adversely affect distant recurrence or overall survival in women aged 65 or older with low-risk, hormone receptor-positive early-stage breast cancer [24]. Similarly, in women aged 55 or older with T1N0, grade 1 or 2 luminal a breast cancer who underwent breast-conserving surgery and received only endocrine therapy, the occurrence of local recurrence at the 5-year mark was minimal, even without radiotherapy [25]. This study introduces a deep learning model focused on predicting breast cancer risk using the InceptionResNetV2 model, demonstrating a 91% accuracy rate in experiments [26].
A hybrid method combining deep learning and genetic algorithms, termed HMB-DLGAHA, has shown promising results in diagnosing breast cancer using ultrasound images. This method uses a CNN to extract features from ultrasound images and a GA to fine-tune the CNN hyperparameters, achieving superior accuracy compared to other advanced methods [27,28]. Moreover, the study discusses the latest advancements in utilizing immunotherapy for breast cancer treatment, outlining the challenges of applying these therapies to a diverse and varied disease. The researchers provide a comprehensive summary of various immunotherapy combinations currently under clinical trials [29,30].
The existing literature showcases significant advancements in breast cancer diagnosis using various machine learning and genetic algorithm techniques. However, several limitations underscore the necessity for further enhancements. For instance, feature selection remains a challenge, with previous methods often struggling to effectively remove irrelevant or redundant features separately, impacting the accuracy of predictive models. Additionally, optimization of model parameters can be challenging due to the multitude of parameters involved. While some studies integrate deep learning and genetic algorithms, they often do not combine these with transfer learning, which can significantly enhance the model’s ability to generalize. Our proposed method addresses these critical limitations by using a genetic algorithm-based optimized transfer learning approach, providing a comprehensive solution that improves accuracy, robustness, and generalization capabilities compared to traditional methods.
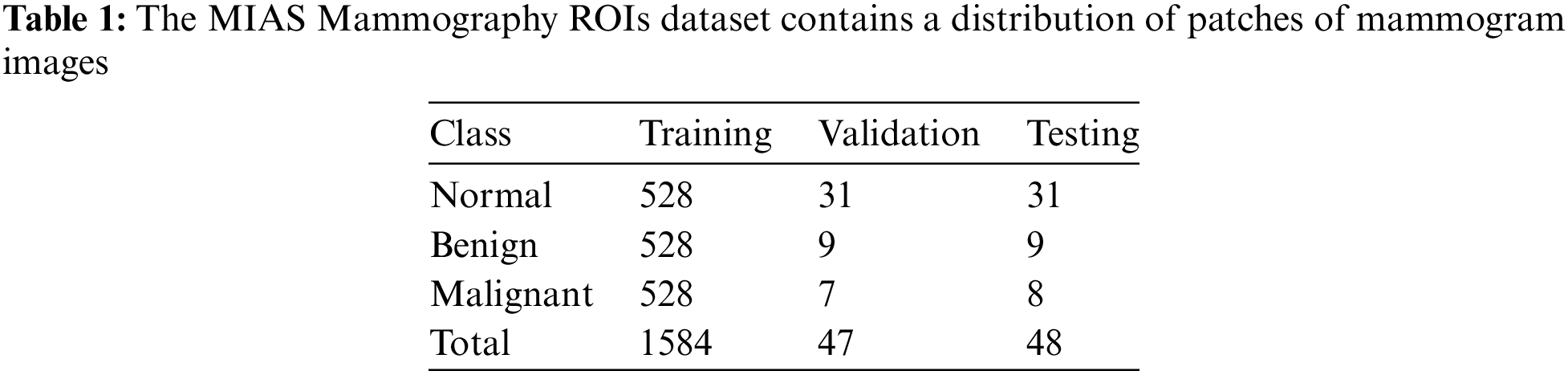
This study utilizes the MIAS Mammography ROIs dataset of breast cancer images, which is divided into training, validation, and testing sets. Various CNNs are employed, including VGG16 and VGG19, which are deep and effective for detailed feature extraction; MobileNet, designed for efficient performance on mobile devices; and DenseNet, which enhances feature propagation and captures detailed features. A Genetic Algorithm (GA) is used for feature extraction and selection, optimizing feature subsets through principles of natural selection. This helps improve model performance by identifying the most relevant features. The baseline approach involves using the built-in capabilities of neural networks for feature extraction and training models like VGG16, VGG19, MobileNet, and DenseNet. Performance is evaluated using accuracy, precision, and recall metrics. The proposed model combines advanced deep learning techniques for early breast cancer detection. It employs GA and a Support Vector Machine (SVM) for feature selection and classification, aiming to enhance diagnostic accuracy and reduce computational complexity.
This study utilizes a dataset consisting of 1679 MIAS Mammography ROIs breast cancer images to classify images into three categories: normal, benign, and malignant. The dataset was divided into training, validation, and testing sets as follows [26,31–33]:
The division of the MIAS Mammography ROIs dataset for the purposes of this study was carefully structured to support the classification objectives. Specifically, the dataset is broken down into three categories—normal, benign, and malignant—with each category utilizing 528 images for training. The distribution for validation and testing varies slightly between categories: the normal class includes 31 images for both validation and testing; the benign class uses 9 images each for validation and testing; and the malignant class employs 7 images for validation and 8 for testing, totaling 1584 images for training, 47 for validation, and 48 for testing.
The methodology for selecting and labeling the regions of interest (ROIs) from these mammograms was meticulous. Researchers cropped the mammograms to accurately locate lesions, obtaining precise ROI patches from the standard dataset. In cases involving normal mammograms, the ROIs were selected randomly, ensuring a diverse representation within the dataset [8]. These preparations facilitated a detailed analysis of the imaging data, as outlined in Table 1, which provides a breakdown of the image patches and the segregated ROIs.
3.2 Pre-Trained Transfer Learning Models
VGG16, a convolutional neural network proposed by the Visual Geometry Group at Oxford, comprises 16 layers with a simple and uniform architecture. It utilizes small 3 × 3 filters across its 13 convolutional layers, which are interspersed with five max-pooling layers, and concludes with three fully connected layers. VGG16 employs the ReLU activation function and culminates in a softmax layer for classification. Pre-trained on large datasets like ImageNet, VGG16 is highly effective for feature extraction and is often employed by removing its fully connected layers to use the convolutional base for generating feature maps from new images.
In breast cancer image analysis, VGG16 is instrumental for detecting malignant or benign tumors. The process involves collecting and preprocessing breast cancer images, applying transfer learning to fine-tune the pre-trained VGG16 on a specific dataset, and using the convolutional layers as fixed feature extractors. The extracted feature maps, which capture spatial hierarchies and texture patterns, are then fed into a classifier to predict cancerous tissues. VGG16 enhances diagnostic accuracy through its deep architecture, which captures intricate details, and reduces training time and resources via transfer learning. However, it is computationally intensive and performs best with large annotated datasets, which can be a limitation in medical imaging [32].
VGG19, a neural network designed by the Visual Geometry Group at Oxford, is deeper than its predecessor, VGG16, comprising 16 convolutional layers and 3 fully connected layers, totaling 19 layers. It employs small 3 × 3 filters, with its architecture organized into five blocks of convolutional layers each followed by a max-pooling layer, using the ReLU activation function after each layer. For feature extraction, the fully connected layers are typically removed, allowing the convolutional base to generate detailed feature maps from new images.
In the context of breast cancer image analysis, VGG19 plays a crucial role in distinguishing between malignant and benign tumors. The workflow involves gathering and preprocessing breast cancer images, utilizing transfer learning to adapt the pre-trained VGG19 model to the specific dataset, and using the convolutional layers to extract essential features. These feature maps, which capture spatial hierarchies and textures, are then input into a classifier to determine if the tissue is cancerous. VGG19’s deeper architecture enhances diagnostic precision by capturing more complex details, and its pre-trained models streamline the training process. Nonetheless, VGG19 is computationally demanding and performs optimally with large annotated datasets, which can be challenging to obtain in medical imaging [34,35].
MobileNet, a convolutional neural network developed by Google, is specifically designed for efficient performance on mobile and embedded vision applications. It utilizes depthwise separable convolutions, which significantly reduce the number of parameters and computational cost. With the introduction of width and resolution multipliers, MobileNet can adjust the network width and input resolution to further optimize performance. The architecture has several versions, including MobileNetV1, V2, and V3, each offering improvements over its predecessor.
For feature extraction, MobileNet is pre-trained on large datasets like ImageNet, making it effective at generating high-level feature maps from new images by removing the fully connected layers. In breast cancer image analysis, MobileNet is employed to classify breast tissue images as malignant or benign. The process involves collecting and preprocessing images, applying transfer learning to fine-tune the pre-trained MobileNet on a specific breast cancer dataset, and using the convolutional layers to extract relevant features. These features are then fed into a classifier to predict the presence of cancer. MobileNet’s efficiency makes it particularly suitable for mobile and embedded applications, providing competitive accuracy while remaining lightweight. Transfer learning enhances its generalization capabilities, making it effective across various medical image datasets. However, despite its efficiency, MobileNet may face limitations on extremely resource-constrained devices and generally performs best with large annotated datasets, which can be challenging to obtain in medical imaging [36].
DenseNet, short for Dense Convolutional Network, is a CNN architecture designed to improve information and gradient flow through the network. Each layer within a dense block is connected to every other layer, a structure that was proposed by Gao Huang and his team. In DenseNet, each layer receives inputs from all preceding layers and passes its outputs to all subsequent layers, enhancing feature propagation and reducing issues related to vanishing gradients. Transition layers, placed between dense blocks, compress and downsample the feature maps. Meanwhile, bottleneck layers utilize 1 × 1 convolutions to reduce computational costs. The growth rate within DenseNet defines the number of feature maps added by each layer, helping to balance model capacity and computational load. DenseNet variants, such as DenseNet121, DenseNet169, DenseNet201, and DenseNet264, indicate the total number of layers.
Pre-trained on extensive datasets like ImageNet, DenseNet is highly effective for feature extraction. This effectiveness is achieved by removing fully connected layers and utilizing dense blocks to generate high-level feature maps from new images. In this work, DenseNet121 is employed to aid in classifying breast tissue images as normal, malignant, or benign. The process involves collecting and preprocessing images, applying transfer learning to fine-tune the pre-trained DenseNet121 on a specific breast cancer dataset, and using dense blocks to extract detailed features. These features are then fed into a classifier to predict the presence of cancerous tissues. The DenseNet121 architecture captures intricate details, enhancing diagnostic accuracy through efficient parameter use and improved feature reuse. While it requires significant computational resources for training and inference, its performance is optimized with large annotated datasets. DenseNet121’s design makes it particularly effective in capturing comprehensive features for medical image analysis, thereby aiding in accurate classification and diagnosis [37].
3.3 Machine Learning Classifiers
Various classifiers are used for classifying databases, especially those related to medical data, including SVM, K-Nearest Neighbor (KNN), Decision Trees (DT), and Random Forest (RF). Each classifier has unique strengths: SVM is effective for high-dimensional data and small datasets; KNN is straightforward and effective but can be slow for large datasets; Decision Trees are easy to interpret and fast but may be prone to overfitting; Random Forest helps reduce overfitting and is both robust and accurate. Each classifier can be effectively applied to breast cancer image classification, depending on specific needs and constraints.
3.3.1 Support Vector Machine (SVM)
The Support Vector Machine (SVM) is highly effective for breast cancer image classification due to its capability to handle high-dimensional data and perform robustly with limited samples. SVM operates by identifying the optimal hyperplane that separates data points of different classes, such as benign and malignant tissue samples. It employs kernel functions to transform the input data into a higher-dimensional space, thereby facilitating the identification of a separating hyperplane. The critical data points, known as support vectors, determine the position and orientation of this hyperplane. In breast cancer classification, features are extracted from mammogram images, and the SVM is trained on these features to distinguish between the classes. For new images, the SVM uses the learned hyperplane to classify them accurately.
3.3.2 K-Nearest Neighbors (KNN)
K-Nearest Neighbors (KNN) is a straightforward, instance-based learning algorithm that classifies data points based on the majority class of their k-nearest neighbors. It uses distance metrics, such as Euclidean or Manhattan, to determine the closest neighbors and assigns a class based on the majority vote among these neighbors. In breast cancer classification, features are extracted from mammogram images, and KNN uses these features to classify the data. Although KNN does not require a traditional training phase and retains all training data, it can be computationally intensive for large datasets. The choice of k is critical and is typically determined through cross-validation to ensure optimal performance.
Decision Trees (DT) classify data by recursively splitting it based on feature values, forming a tree where each node represents a feature and each branch represents a decision rule. The tree is constructed by selecting features that offer the highest information gain or lowest Gini impurity. In breast cancer classification, features extracted from mammogram images are used as inputs. The decision tree is then trained by recursively splitting the data, creating a tree structure that classifies the images. For new images, the decision tree navigates from the root to a leaf node based on the feature values, ultimately assigning a class label. While decision trees are easy to interpret, they can be prone to overfitting, making them potentially less robust compared to ensemble methods.
Random Forest (RF) is an ensemble learning method that builds multiple decision trees and combines their predictions to enhance robustness and reduce overfitting. It constructs a multitude of decision trees during training, each trained on a bootstrap sample of the data and considering random subsets of features at each split. The final prediction is made by aggregating the predictions of all individual trees, typically through majority voting. For breast cancer classification, features extracted from mammogram images are used, and multiple decision trees are trained on different subsets of the data. For new images, the Random Forest aggregates predictions from all trees, thereby enhancing both robustness and accuracy. This ensemble approach makes Random Forest highly effective for medical image classification.
A genetic algorithm operates as a search heuristic that mimics natural evolution. This method is particularly well-suited for solving optimization problems such as feature selection, where a vast feature space must be effectively reduced without compromising the predictive power of the model.
a) Chromosome Representation: Each chromosome in our GA represents a potential subset of features extracted from the CNN models. Mathematically, a chromosome is encoded as a binary vector
b) Initial Population: The GA begins with an initial population of PPP chromosomes, each representing a different feature subset. The initial population is generated randomly to ensure diversity in the feature subsets explored.
c) Fitness Function: The fitness of each chromosome is evaluated using a Support Vector Machine (SVM) classifier. The fitness function
d) Selection: Selection is performed using tournament selection, a method where k chromosomes are chosen at random, and the fittest among them (based on their fitness scores) is selected for reproduction. This can be mathematically described as:
e) Crossover: Crossover combines pairs of parent chromosomes to produce offspring, introducing genetic variation. We employ single-point crossover, where a random crossover point ppp is chosen, and two offspring are generated by exchanging the segments after ppp between the parents. Let c1 and c2 be two parent chromosomes. The offspring o1 and o2 are created as follows:
f) Mutation: Mutation introduces random changes to individual chromosomes, enhancing diversity and preventing premature convergence. Each bit in the chromosome has a small probability μ\muμ of being flipped (from 0 to 1 or from 1 to 0). For a chromosome c, the mutated chromosome c′ is given by:
g) Iterative Process: The GA iteratively evolves the population over multiple generations. In each generation, the processes of selection, crossover, and mutation are applied to create a new population. The algorithm terminates when a stopping criterion is met, such as a maximum number of generations G or convergence to a stable fitness value.
The baseline approach uses the four popular pre-trained convolutional neural network (CNN) models, explained in Section 3.2 as a starting point to extract the features from breast ROIs images. In the baseline approach features are extracted using the built-in capabilities of a number of CNN models such as VGG16, VGG19, MobileNet, and DenseNet. After each of the models is trained using training datasets, features are extracted from the last layer of each trained model. Before the training data is fed int o the models, the input images undergo preprocessing. As illustrated in Fig. 1, the processes begin with an input image from a comprehensive dataset of breast cancer images. These images are rigorously preprocessed and enhanced using various image processing methos aimed at improving image quality and ensuring suitability for further analysis. The preprocessing steps emphasize noise reduction, contrast enhancement, resizing, and normalization.
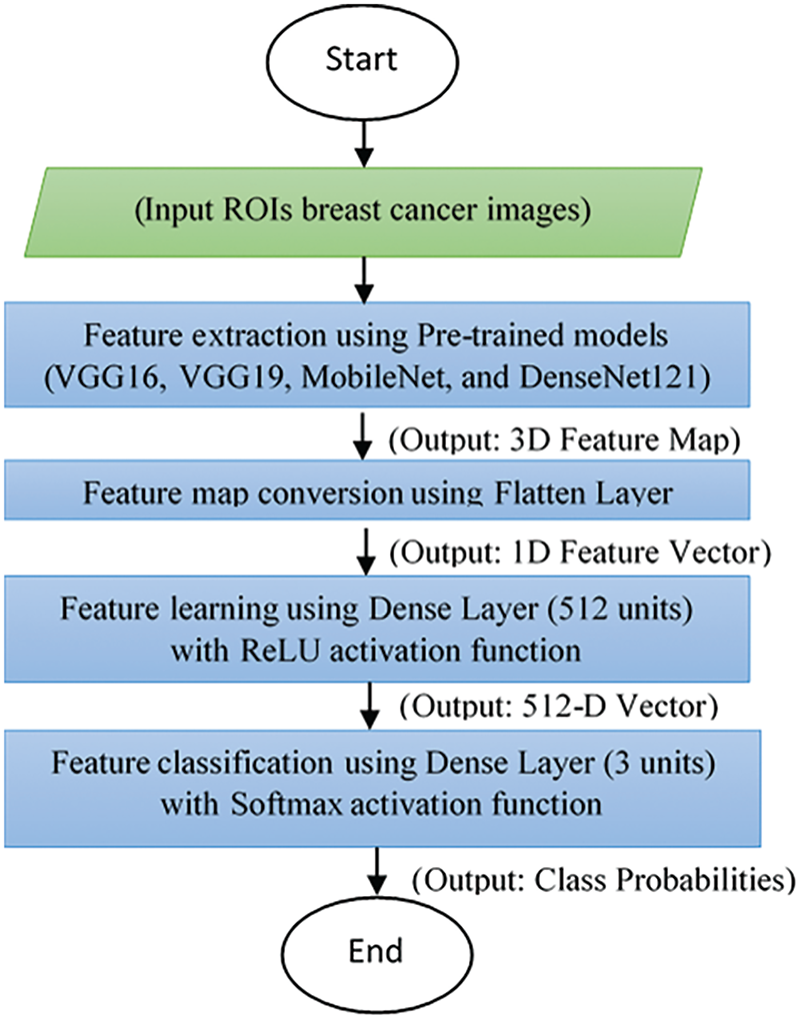
Figure 1: A flowchart architecture of baseline approach
Following the preprocessing, the model extracts feature from the images using the pre-trained deep learning models. These extracted features are then subjected to validation and testing, using a portion of the dataset reserved for these purposes. Validation aids in fine-tuning model parameters, while testing provides an unbiased evaluation of the model’s performance. The final phase involves a comprehensive performance evaluation of the model, where the evaluation metrics include classifying images into three categories: normal, malignant, or benign. The model’s performance is assessed using accuracy, which measures the proportion of correctly classified instances among the total instances. Additionally, precision and recall metrics are utilized to evaluate the model’s effectiveness in identifying true positive instances among those classified as positive and the actual positive instances, respectively. An overall result or aggregate measure of the model’s performance on the test data is also considered reflecting the goodness of the features extracted from the layer immediately before the output layer. These extracted features are further refined in subsequent steps of our proposed model.
The graphical abstract of proposed approach for automatic early breast cancer classification is presented in Fig. 2. Initially, the model preprocesses the input images using standard procedures, followed by employing multiple pre-trained CNNs to extract features. In this critical phase, each CNN is specifically trained on the input images to classify the data into malignant, benign, or normal categories. The features extracted from all the CNNs are then aggregated into a comprehensive feature set. Subsequently, a Genetic Algorithm (GA) is used to optimize these features further, aiming to identify the most effective subset for detection. After optimizing the features, various classifiers are employed to categorize the data accurately. This integration of advanced techniques, including the GA for feature optimization, is a key aspect of the model as it enhances the precision of cancer detection while minimizing computational complexity. This model holds significant potential to improve early breast cancer detection substantially. In the following subsections, we describe the steps of proposed approach in more details.
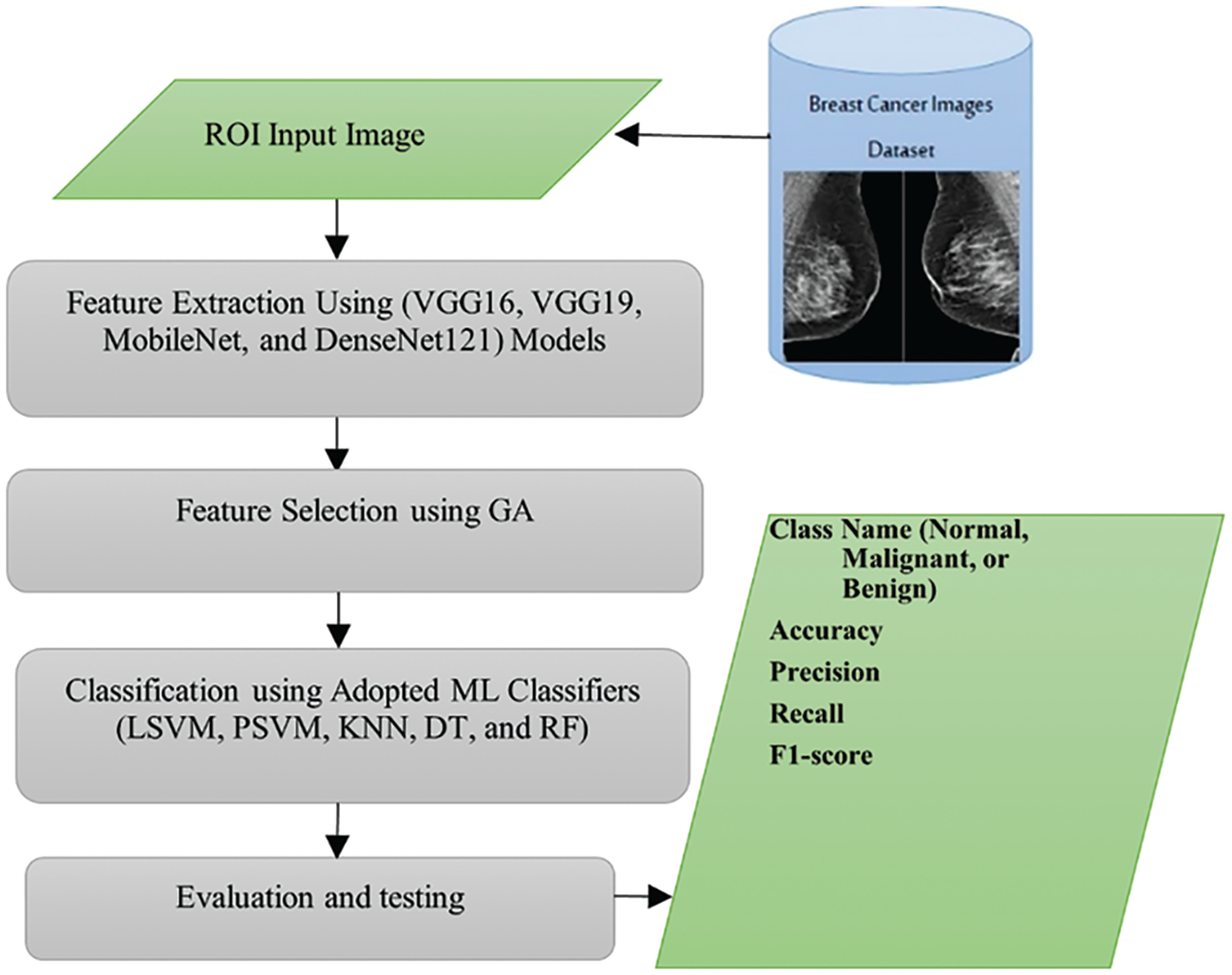
Figure 2: Graphical abstract of proposed approach of automatic breast cancer classification
3.6.1 Feature Extraction Using Pre-Trained Models
As mentioned earlier, in the proposed approach, features are extracted using the built-in capabilities of several CNN models, including VGG16, VGG19, MobileNet, and DenseNet. Each CNN model produces a feature vector
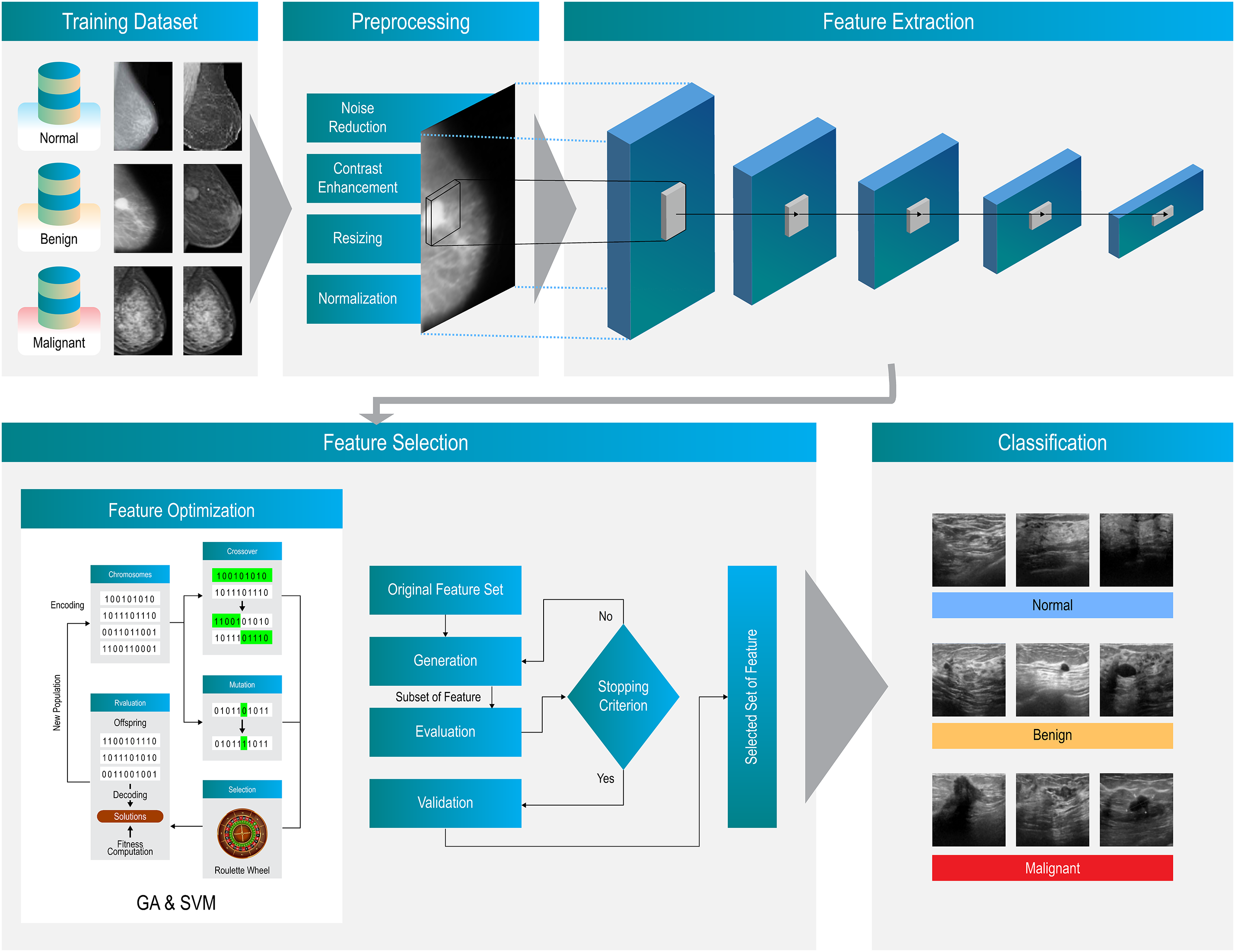
Figure 3: The architecture detail of proposed approach
3.6.2 Feature Selection Using GA
The Genetic Algorithm (GA) operates by refining a composite feature set X derived from four distinct deep learning architectures: VGG16, VGG19, MobileNet, and DenseNet. This feature set is critical for the accurate classification of input images into one of three categories: Malignant, Benign, or Normal. Within the GA framework, a classifier assesses the effectiveness of each feature subset, guiding the selection process towards the most promising configurations.
Genetic Algorithms are a class of search heuristics that emulate the principles of natural evolution, proving particularly adept at tasks such as feature extraction and selection. The process initiates with the generation of an initial population of potential solutions. Each solution, commonly termed an individual or chromosome, comprises a binary-encoded subset of features, where each bit indicates the presence (1) or absence (0) of a specific feature.
The effectiveness of each chromosome is assessed via a fitness function—predetermined to evaluate how well the subset aids in performing classification or regression tasks. In our methodology, the well-established Support Vector Machine (SVM) calculates the fitness score for each individual, focusing on classification accuracy as the primary metric.
To ensure the propagation of superior solutions, chromosomes with higher fitness scores are chosen to create a mating pool. We employ tournament selection as our method of choice to encourage competition among the most effective chromosomes. During the crossover phase, feature subsets from two high-performing chromosomes are recombined to generate offspring, thereby fostering diversity and amalgamating beneficial attributes from different progenitors. The recombination can occur through various techniques, including single-point, multi-point, or uniform crossover.
Following crossover, mutations are introduced to maintain genetic diversity and explore novel feature combinations. This process involves randomly flipping bits in the chromosomes, thus altering the selected features within the extensive feature set. The newly formed generation of individuals, created via crossover and mutation, supersedes the existing population through strategies such as generational replacement, where the entire population is renewed, or steady-state replacement, where only select individuals are replaced.
This iterative cycle persists over numerous generations, each progressively enhancing the efficacy of the feature subsets and, consequently, the overall performance of the model. The iterative process concludes either when no new feature subset is selected for a predefined number of consecutive iterations or when a specific number of iterations has been completed. The final feature subset, at the culmination of this process, is deemed the optimal configuration for the classifier. A simplified pseudocode of the proposed approach is shown below:
By integrating the GA with CNN models, our approach effectively reduces the dimensionality of the feature space while retaining the most informative features for accurate breast cancer diagnosis. This synergistic combination leverages the feature extraction capabilities of deep learning and the optimization power of genetic algorithms, resulting in a robust and efficient diagnostic system.
3.6.3 Classification Using Adopted Machine Learning Classifiers
In this step, the ML classifiers outlined in Section 3.4 are trained using the selected features from Section 3.6.2. The training utilizes a set of breast images and is evaluated using a separate test set of breast images. This process involves several key steps to transform extracted and selected features into actionable insights through predictive modeling. In the training process, two versions of SVM used in this study Localized Support Vector Machine (LSVM) and Proximal Support Vector Machine (PSVM), along with KNN, DT, and RF models, learn the relationship between features and target labels. Hyper-parameter tuning is performed using the validation set to optimize the model parameters.
Classification of new samples in the test set is conducted using the decision functions derived from the classifiers during the training phase. The class label is assigned based on the sign of the decision function, f(x), as follows:
Eq. (1) assigns the sample to one class if
4 Experimental Results and Discussion
This section explains the experiment sequence according to data preparation, experimental steps, comparison methods, and evaluation metrics. The experiment targets are clearly defined, and the steps for carrying out the experiment are detailed, ensuring a comprehensive evaluation of both the baseline and proposed methods. We detail the step-by-step outline for encompassing the phenomena observed, their causes, and recommendations based on the results. First, the results of baseline approach will be given; then, the results of proposed approach will be described to show the effectiveness of proposed approach to classify the breast images into normal, malignant or benign classes. In data preparation, the data source used in the experimental results is a publicly available breast cancer imaging dataset, namely MIAS Mammography ROIs, (www.kaggle.com/datasets/annkristinbalve/mias-mammography-rois, accessed on 05 April 2024), described in Section 3.1. It is a preprocessed version of the original MIAS dataset and contains a sufficient number of labeled images available in NPY format, ensuring adequate evaluation for the research models. All images in the dataset are preprocessed by removing artifacts such as labels and improving them by using Contrast Limited Adaptive Histogram Equalization (CLAHE) method. The region of interests (ROI) of abnormal images (malignant and benign) are extracted using
For the step-by-step experimental procedure of the baseline approach, we start with pre-trained model selection in which some of common and effective models like VGG16, VGG19, MobileNet, and DenseNet are chosen. Then, we execute the feature extraction step by freezing early layers of the pre-trained model to use them as feature extractors and adding a flatten layer with two custom dense layers on top of the pre-trained models for classification. The first dense layer consists of 512 neurons and the second dense layer contains three neurons equal to the number of classes. After that, we apply training and validation step by using a default learning rate (0.001), batch size (32), and number epochs (50); as well as, utilizing Adam optimizer and categorical cross-entropy loss function. The training and validation step is done on the classification layer using training and validation sets. Finally, evaluation step of trained models is applied on the test set using precision, accuracy, recall, and F1-score metrics. Similarly, the step-by-step experimental procedure of the proposed approach includes pre-trained model selection, feature extraction, classification models selection and building, genetic algorithm design for training and validating the classification models, and evaluation. In the feature extraction step of proposed approach, we add a 2D global average pooling layer on top of the pre-trained models for down-sampling the spatial dimensions of features with the average value of each small region. After that, we reshape the output of 2D global average pooling layer into one dimension vector. For the classification models selection and building, we select and build a four classifiers described in Section 3.3. These classifiers are built with the default values of their hyper-parameters. The genetic algorithm design step consists of a number of sub-steps includes initializing the values of genetic algorithm hyperparameters, such as population size with 3, the number of generations with 25, mutation rate with 0.03, crossover rate with 0.7. Then, the chromosome encoding sub-step encodes the input reshaped features obtained from the 2D global average pooling layer. After that, we train the selected classifiers on a subset of the input features and the fitness function is evaluated based on the validation accuracy. In the selection sub-step, we select top-performing models based on fitness scores. The crossover and mutation sub-step generates the offspring by applying crossover to combine the selected features of trained models and performs mutation by introducing random changes to some offspring to maintain diversity. The iteration sub-step repeats fitness evaluation, selection, crossover, and mutation for several generations according to the number of generations’ value. The final trained models with the best average validation accuracy of the final population are selected for evaluation step on the test set using the evaluation metrics indicted in the baseline approach. The experiments are implemented using TensorFlow and Keras libraries on a graphics processing unit (GPU) hardware to accelerate training process of models. In the following subsections, we describe the experimental results with graphs and numbers.
4.1 Results of Baseline Approach
The baseline approach (BL) results for breast cancer image classification were evaluated using various metrics, including precision, recall, F1-score, and accuracy. It should be noted that the BL approach for feature extraction relied on the inherent capabilities of each neural network model without utilizing feature selection optimization techniques. These results were assessed separately on validation and test sets across different models: VGG16, VGG19, MobileNetV2, and DenseNet121.
Fig. 4 illustrates the training and validation progress of four different models over 50 epochs, evaluated using two metrics: accuracy and loss. The subfigures (a, b, c, and d) are organized into pairs, with each pair depicting accuracy progress and loss progress. Fig. 4a shows the accuracy progress, where both training and validation accuracy improve over the epochs, though there is noticeable fluctuation in the validation accuracy.

Figure 4: Accuracy and loss of training and validation progress over 50 epochs for the four different models: (a) VGG16, (b) VGG19, (c) MobileNetV2, and (d) DenseNet121
The loss progress demonstrates that both training and validation losses decrease over the epochs, although the validation loss exhibits fluctuations. Fig. 4b presents a similar trend in accuracy progress to Fig. 4a, but with reduced fluctuations in validation accuracy. Here, the loss progress indicates a steady decrease in both training and validation loss, with some fluctuations in validation loss, particularly noticeable after epoch 20.
Fig. 4c shows a steady increase in training accuracy, while validation accuracy exhibits more pronounced fluctuations compared to Fig. 4b. The loss progress reveals a steady decrease in training loss, but considerable fluctuations in validation loss, suggesting potential overfitting. Fig. 4d indicates that training accuracy reaches nearly 100%, while validation accuracy stabilizes after initial fluctuations. The loss progress reveals a sharp decline and stabilization of training loss at a very low value, whereas validation loss initially decreases but then begins to increase and fluctuate, indicating strong overfitting.
The confusion matrices in Fig. 5 provide a clear visual representation of the performance of the four different models in classifying instances into three classes: Normal, Benign, and Malignant. Overall, Models (a) and (b) demonstrate similar performance, showing higher accuracy in predicting the Benign and Malignant classes compared to Models (c) and (d). Models (c) and (d) display more misclassifications, indicating areas for improvement. The consistent perfect classification of the Normal class across all models suggests that this class is the easiest to predict, while the variability in the Benign and Malignant classes highlights the challenges in distinguishing between these categories.


Figure 5: Confusion matrices of baseline approach on validation set for the four different models: (a) VGG16, (b) VGG19, (c) MobileNetV2, and (d) DenseNet121
Fig. 6 compares the performance metrics of four models (VGG16, VGG19, MobileNetV2, and DenseNet121) on the validation set, focusing on Accuracy, Precision, and Recall. Overall, VGG16 outperforms the other models across all three metrics. VGG19 also shows strong performance but slightly lags behind VGG16. DenseNet121 and MobileNetV2 exhibit similar performance metrics, with both models scoring lower in accuracy, precision, and recall compared to VGG16 and VGG19. These results suggest that VGG16 is the most effective model among the four for the evaluated task.

Figure 6: Pre-trained transfer learning models on validation set
Fig. 7 presents the four confusion matrices from the test set, illustrating the performance of classification models on a three-class problem (Normal, Benign, Malignant). All models excel in identifying Normal instances, with variations in misclassification rates for the other two classes. Model (a) shows some misclassification of Malignant as Benign, Model (b) has balanced misclassification between Benign and Malignant, Model (c) perfectly classifies Normal instances but confuses Malignant and Benign, and Model (d) performs similarly to Model (b) but with more frequent misclassification of Malignant as Benign.

Figure 7: Confusion matrices of baseline approach on test set for the four different models: (a) VGG16, (b) VGG19, (c) MobileNetV2, and (d) DenseNet121
Overall, while all models perform well in classifying Normal instances, their performance varies for the Benign and Malignant classes. Model (c) shows the best performance for Normal but has more misclassifications in the Malignant category.
Fig. 8 compares the performance of the four models—VGG16, VGG19, MobileNetV2, and DenseNet121—across accuracy, precision, and recall metrics. VGG16 shows the lowest performance with an accuracy of 0.7708, precision of 0.7872, and recall of 0.7708. VGG19 performs better with an accuracy of 0.8125, precision of 0.8085, and recall of 0.7917. MobileNetV2 achieves the highest performance across all metrics, with an accuracy, precision, and recall of 0.8333. DenseNet121 has consistent performance with an accuracy, precision, and recall of 0.7917, slightly outperforming VGG16 but trailing behind VGG19 and MobileNetV2. Overall, MobileNetV2 demonstrates the best performance, followed by VGG19, DenseNet121, and VGG16.

Figure 8: Pre-trained transfer learning models on test set
4.2 Results of Proposed Approach
Fig. 9 compares the performance of VGG16, VGG19, MobileNetV2, and DenseNet121 across five classification algorithms: LSVM, PSVM, KNN, DT, and RF, measured by percentage accuracy. VGG16 shows consistent performance, achieving the highest accuracy with RF (85.42%) and the lowest with PSVM (79.17%). Similarly, VGG19 achieves its highest accuracy with DT (83.33%) and its lowest with PSVM (79.17%). MobileNetV2 exhibits the best overall performance, excelling with LSVM (89.58%) and DT (87.50%), but showing its lowest performance with PSVM (83.33%). DenseNet121 also demonstrates strong performance, particularly with LSVM (85.42%) and DT (87.50%), and records its lowest accuracy with PSVM (83.33%).

Figure 9: The proposed approach on test set using GA with adopted classifiers
Overall, MobileNetV2 stands out as the top performer across most algorithms, especially in LSVM and DT, while VGG16 and VGG19 maintain consistent but slightly lower performance. DenseNet121 shows strong results that are comparable to MobileNetV2, albeit with lower accuracy in PSVM.
Fig. 10 shows the “Average Accuracy of Each Generation” over 25 generations. The generation number ranges from 0 to 25, with average accuracy fluctuating between approximately 0.75 and 0.85. The initial generation (0) starts at about 0.81. Across the generations, for example, from generation 1 to 5, accuracy oscillates between 0.77 and 0.81. The highest accuracy occurs around generation 10, peaking above 0.83, while a significant drop is observed around generation 15, where accuracy dips below 0.76, marking the lowest point on the graph. Despite these fluctuations, the overall trend indicates that average accuracy does not improve significantly over the generations, generally hovering around the 0.79 to 0.81 range. Toward the later generations (20 to 25), accuracy stabilizes around 0.80 to 0.81.
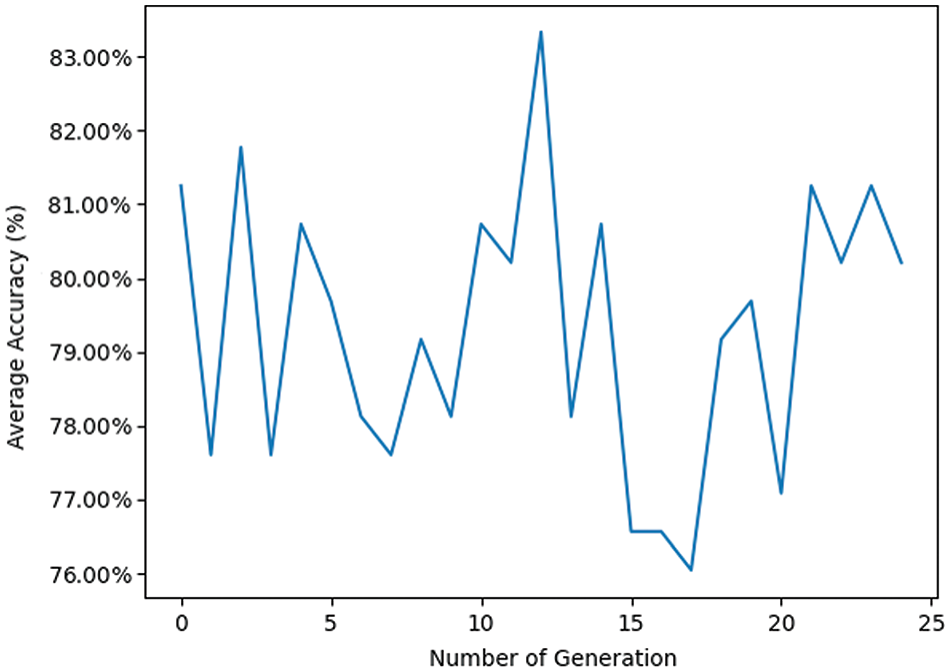
Figure 10: Pre-trained VGG16 transfer learning model with GA using RF classifier
The high fluctuation in average accuracy indicates variability in the performance of each generation, which could be due to various factors such as changes in the data, algorithm adjustments, or randomness in the process. The peaks and troughs, such as those observed in generations 10 and 15, highlight moments of particularly high and low performance, respectively. These could be investigated further to understand what caused these significant changes. The lack of a clear upward trend suggests that the method or algorithm used may not be consistently improving with each generation, indicating the need for refinement in the approach, such as better optimization techniques, parameter tuning, or additional data preprocessing. The line graph provides a visual representation of the average accuracy across 25 generations. While there are moments of high accuracy, the overall lack of consistent improvement suggests areas for further investigation and potential optimization in the process being analyzed.
Figs. 11 and 12 provide further insights into classification performance. Fig. 11 shows examples of classifying some samples selected randomly from the test set (Red color for the misclassified samples, the correct labels are Malignant, Normal, and Malignant; however, the model classified them as Benign, Benign, and Normal). Fig. 12 presents the “Average Accuracy of Each Generation,” illustrating the performance of a pre-trained VGG19 model combined with a Genetic Algorithm (GA) using a Random Forest (RF) classifier over 25 generations. The graph displays significant fluctuations in accuracy, for instance, a peak just above 0.82 around generation 10 and a dramatic decrease to just below 0.77 at generation 15. Subsequently, accuracy rises sharply again to approximately 0.82 at generation 20 before showing a downward trend toward the end, stabilizing around 0.79 at generation 25. These fluctuations suggest that the performance of each generation is quite variable, which could be attributed to different factors like the stochastic nature of the genetic algorithm, the selection of features, or the tuning parameters of the random forest classifier. The peaks and troughs observed might indicate that certain generations are finding more optimal solutions or encountering overfitting issues. The lack of a clear upward trend in improvement across generations implies that while the model occasionally reaches higher accuracies, it does not consistently enhance its performance over time. This graph provides useful insights into the behavior of the transfer learning model combined with GA and RF across multiple generations, highlighting the variability in performance and the need for further optimization and stability in the model’s learning process. Fig. 13 displays nine randomly selected mammogram images from the test set, each classified into three categories: Normal, Benign, and Malignant.

Figure 11: Examples of classifying some samples selected randomly from the test set
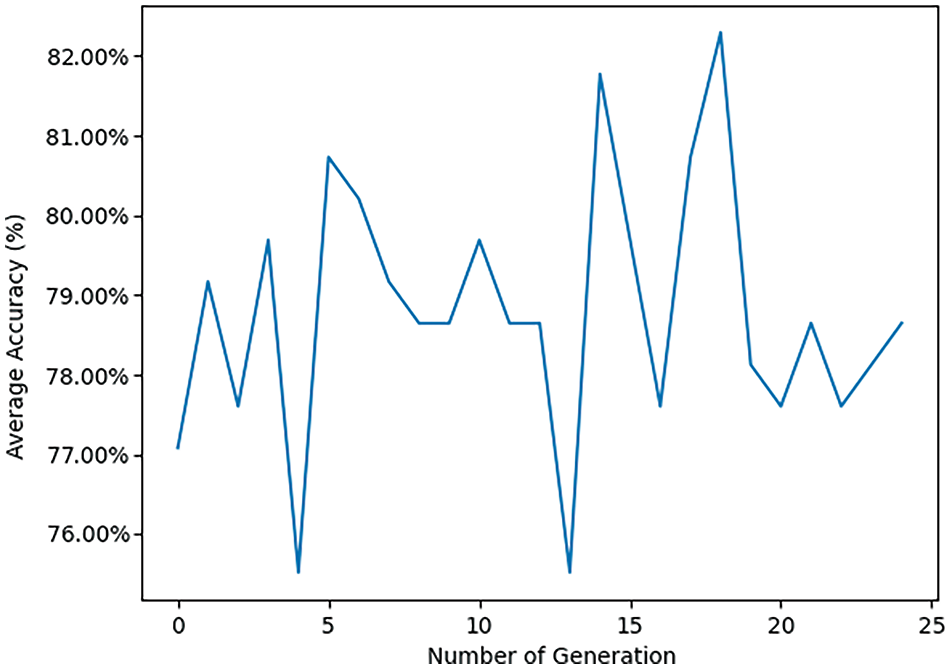
Figure 12: Pre-trained VGG19 transfer learning model with GA using RF classifier

Figure 13: Examples of classifying some samples selected randomly from the test set
Fig. 14 illustrates how the average accuracy of MobileNetV2 changes across 25 generations, marking the evolution of accuracy over time. Starting at around 0.81 in the first generation, the trend shows an overall upward trajectory in accuracy, with fluctuations reflecting the learning or optimization process. The highest accuracy achieved is slightly above 0.88 around the 21st generation, marking peak performance. After reaching this peak, accuracy slightly drops and stabilizes around 0.87 by the 24th generation, suggesting a plateau in performance. This trend indicates that the process or model is effectively learning or optimizing over time, thereby increasing its performance. The fluctuations suggest that there may be variability or instability in the process, but the general trend is positive, showing consistent improvement. The peak at generation 21 indicates a significant improvement, followed by a slight decline, which could imply reaching a local optimum before settling to a more stable performance level, highlighting areas for potential further optimization.
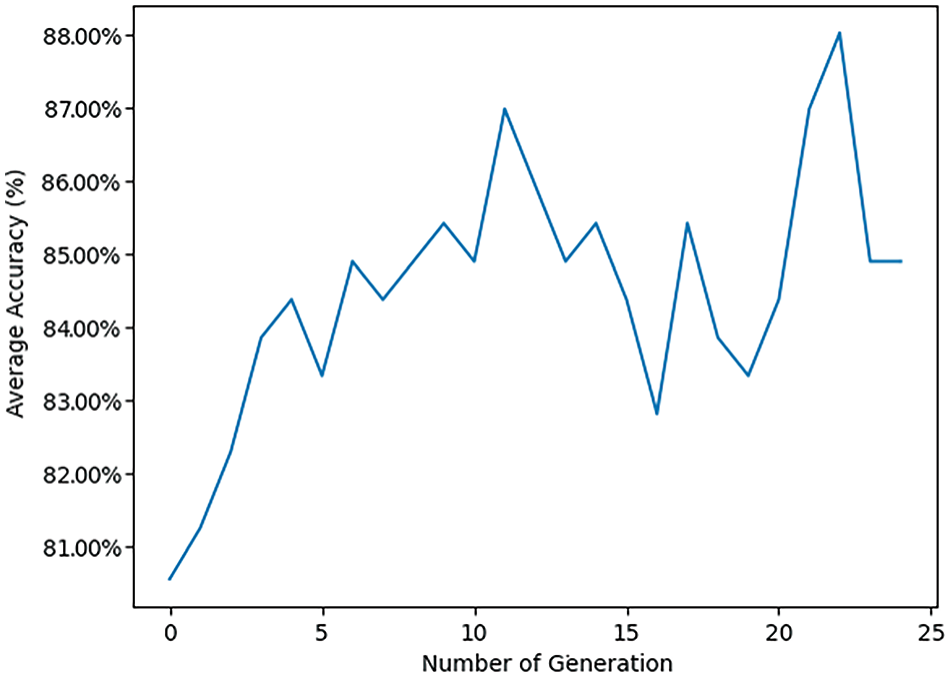
Figure 14: Pre-trained MobileNetV2 transfer learning model with GA using LSVM classifier
Fig. 15 provides examples of classifying some samples selected randomly from the test set (Red color for the misclassified sample, the correct label is Malignant and the model classified it as Normal).

Figure 15: Examples of classifying some samples selected randomly from the test set
Fig. 16 illustrates the evolution of accuracy over time for a pre-trained DenseNet121 transfer learning model with GA using an RF classifier across 25 generations. The accuracy begins at around 0.73 in the first generation, indicating the initial performance level. There is an overall upward trend in accuracy, with fluctuations throughout the generations that reflect the learning or optimization process. The highest accuracy is achieved slightly above 0.88 around the 21st generation, marking the peak performance. After reaching this peak, the accuracy drops slightly and stabilizes around 0.85 by the 24th generation, suggesting a plateau in performance. This trend indicates that the average accuracy improves as the generations progress, suggesting that the model is effectively learning or optimizing over time, thereby increasing its performance. The fluctuations suggest variability or instability in the process, but the general trend is positive, showing consistent improvement. The peak at generation 21 signifies a significant improvement, followed by a slight decline, which could imply reaching a local optimum before settling to a more stable performance level, highlighting areas for potential further optimization.

Figure 16: Pre-trained DenseNet121 transfer learning model with GA using RF classifier
Fig. 17 shows examples of classifying some samples selected randomly from the test set (Red color for the misclassified samples, with correct labels being Benign; however, the model classified them as Malignant).
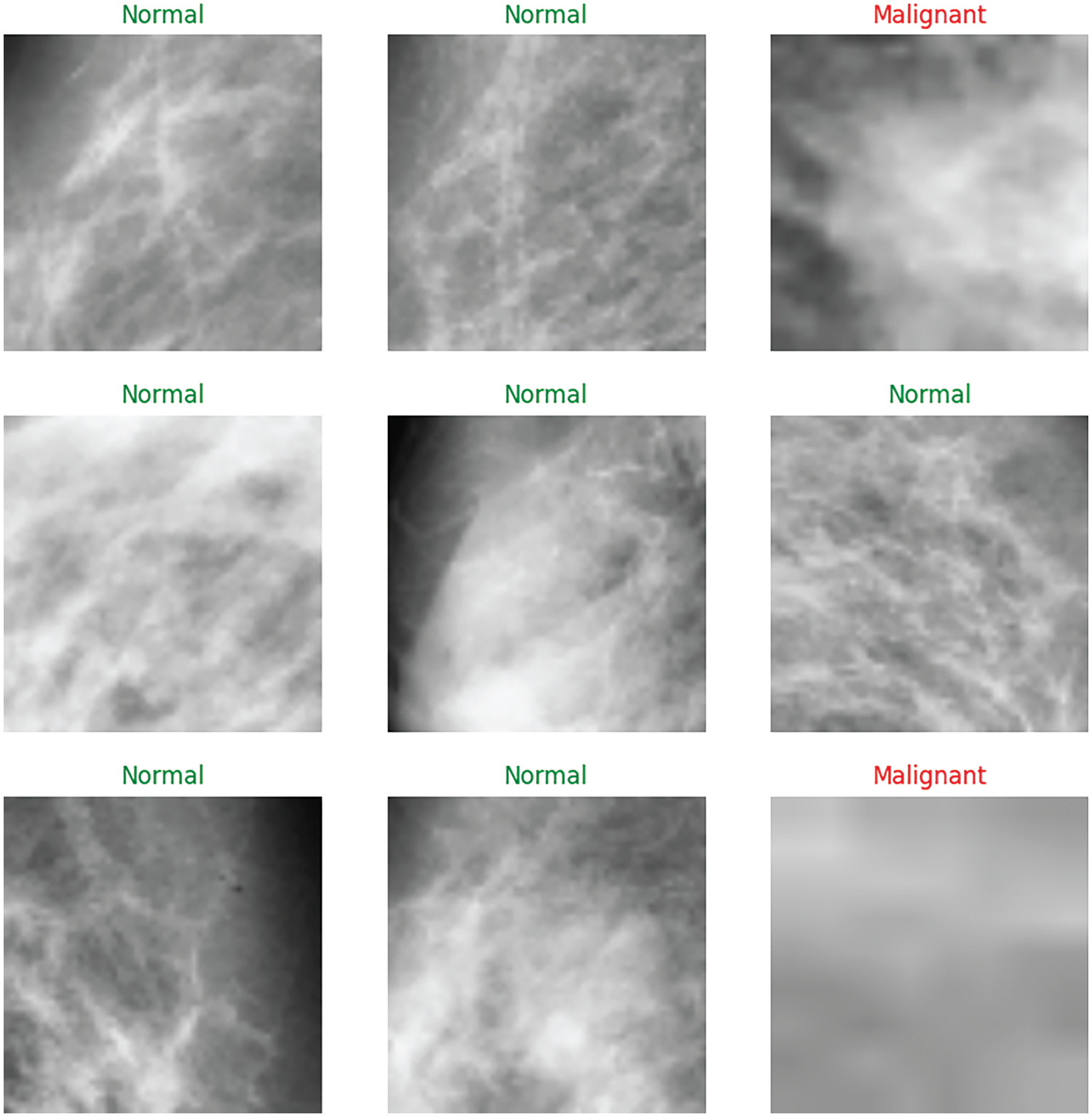
Figure 17: Examples of classifying some samples selected randomly from the test set
Fig. 18 displays four confusion matrices labeled (a), (b), (c), and (d), each representing the performance of a classification model on three classes: Normal, Benign, and Malignant. All four confusion matrices consistently show perfect classification for the Normal class, indicating the model’s effectiveness in distinguishing Normal cases. However, there is significant variability in the classification of Benign and Malignant cases. Common observations include frequent confusion between Benign and Malignant classes across all matrices, indicating difficulty in distinguishing these classes accurately. Misclassifications typically occur between Benign and Malignant, with varying degrees of accuracy depending on the model used for each matrix. These results suggest that while the classification model performs well for Normal cases, there is room for improvement in distinguishing between Benign and Malignant cases. Enhancements in the model, such as feature selection, parameter tuning, or more sophisticated algorithms, may help improve classification accuracy for these challenging cases.
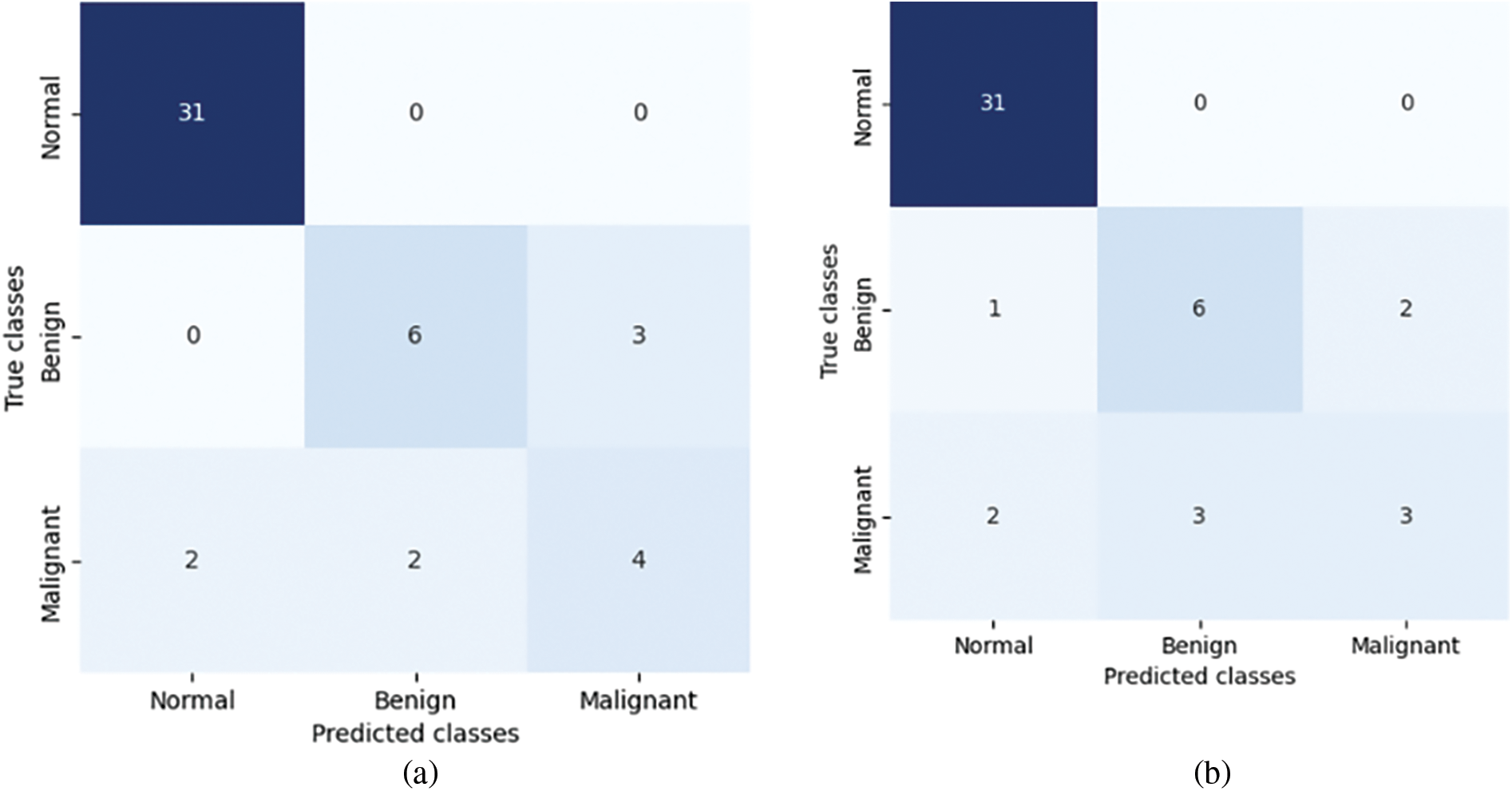
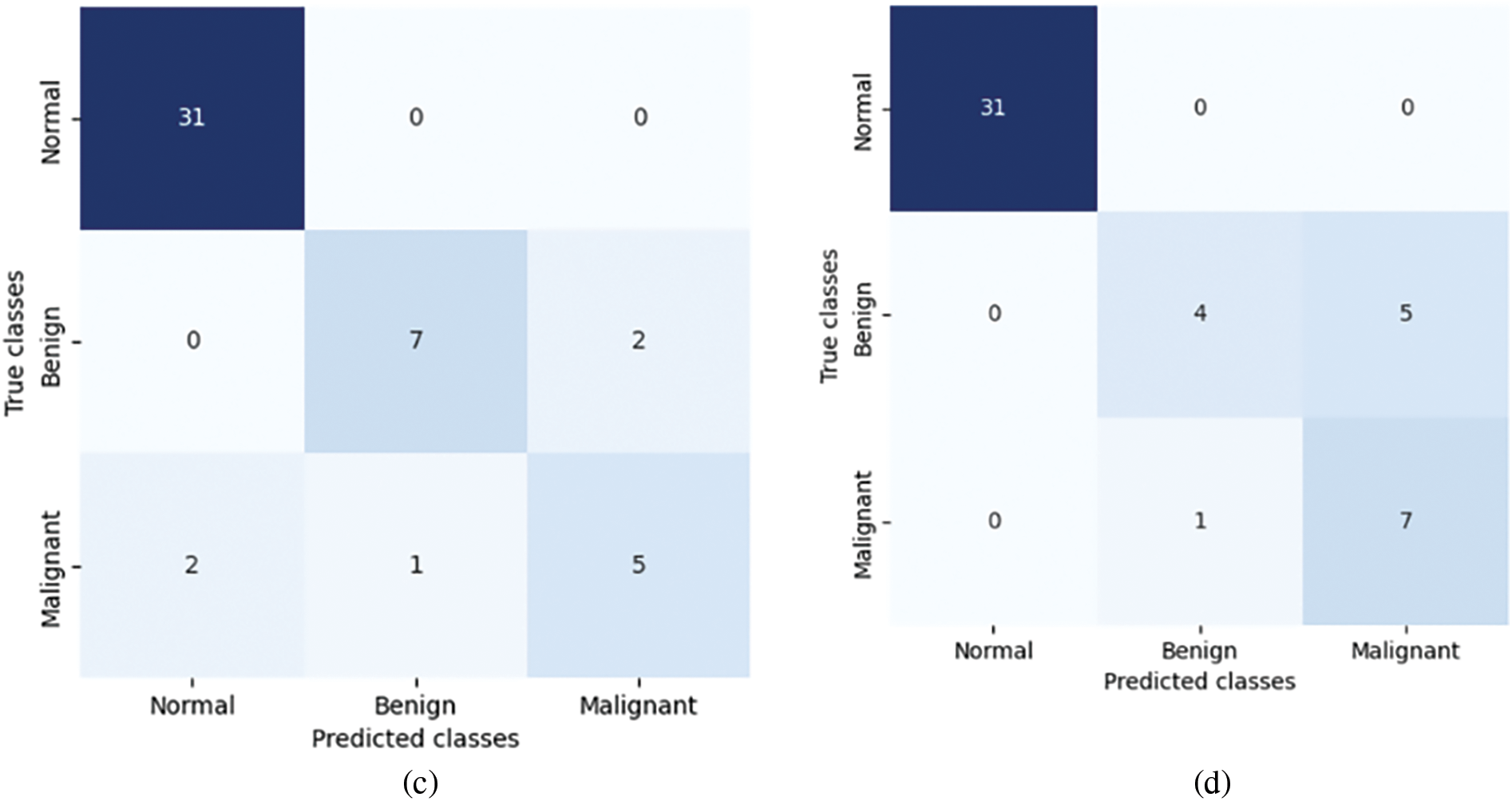
Figure 18: Confusion matrices of proposed approach on test set for the four different models: (a) VGG16-GA-RF, (b) VGG19-GA-RF, (c) MobileNetV2-GA-LSVM, and (d) DenseNet121-GA-RF
Fig. 19 compares the performance metrics (Accuracy, Precision, and Recall) of four different models: VGG16-GA-RF, VGG19-GA-RF, MobileNetV2-GA-LSVM, and DenseNet121-GA-RF. Each metric is represented by a different color bar for each model. The DenseNet121-GA-RF model achieves the highest accuracy at 0.8958, followed by the MobileNetV2-GA-LSVM model at 0.875. The VGG16-GA-RF model has an accuracy of 0.8542, while the VGG19-GA-RF model has the lowest accuracy at 0.8333. For Precision, the DenseNet121-GA-RF model again performs the best with a precision of 0.8931. The VGG16-GA-RF model has a precision of 0.8426, and the VGG19-GA-RF model has a lower precision of 0.8138. The MobileNetV2-GA-LSVM model has a precision of 0.8448. In terms of Recall, both the VGG16-GA-RF and DenseNet121-GA-RF models have a recall of 0.8542. The MobileNetV2-GA-LSVM model has a recall of 0.875, while the VGG19-GA-RF model has the lowest recall at 0.8333. These results indicate that the DenseNet121-GA-RF model consistently performs well across all three metrics, achieving the highest accuracy and precision, and sharing the highest recall with the VGG16-GA-RF model. The MobileNetV2-GA-LSVM model also shows strong performance, particularly in recall. The VGG16-GA-RF model performs moderately across all metrics. In contrast, the VGG19-GA-RF model exhibits the lowest performance in all three metrics, suggesting it may not be as effective as the other models for this particular task. The bar chart visually highlights the performance differences between these models, providing clear evidence of the superior performance of the DenseNet121-GA-RF model and identifying areas where the VGG19-GA-RF model may need improvement.
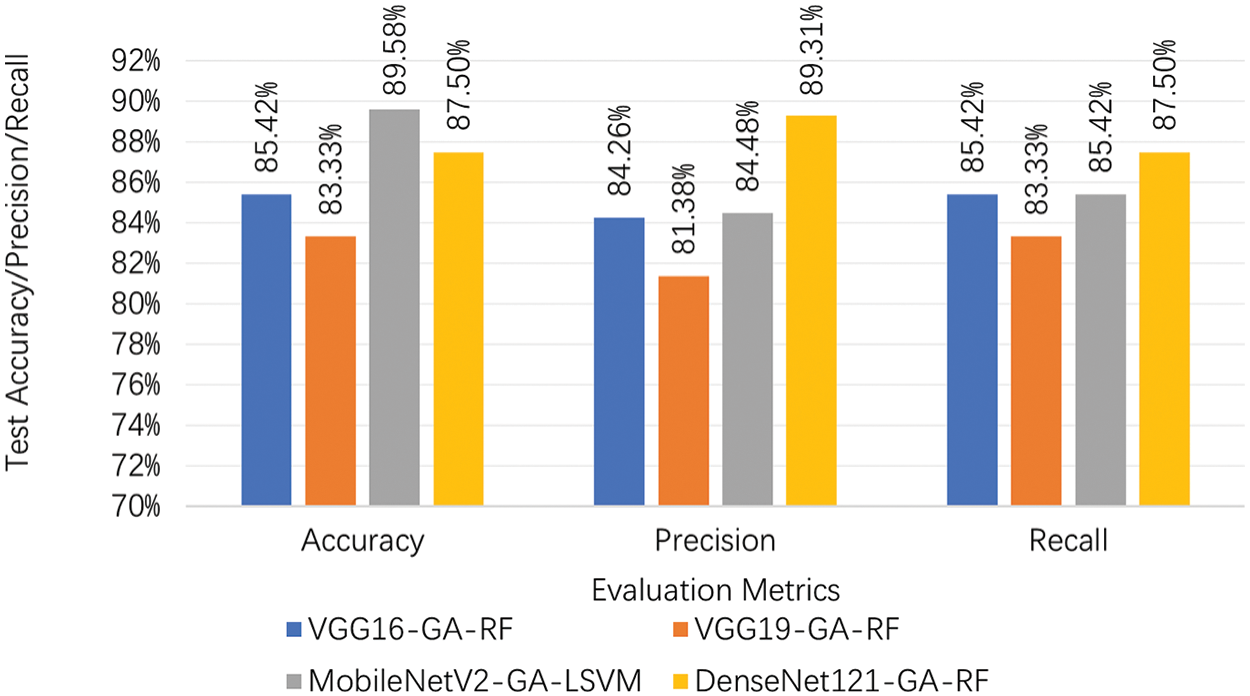
Figure 19: Pre-trained transfer learning models on test set of the proposed approach
Fig. 20 compares the F1-score and Accuracy of four models: DenseNet121, MobileNetV2, VGG19, and VGG16. These models are evaluated under two approaches: the Proposed Approach and the Baseline Approach. Each metric is represented by different colored bars: orange for F1-score and blue for Accuracy.

Figure 20: Comparison between the baseline approach and the proposed approach
Under the Proposed Approach, the DenseNet121 model achieves an F1-score of 86.96% and an Accuracy of 87.50%. The MobileNetV2 model has the highest accuracy at 89.58% and a slightly lower F1-score of 89.12%. The VGG19 model has an F1-score of 81.79% and an Accuracy of 83.33%. The VGG16 model exhibits an F1-score of 84.69% and an Accuracy of 85.42%.
Under the Baseline Approach, the DenseNet121 model has an F1-score of 75.81% and an Accuracy of 79.17%. The MobileNetV2 model shows an F1-score of 81.25% and an Accuracy of 83.33%. The VGG19 model presents an F1-score of 80.61% and an Accuracy of 81.25%. The VGG16 model records an F1-score of 77.47% and an Accuracy of 77.08%.
These results indicate that under the Proposed Approach, all models show improved performance compared to the Baseline Approach. DenseNet121 and MobileNetV2 particularly stand out, with MobileNetV2 achieving the highest accuracy of 89.58%. DenseNet121 also shows significant improvement in both F1-score and accuracy under the Proposed Approach compared to the Baseline Approach.
The VGG16 and VGG19 models also display notable improvements under the Proposed Approach. VGG16 achieves an F1-score of 84.69% and accuracy of 85.42%, while VGG19 records an F1-score of 81.79% and accuracy of 83.33%. Under the Baseline Approach, both models perform lower, with VGG16 having the lowest scores of 77.47% (F1-score) and 77.08% (accuracy).
This bar chart effectively highlights the performance improvements of all models under the Proposed Approach compared to the Baseline Approach, particularly showcasing the strong performance of MobileNetV2 and DenseNet121. The consistent improvement across all models under the Proposed Approach suggests the effectiveness of the proposed modifications or enhancements in boosting the models’ performance.
Detailed analysis of the experimental results can be clarified in terms of phenomena, cause, and recommendations. The phenomena are the observation for improved performance metrics in the proposed approach compared to the baseline and the cause is the optimization of selected features by the genetic algorithm that leads to better training of the models. The recommendations are incorporating genetic algorithms in the optimization of other machine learning models for medical diagnosis, exploring the application of this approach to other types of cancer and medical imaging tasks for scalability, and investigating the integration of other optimization techniques such as Bayesian optimization for further research and improvements.
The experimental results demonstrate the effectiveness of the proposed diagnostic method in classifying breast cancer images into normal, benign, and malignant categories, utilizing a robust dataset and advanced image processing techniques. The method involved a meticulous data preparation process using the MIAS Mammography ROIs dataset, where images were enhanced using the CLAHE method and augmented to increase data diversity and balance the training set. In the experimental analysis, both baseline and proposed methods were meticulously evaluated. The baseline method utilized conventional pre-trained models without feature selection optimization, revealing a competent performance, particularly in distinguishing normal images with high accuracy. However, the variability in precision between benign and malignant classifications suggested areas for improvement.
The proposed method, integrating genetic algorithms with pre-trained models, demonstrated significant advancements over the baseline. It was particularly effective in optimizing feature selection, which led to noticeable improvements in classifying challenging malignant cases, thereby potentially reducing the rate of false negatives—a critical factor in medical diagnostics. Figures comparing the training and validation progress indicated that while the baseline models were effective, the proposed method excelled in stability and accuracy across multiple metrics. This method not only enhanced the efficiency of the classification process but also proved scalable for larger datasets, outperforming traditional methods which typically require longer processing times.
The use of advanced analytics to dissect the performance of each model across different epochs showcased how specific models like MobileNetV2 and DenseNet121 consistently provided superior performance, adapting effectively to the nuances of medical image classification. As a whole, the proposed approach marks a significant improvement in the field of medical imaging diagnostics, offering a more accurate, efficient, and scalable method for the early detection of breast cancer. The integration of genetic algorithms has proven particularly valuable, optimizing the feature selection process and significantly boosting the classification accuracy of the system. This methodology not only sets a new standard for clinical diagnostic practices but also paves the way for further research into its application across different forms of medical diagnostics.
The proposed method can be integrated into clinical settings by deploying it alongside radiologists’ assessments, requiring compatible hardware and software systems. Ensuring interoperability with existing medical imaging systems and electronic health records is crucial. Potential obstacles include the need for high-performance computing infrastructure and regulatory approval, which involves demonstrating safety, efficacy, and addressing ethical issues like patient data privacy and model transparency. Our approach improves diagnostic accuracy from 83.33% to 89.58%, reducing unnecessary biopsies and healthcare costs. Automating preliminary mammogram analysis can streamline diagnostics, allowing radiologists to focus on complex cases. Clinicians will need training to effectively use and trust AI-supported tools, understanding their capabilities and limitations. By addressing these aspects, we demonstrate the method’s potential for improving breast cancer diagnosis and its practical implementation in clinical settings.
The study presented a GA-based optimized transfer learning approach for breast cancer diagnosis, utilizing various CNNs combined with GA for feature selection. The proposed approach aimed to enhance the diagnostic accuracy of detecting breast cancer from mammography images by optimizing the feature selection process and classifier performance.
Key findings and conclusions indicate that the proposed approach significantly improved the accuracy, precision, and recall of breast cancer diagnosis models compared to baseline methods. DenseNet121 and MobileNetV2 models, in particular, demonstrated the highest performance metrics, underscoring their suitability for this task. The use of a Genetic Algorithm for feature selection proved effective in optimizing the CNN models, leading to enhanced performance in classifying breast cancer images. The GA successfully identified the most relevant features that contributed to the improved performance of the models.
The DenseNet121-GA-RF model consistently performed well across all metrics, achieving the highest accuracy and precision. MobileNetV2-GA-LSVM also showed strong performance, particularly in the recall. Meanwhile, VGG16 and VGG19 models exhibited moderate performance but still benefited from the proposed approach compared to the baseline.
The study suggests further optimization and refinement of the GA and CNN models to enhance stability and reduce performance fluctuations across generations. Additional data preprocessing, parameter tuning and the use of more sophisticated algorithms may further improve the diagnostic accuracy. The improved accuracy and efficiency of the proposed method could lead to better diagnostic tools for breast cancer, potentially reducing the need for invasive biopsies and lowering healthcare costs. The approach holds promise for integration into clinical practice, providing radiologists with more reliable and accurate diagnostic support.
In conclusion, the GA-based optimized transfer learning approach presents a robust method for enhancing breast cancer diagnosis through effective feature selection and model optimization. The study’s findings underscore the potential of combining advanced deep learning techniques with evolutionary algorithms to achieve superior diagnostic performance in medical imaging.
Future research could focus on further optimizing the genetic algorithm by investigating advanced techniques such as adaptive genetic algorithms or hybrid approaches that combine GA with other optimization methods like particle swarm optimization or simulated annealing. Additionally, integrating the genetic algorithm with other machine learning techniques, such as ensemble learning methods or reinforcement learning, may further enhance the robustness and accuracy of the diagnostic models. Exploring the performance of newer or less commonly used CNN architectures in combination with GA could offer additional improvements in diagnostic accuracy. Implementing more sophisticated data augmentation and preprocessing techniques would increase the variability and quality of the training data, improving the generalization capability of the models. Finally, developing and testing the proposed method on diverse and larger datasets to ensure generalization and scalability, confirming its robustness across different imaging conditions and populations.
Acknowledgement: Not applicable.
Funding Statement: The authors extend their appreciation to the Deputyship for Research & Innovation, “Ministry of Education” in Saudi Arabia for funding this research work through the project number (IFKSUDR_D127).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Hussain AlSalman and Mohammad Mehedi Hassan; data collection: Mabrook AlRakhami and Taha Alfakih; analysis and interpretation of results: Mabrook AlRakhami, Mohammad Mehedi Hassan, Hussain AlSalman and Amerah Alabrah; draft manuscript preparation: Mohammad Mehedi Hassan, Hussain AlSalman, Mabrook AlRakhami, Taha Alfakih and Amerah Alabrah. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available at www.kaggle.com/datasets/annkristinbalve/mias-mammography-rois, accessed on 05 April 2024.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Alshareef B, Yaseen W, Jawa W, Barnawe Y, Alshehry W, Alqethami H, et al. Breast cancer awareness among female school teachers in Saudi Arabia: a population based survey. Asian Pac J Cancer Prev. 2020;21(2):337. doi:10.31557/APJCP.2020.21.2.337 [Google Scholar] [PubMed] [CrossRef]
2. Perović A, Pavlović-Stojanović J, Lazić L, Antonijević-Đorđević D, Bjelica M, Popov I, et al. The significance of colorectal cancer in the morbidity and mortality of the adult population of the South Banat District in the period from 2010 to 2019. Zdravstvena zaštita. 2020;49(4):1–16 (In Serbian). doi:10.5937/zdravzast49-27293. [Google Scholar] [CrossRef]
3. Hartmann LC, Degnim AC, Santen RJ, Dupont WD, Ghosh K. A typical hyperplasia of the breast—risk assessment and management options. N Engl J Med. 2015;372(1):78–89. doi:10.1056/NEJMsr1407164 [Google Scholar] [PubMed] [CrossRef]
4. Tuggener L, Satyawan YP, Pacha A, Schmidhuber J, Stadelmann T. The DeepScoresV2 dataset and benchmark for music object detection. In: 2020 25th International Conference on Pattern Recognition (ICPR), 2021 Jan 10–15; Milan, Italy: IEEE. p. 9188–95. [Google Scholar]
5. Wang D, Khosla A, Gargeya R, Irshad H, Beck AH. Deep learning for identifying metastatic breast cancer. arXiv:1606.05718. 2016. [Google Scholar]
6. Balasubramaniam S, Velmurugan Y, Jaganathan D, Dhanasekaran S. A modified LeNet CNN for breast cancer diagnosis in ultrasound images. Diagnostics. 2023;13(17):2746. doi:10.3390/diagnostics13172746 [Google Scholar] [PubMed] [CrossRef]
7. Tabakhi S, Moradi P. Universal feature selection tool (UniFeatan open-source tool for dimensionality reduction. Neurocomputing. 2023;535:156–65. doi:10.1016/j.neucom.2023.03.037. [Google Scholar] [CrossRef]
8. Elkorany AS, Elsharkawy ZF. Efficient breast cancer mammograms diagnosis using three deep neural networks and term variance. Sci Rep. 2023;13(1):2663. doi:10.1038/s41598-023-29875-4 [Google Scholar] [PubMed] [CrossRef]
9. Maldonado J, Riff MC, Neveu B. A review of recent approaches on wrapper feature selection for intrusion detection. Expert Syst Appl. 2022;198:116822. doi:10.1016/j.eswa.2022.116822. [Google Scholar] [CrossRef]
10. Oza P, Sharma P, Patel S, Kumar P. Deep convolutional neural networks for computer-aided breast cancer diagnostic: a survey. Neural Comput Appl. 2022;34(3):1815–36. doi:10.1007/s00521-021-06804-y. [Google Scholar] [CrossRef]
11. Oza P, Sharma P, Patel S, Bruno A. A bottom-up review of image analysis methods for suspicious region detection in mammograms. J Imaging. 2021;7(9):190. doi:10.3390/jimaging7090190 [Google Scholar] [PubMed] [CrossRef]
12. Bressan RS, Bugatti PH, Saito PT. Breast cancer diagnosis through active learning in content-based image retrieval. Neurocomputing. 2019;357:1–10. doi:10.1016/j.neucom.2019.05.041. [Google Scholar] [CrossRef]
13. Tiryaki VM, Tutkun N. Breast cancer mass classification using machine learning, binary-coded genetic algorithms and an ensemble of deep transfer learning. Comput J. 2024;67(3):1111–25. doi:10.1093/comjnl/bxad046. [Google Scholar] [CrossRef]
14. Shukla AK. Simultaneously feature selection and parameters optimization by teaching-learning and genetic algorithms for diagnosis of breast cancer. Int J Data Sci Anal. 2024;1–22. doi:10.1007/s41060-024-00513-0. [Google Scholar] [CrossRef]
15. Talebzadeh H, Talebzadeh M, Satarpour M, Jalali F, Farhadi B, Vahdatpour MS, et al. Enhancing breast cancer diagnosis accuracy through genetic algorithm-optimized multilayer perceptron. Multiscale Multidiscip Model Exp Des. 2024;7(4):1–17. doi:10.1007/s41939-024-00487-3. [Google Scholar] [CrossRef]
16. Dalal S, Onyema EM, Kumar P, Maryann DC, Roselyn AO, Obichili MI, et al. A hybrid machine learning model for timely prediction of breast cancer. Int J Model Simul Sci Comput. 2023;14(4):2341023. doi:10.1142/S1793962323410234. [Google Scholar] [CrossRef]
17. Soulami KB, Saidi MN, Honnit B, Anibou C, Tamtaoui A. Detection of breast abnormalities in digital mammograms using the electromagnetism-like algorithm. Multimed Tools Appl. 2019;78:12835–63. [Google Scholar]
18. Hassan SAA, Sayed MS, Abdalla MI, Rashwan MA. Detection of breast cancer mass using MSER detector and features matching. Multimed Tools Appl. 2019;78:20239–62. doi:10.1007/s11042-019-7358-1. [Google Scholar] [CrossRef]
19. Patil RS, Biradar N. Automated mammogram breast cancer detection using the optimized combination of convolutional and recurrent neural network. Evol Intell. 2021;14:1459–74. [Google Scholar]
20. Zhang Y-D, Satapathy SC, Guttery DS, Górriz JM, Wang S-H. Improved breast cancer classification through combining graph convolutional network and convolutional neural network. Inf Process Manag. 2021;58(2):102439. doi:10.1016/j.ipm.2020.102439. [Google Scholar] [CrossRef]
21. Botlagunta M, Botlagunta MD, Myneni MB, Lakshmi D, Nayyar A, Gullapalli JS, et al. Classification and diagnostic prediction of breast cancer metastasis on clinical data using machine learning algorithms. Sci Rep. 2023;13(1):485. doi:10.1038/s41598-023-27548-w [Google Scholar] [PubMed] [CrossRef]
22. Gupta S, Panwar A, Yadav R, Aeri M, Manwal M. Employing deep learning feature extraction models with learning classifiers to diagnose breast cancer in medical images. In: 2022 IEEE Delhi Section Conference (DELCON), 2022 Feb 11–13; New Delhi, India: IEEE. p. 1–6. [Google Scholar]
23. Alhasani AT, Alkattan H, Subhi AA, El-Kenawy E-SM, Eid MM. A comparative analysis of methods for detecting and diagnosing breast cancer based on data mining. Methods. 2023;7(9). doi:10.54216/JAIM.040201. [Google Scholar] [CrossRef]
24. Kunkler IH, Williams LJ, Jack WJ, Cameron DA, Dixon JM. Breast-conserving surgery with or without irradiation in early breast cancer. New Eng J Med. 2023;388(7):585–94. doi:10.1056/NEJMoa2207586 [Google Scholar] [PubMed] [CrossRef]
25. Whelan TJ, Smith S, Parpia S, Fyles AW, Bane A, Liu F-F, et al. Omitting radiotherapy after breast-conserving surgery in luminal A breast cancer. New Eng J Med. 2023;389(7):612–9. doi:10.1056/NEJMoa2302344 [Google Scholar] [PubMed] [CrossRef]
26. Humayun M, Khalil MI, Almuayqil SN, Jhanjhi NZ. Framework for detecting breast cancer risk presence using deep learning. Electronics. 2023;12(2):403. doi:10.3390/electronics12020403. [Google Scholar] [CrossRef]
27. Davoudi K, Thulasiraman P. Evolving convolutional neural network parameters through the genetic algorithm for the breast cancer classification problem. Simulation. 2021;97(8):511–27. doi:10.1177/0037549721996031 [Google Scholar] [PubMed] [CrossRef]
28. Alhussan AA, Abdelhamid AA, Towfek S, Ibrahim A, Abualigah L, Khodadadi N, et al. Classification of breast cancer using transfer learning and advanced al-biruni earth radius optimization. Biomimetics. 2023;8(3):270. doi:10.3390/biomimetics8030270 [Google Scholar] [PubMed] [CrossRef]
29. Balaha HM, Saif M, Tamer A, Abdelhay EH. Hybrid deep learning and genetic algorithms approach (HMB-DLGAHA) for the early ultrasound diagnoses of breast cancer. Neural Comput Appl. 2022;34(11):8671–95. [Google Scholar]
30. Debien V, De Caluwé A, Wang X, Piccart-Gebhart M, Tuohy VK, Romano E, et al. Immunotherapy in breast cancer: an overview of current strategies and perspectives. npj Breast Cancer. 2023;9(1):7. doi:10.1038/s41523-023-00508-3 [Google Scholar] [PubMed] [CrossRef]
31. Voon W, Hum YC, Tee YK, Yap W-S, Salim MIM, Tan TS, et al. Performance analysis of seven Convolutional Neural Networks (CNNs) with transfer learning for Invasive Ductal Carcinoma (IDC) grading in breast histopathological images. Sci Rep. 2022;12(1):19200. doi:10.1038/s41598-022-21848-3 [Google Scholar] [PubMed] [CrossRef]
32. Melekoodappattu JG, Dhas AS, Kandathil BK, Adarsh K. Breast cancer detection in mammogram: combining modified CNN and texture feature based approach. J Ambient Intell Humaniz Comput. 2023;14(9):11397–406. doi:10.1007/s12652-022-03713-3. [Google Scholar] [CrossRef]
33. Yerushalmy J. Statistical problems in assessing methods of medical diagnosis, with special reference to X-ray techniques. Public Health Rep. 1947;62:1432–49. doi:10.2307/4586294. [Google Scholar] [CrossRef]
34. Shen X, Wei L, Tang SJS. Dermoscopic image classification method using an ensemble of fine-tuned convolutional neural networks. Sensors. 2022;22(11):4147. doi:10.3390/s22114147 [Google Scholar] [PubMed] [CrossRef]
35. Nguyen T-H, Nguyen T-N, Ngo B-V. A VGG-19 model with transfer learning and image segmentation for classification of tomato leaf disease. AgriEngineering. 2022;4(4):871–87. doi:10.3390/agriengineering4040056. [Google Scholar] [CrossRef]
36. Ogundokun RO, Misra S, Akinrotimi AO, Ogul HJS. MobileNet-SVM: a lightweight deep transfer learning model to diagnose BCH scans for IoMT-based imaging sensors. Sensors. 2023;23(2):656. doi:10.3390/s23020656 [Google Scholar] [PubMed] [CrossRef]
37. Zhou T, Ye X, Lu H, Zheng X, Qiu S, Liu Y. Dense convolutional network and its application in medical image analysis. Biomed Res Int. 2022;2022(1):2384830. doi:10.1155/2022/2384830 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools