 Open Access
Open Access
REVIEW
A Comprehensive Systematic Review: Advancements in Skin Cancer Classification and Segmentation Using the ISIC Dataset
1 Department of Computer Sciences, Pakistan Institute of Engineering and Applied Sciences, Nilore, 50700, Islamabad
2 Future Technology Research Center, National Yunlin University of Science and Technology, Douliou, 64002, Taiwan
3 Directorate of IT Services, University of Gujrat, Gujrat, 50700, Pakistan
* Corresponding Author: Aneela Zameer. Email:
Computer Modeling in Engineering & Sciences 2024, 140(3), 2131-2164. https://doi.org/10.32604/cmes.2024.050124
Received 28 January 2024; Accepted 26 April 2024; Issue published 08 July 2024
Abstract
The International Skin Imaging Collaboration (ISIC) datasets are pivotal resources for researchers in machine learning for medical image analysis, especially in skin cancer detection. These datasets contain tens of thousands of dermoscopic photographs, each accompanied by gold-standard lesion diagnosis metadata. Annual challenges associated with ISIC datasets have spurred significant advancements, with research papers reporting metrics surpassing those of human experts. Skin cancers are categorized into melanoma and non-melanoma types, with melanoma posing a greater threat due to its rapid potential for metastasis if left untreated. This paper aims to address challenges in skin cancer detection via visual inspection and manual examination of skin lesion images, processes historically known for their laboriousness. Despite notable advancements in machine learning and deep learning models, persistent challenges remain, largely due to the intricate nature of skin lesion images. We review research on convolutional neural networks (CNNs) in skin cancer classification and segmentation, identifying issues like data duplication and augmentation problems. We explore the efficacy of Vision Transformers (ViTs) in overcoming these challenges within ISIC dataset processing. ViTs leverage their capabilities to capture both global and local relationships within images, reducing data duplication and enhancing model generalization. Additionally, ViTs alleviate augmentation issues by effectively leveraging original data. Through a thorough examination of ViT-based methodologies, we illustrate their pivotal role in enhancing ISIC image classification and segmentation. This study offers valuable insights for researchers and practitioners looking to utilize ViTs for improved analysis of dermatological images. Furthermore, this paper emphasizes the crucial role of mathematical and computational modeling processes in advancing skin cancer detection methodologies, highlighting their significance in improving algorithmic performance and interpretability.Keywords
As per the World Health Organization (WHO), cancer stands out as a leading cause of death [1]. Statistical analyses reveal a concerning trend, with over two individuals succumbing to skin cancer every hour in the United States alone. The incidence of various skin cancer types has witnessed a rise over the past decade, with approximately 3 million non-melanoma skin cancer cases and 132,000 melanoma skin cancer cases diagnosed globally each year [2]. The primary factors contributing to skin cancer include UV radiation, genetic predisposition, unhealthy lifestyle choices, and smoking. UV radiation is singled out as the predominant cause, and the swift depletion of ozone layers exacerbates the issue by allowing more harmful UV radiation to reach the Earth’s surface. Consequently, an upsurge in skin cancer cases is anticipated [3]. Early detection of skin cancer holds immense importance, offering heightened chances of successful treatment and prevention of metastasis to other organs. Dermoscopy, a noninvasive technique for examining pigmented skin lesions at the surface level, proves valuable for early diagnosis. Despite this, early detection remains a formidable task, even for seasoned dermatologists, owing to the diverse manifestations of skin cancer. Challenges arise from deciphering whether a manifestation like swelling is benign or malignant. Furthermore, the inconspicuous nature of small forms, such as moles, makes detection difficult with the naked eye. Therefore, there is a pressing need for more precise and reliable detection methods.
As per data provided by the WSRF for the year 2020 [4], Australia exhibited the highest incidence rate, recording an estimated 161,171 new cases. New Zealand reported 2801 cases, while Denmark, Netherlands, Norway, Sweden, Switzerland, Germany, Slovenia, and Finland each had fewer than 10,000 new cases. Fig. 1 visually represents the global skin cancer index. Skin cancer affects both men and women on a global scale, with men accounting for 173,844 new cases and women for 150,791 new cases. In terms of male incidence rates, Australia led with an estimated 9462 new cases, followed by Germany with 17,260 cases. New Zealand, Denmark, Netherlands, Norway, Sweden, Slovenia, Switzerland, and Finland each reported fewer than 5000 cases. For women, Slovenia reported the highest incidence rate of melanoma skin cancer, with approximately 14,208 new cases, followed by Norway with 3890 cases. New Zealand, Sweden, Switzerland, Denmark, Germany, Netherlands, and Finland each reported fewer than 3000 new cases. The data provided by WCRF highlights the global distribution of melanoma skin cancer cases and the varying incidence rates across different countries [5].
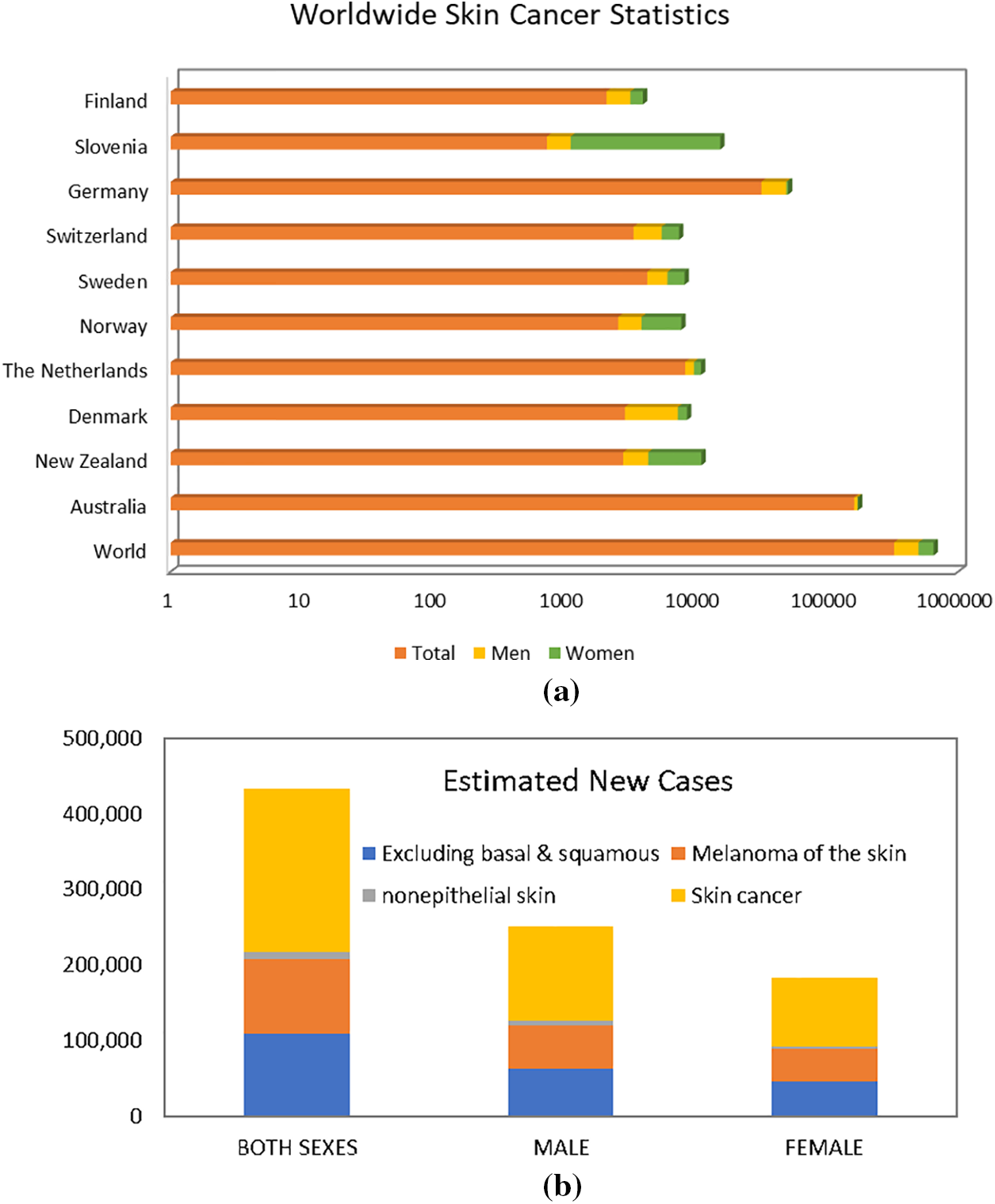
Figure 1: Worldwide skin cancer statistics
Section 1 serves as the introduction, setting the stage by introducing the ISIC dataset and its pivotal role in collaborative efforts for skin cancer analysis. In Section 2, a concise overview of skin cancer classification and segmentation utilizing the ISIC dataset is presented, providing readers with foundational knowledge. The methodology employed in these processes is detailed in Section 3. Section 3 delves into the dataset’s characteristics, offering insights into its composition and complexities. Challenges such as data quality and labeling accuracy are thoroughly discussed in Section 4, alongside proposed solutions. Section 5 explores future directions for advancing skin cancer diagnostics. The paper concludes in Section 6, summarizing findings and highlighting the significant contributions of vision transformers in enhancing skin cancer analysis using the ISIC dataset. This structured approach efficiently guides researchers and clinicians through the dataset introduction, challenges, and future directions in skin cancer diagnostics. Fig. 2 presents the overall layout of the survey paper.
Figure 2: Layout of survey paper on ISIC dataset
This structured approach aims to efficiently guide researchers and clinicians through dataset introduction, challenges, and future directions in advancing skin cancer diagnostics.
The significance of mathematical and computational modeling processes cannot be overstated. This paper emphasizes their pivotal role, particularly in advancing methodologies for skin cancer detection. It underscores the crucial intersection between computer science and mathematical modeling, illustrating how these disciplines collaborate to improve algorithmic performance and interpretability. In Section 4, the paper delves into the intricate mathematical equations that underpin the algorithms utilized in the ISIC, SCC, and SCS tasks. Detailed discussions on the mathematical modeling processes are provided for well-established architectures such as VGG-16, ResNet50, AlexNet, and ShuffleNet within the context of SCC. Additionally, comprehensive explanations of the mathematical equations governing the operation of advanced segmentation architectures, including U-Net and SegNet, are meticulously outlined. This rigorous examination illuminates the computational and mathematical foundations of these algorithms, enriching our understanding of their functioning and facilitating their application in dermatological image analysis using the ISIC dataset. All abbreviations are presented in Table 1.
This section provides a comprehensive overview of the application of medical datasets, specifically focusing on the widely acclaimed ISIC image datasets. It delves into crucial issues pertinent to their utilization, encompassing concerns such as the prevalence of duplicate images, class imbalances, variations in image resolution, and potential label inaccuracies. Moreover, it emphasizes the primary focus of this paper, which is centered around SCC and SCS utilizing the ISIC dataset. By synthesizing these observations, this paper underscores the pivotal role of CNNs as formidable tools in the realm of dermatology, offering promising avenues for advancing clinical decision-making processes. Furthermore, it underscores the significance of leveraging the ISIC dataset as a cornerstone in the pursuit of enhancing skin cancer diagnosis and treatment through innovative machine-learning methodologies.
The primary method for diagnosing skin cancer is visual examinations by dermatologists, which demonstrate an accuracy of approximately 60%. Dermoscopy, an enhanced imaging technique, increases diagnostic accuracy to 89%. Dermoscopy also exhibits specific sensitivity rates, such as 82.6% for melanocytic lesions, 98.6% for basal cell carcinoma, and 86.5% for squamous cell carcinoma [6]. Despite its effectiveness, dermoscopy faces challenges in accurately diagnosing certain lesions, especially early melanomas lacking distinct features. While it significantly improves melanoma diagnosis, there remains a need to enhance accuracy, particularly for featureless melanomas, to further improve patient survival rates. The limitations of dermoscopy and the quest for higher diagnostic accuracy have led to the development of computer-aided detection methods for skin cancer diagnosis [7].
The utilization of ISIC datasets is diverse, with a predominant focus on classification and segmentation tasks. Binary classification tasks have garnered significant attention, given their capacity to offer a larger pool of images for algorithm training. The advent of ISIC 2018 and ISIC 2019 marked a shift towards exploring multiclass classification, primarily leveraging the ISIC 2020 dataset. It is noteworthy that the ISIC 2020 challenge centered specifically on melanoma detection, indicating a potential surge in additional binary classification studies in subsequent research. While segmentation tasks have not gained as much popularity as lesion diagnosis, especially beyond 2019 when ISIC discontinued this challenge type, the ISIC 2016 to 2018 datasets stand out for providing delineated segmentation masks. However, the number of segmentation tasks is relatively limited compared to the abundance found in classification tasks [8].
Beyond classification and segmentation, researchers have delved into other dimensions of ISIC datasets. For instance, a study by [9] investigated the impact of color constancy, while reference [10] explored data augmentation using generative adversarial networks. These diverse applications highlight the versatility of ISIC datasets for addressing various aspects of skin image analysis beyond traditional classification and segmentation tasks [11]. The utilization of the ISIC dataset has significantly propelled the field of skin cancer analysis, particularly in the realms of classification and segmentation. The dataset’s richness and diversity have enabled researchers to explore and implement state-of-the-art techniques, leveraging advanced machine learning algorithms and deep neural networks. The collective efforts within the research community, as showcased in this paper, underscore the dataset’s pivotal role in enhancing diagnostic precision and efficacy. Through a synthesis of the latest developments and strategies, this study strives to not only provide a comprehensive overview but also to equip researchers and clinicians with the necessary insights to navigate the complexities of skin cancer analysis. By delving into the intricacies of image segmentation methodologies and the nuanced landscape of classification techniques, this paper seeks to foster a deeper understanding of the ISIC dataset’s potential, fostering advancements that hold promise for more accurate, reliable, and clinically relevant outcomes in the diagnosis and treatment of skin cancer [12].
In recent times, advancements in deep learning techniques within artificial intelligence have led to the development of novel solutions for detecting abnormalities across various medical domains, including breast cancer, lung cancer, skin cancer, brain tumors, liver cancer, and colon cancer, utilizing medical imaging technologies. CNNs have emerged as pivotal tools, particularly in image processing applications such as CT, MRI, histology images, and pathology images, among others, owing to their remarkable performance. In dermatology, CNNs have demonstrated significant potential in assisting physicians with the diagnosis of SCC and SCS.
This review employs the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) strategy for article selection. Using a Google Scholar search with the keywords “skin cancer classification” and “skin cancer segmentation,” 699 SCC and 621 SCS publications were initially identified. After removing duplicates, there are 549 SCC and 520 SCS articles. The first screening round, excluding the ISIC dataset and those categorized as reviews, database entries, or letters, results in 282 SCC and 224 SCS articles from 2013 to 2023. Further exclusions are made based on relevance to the review’s objectives and suitability of schemes, leaving 232 SCC and 194 SCS articles. Subsequently, articles not explicitly illustrating SC and SCC systems or similar strategies are deleted, reducing the count to 197 SCC and 184 SCS articles. Fig. 2 presents the overall structure of paper and Fig. 3 demonstrates the PRISMA process extensively.
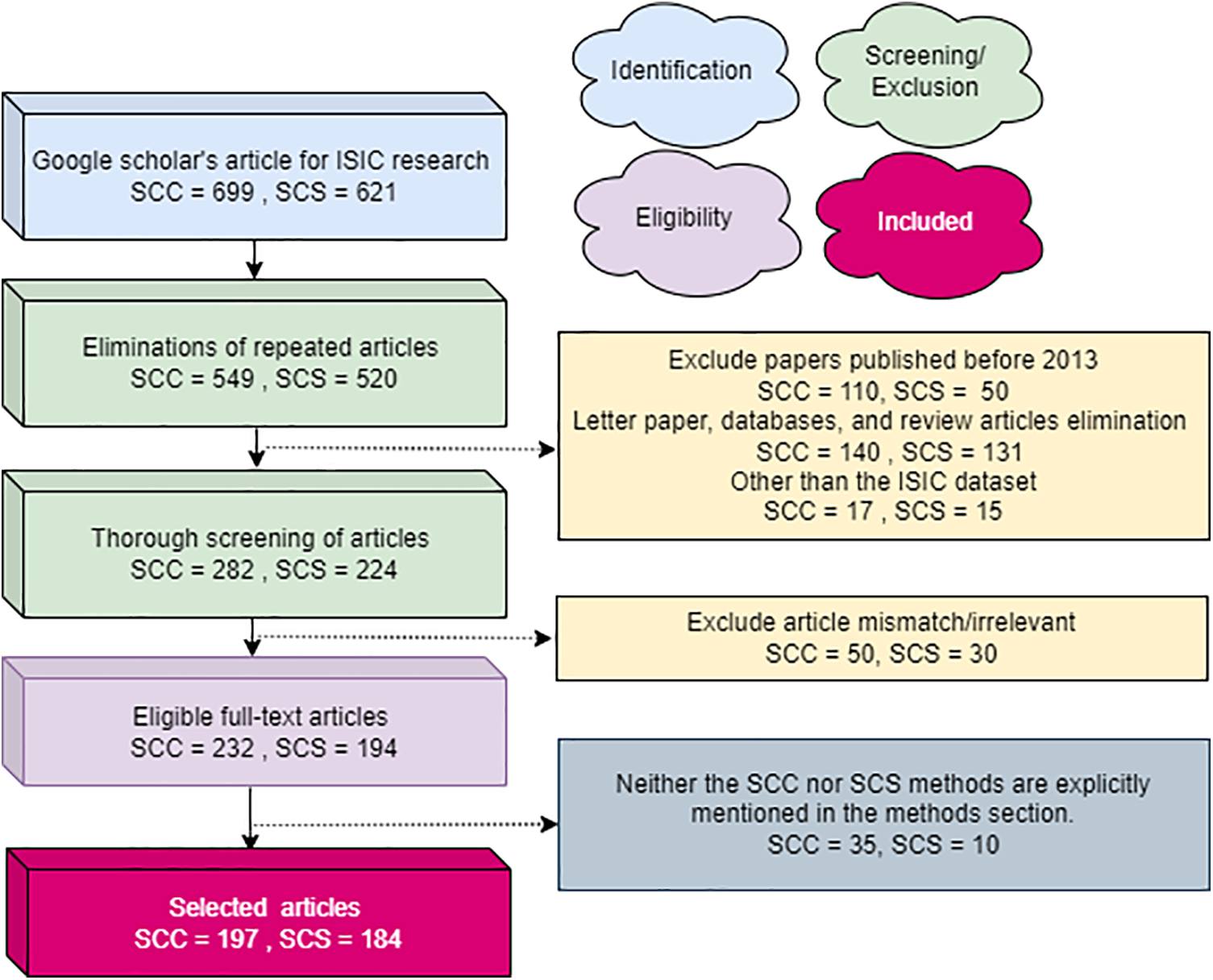
Figure 3: The PRISMA process utilized for articles entails detailing the criteria for inclusion and exclusion of evidence within the article
This section provides an in-depth overview of the most prevalent forms of skin cancers, exploring their characteristics, impact on individuals, and the role of datasets like ISIC in enhancing research and diagnosis. Fig. 4 presents the graphical view of all skin cancer types.
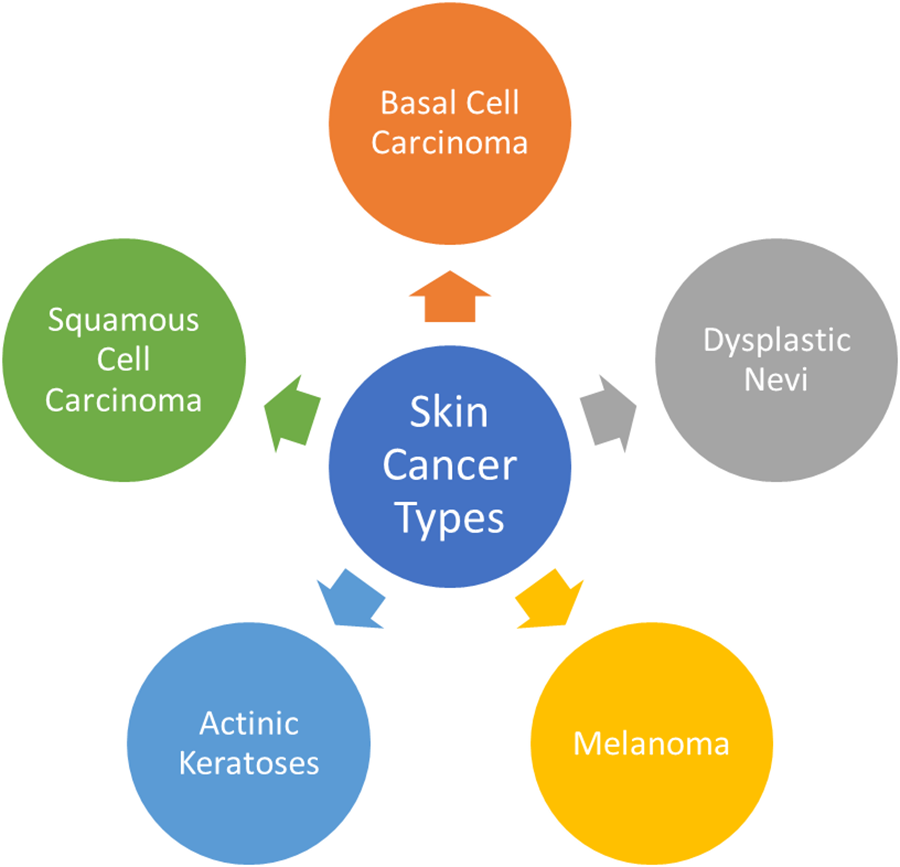
Figure 4: Most common skin cancer types
Melanoma, often referred to as “the most serious skin cancer,” is notorious for its potential to metastasize throughout the body. It can arise in normal, healthy skin or develop within existing moles [13,14]. Men are prone to developing melanoma on their faces or trunks, with occurrences even in sun-protected skin for both genders [15]. Early detection is paramount, and the ISIC dataset has played a pivotal role in advancing research for the timely identification and treatment of melanoma. Studies leveraging the dataset have contributed to improving diagnostic accuracy and enhancing our understanding of melanoma’s diverse manifestations [16]. Recent investigations utilizing the ISIC dataset have delved into molecular markers associated with melanoma progression, shedding light on potential therapeutic targets for personalized treatment strategies [17]. Moreover, advancements in machine learning algorithms, fueled by the wealth of data in the ISIC dataset, have enabled the development of predictive models for melanoma prognosis [18]. These models not only aid in risk stratification but also contribute to optimizing treatment plans for affected individuals. The collaborative nature of data sharing within the ISIC community has fostered a global exchange of insights, accelerating the pace of melanoma research and paving the way for innovative approaches in precision medicine [19]. Fig. 5 has an image annotation of melanoma.
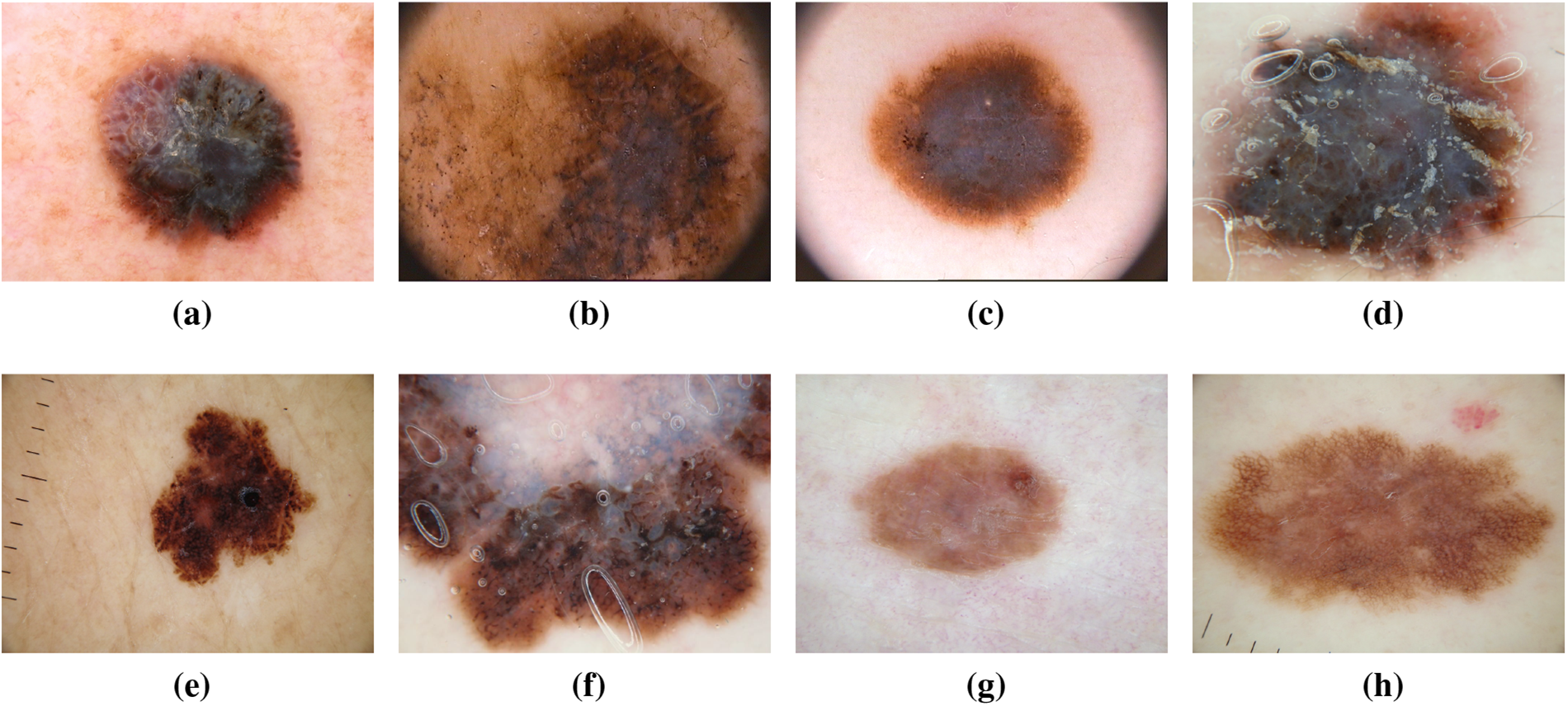
Figure 5: From (a) to (h): different states of Melanoma images of ISIC dataset
Dysplastic nevi, recognized as atypical moles, exhibit characteristics resembling normal moles but possess melanoma-like traits. With irregular shapes or colors and larger sizes than typical moles, they can emerge on various skin areas, including those typically covered or exposed to the sun. The ISIC dataset has been instrumental in studying and classifying dysplastic nevi, aiding researchers in identifying patterns and features associated with their progression towards malignancy [20,21]. In recent years, the wealth of data within the ISIC dataset has facilitated in-depth analyses exploring the genetic markers linked to dysplastic nevi. This has not only refined our understanding of the molecular underpinnings of atypical moles but has also contributed to the development of more accurate diagnostic tools [22]. Machine learning models trained on the ISIC dataset have showcased promising capabilities in distinguishing between benign and potentially malignant nevi, providing clinicians with valuable insights for early intervention [23]. Moreover, collaborative efforts within the dermatology community, fueled by the shared ISIC dataset, have led to the identification of novel biomarkers associated with dysplastic nevi progression [24]. This collaborative approach has fostered a collective understanding of atypical mole biology, paving the way for more targeted and effective strategies in the prevention and early detection of melanoma. The ISIC dataset continues to be a cornerstone in ongoing research endeavors focused on unraveling the complexities of dysplastic nevi, guiding advancements in both clinical practice and scientific understanding [25]. Fig. 6 has an image annotation of dysplastic nevi.
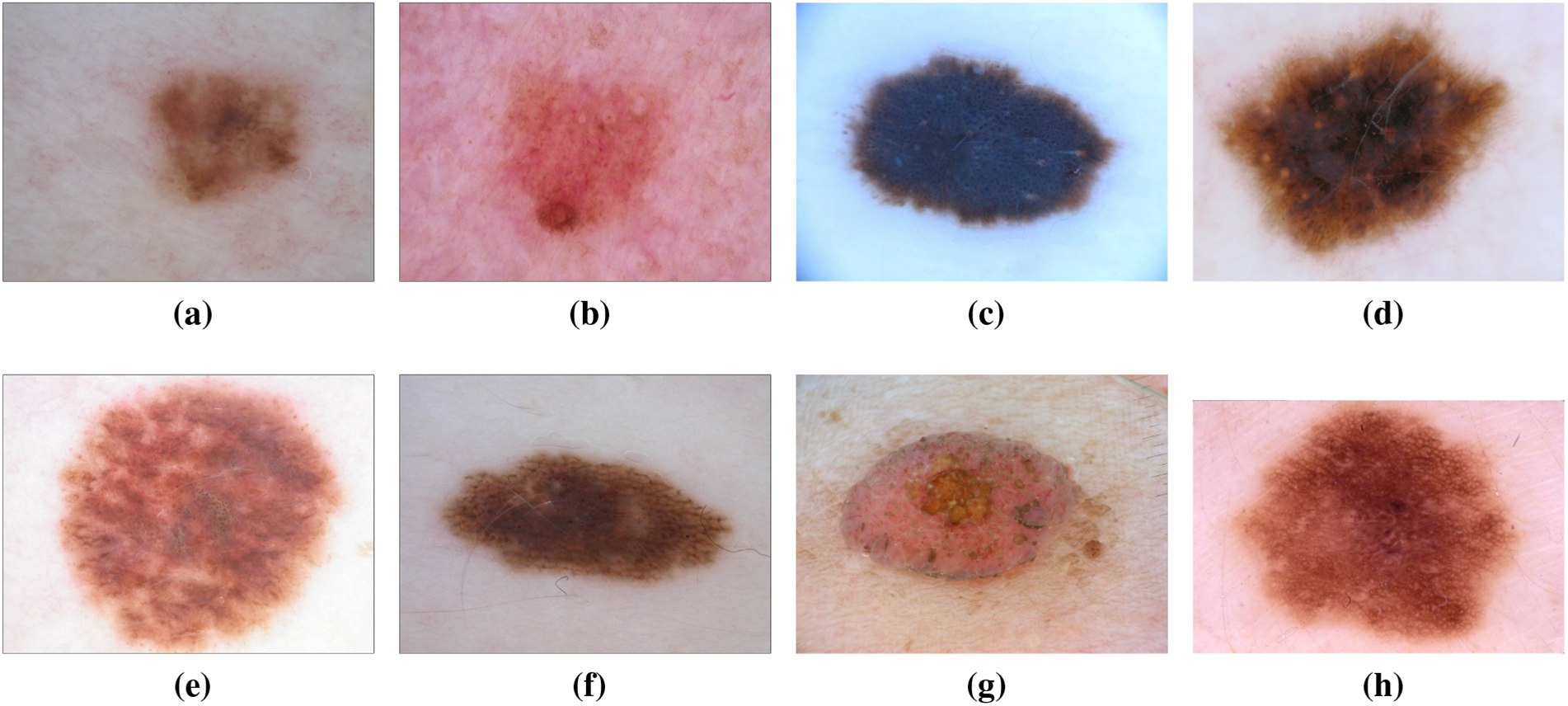
Figure 6: From (a) to (h): different states of Dysplastic Nevi images of ISIC dataset
As the most prevalent form of skin cancer, BCC often manifests as a flesh-colored growth, a pearl-shaped bump, or a pinkish skin patch. Linked to prolonged indoor tanning or frequent sun exposure, it primarily affects individuals with fair skin but can also impact those with darker skin tones [26]. The ISIC dataset has significantly contributed to BCC research, providing a wealth of diverse images for analysis. This dataset has been instrumental in developing and validating machine learning models for early detection, preventing the spread of BCC and associated complications [27,28]. Recent studies leveraging the ISIC dataset have explored the molecular signatures associated with different subtypes of BCC. This deeper understanding has not only refined diagnostic accuracy but has also paved the way for targeted therapeutic interventions [29]. Additionally, the diverse representation of BCC cases in the ISIC dataset has facilitated the development of robust classification algorithms capable of distinguishing BCC from other skin lesions with high precision [30]. Fig. 7 has an image annotation of BCC. Furthermore, collaborative initiatives fueled by the ISIC dataset have allowed researchers to investigate the impact of environmental factors on BCC development, providing insights into potential preventive measures [31]. The shared dataset has acted as a catalyst for international collaboration, fostering a collective effort to mitigate the impact of BCC on public health. As a result, the ISIC dataset stands as a cornerstone in ongoing efforts to advance our understanding of BCC, with implications for improved diagnosis, treatment, and prevention strategies [32].
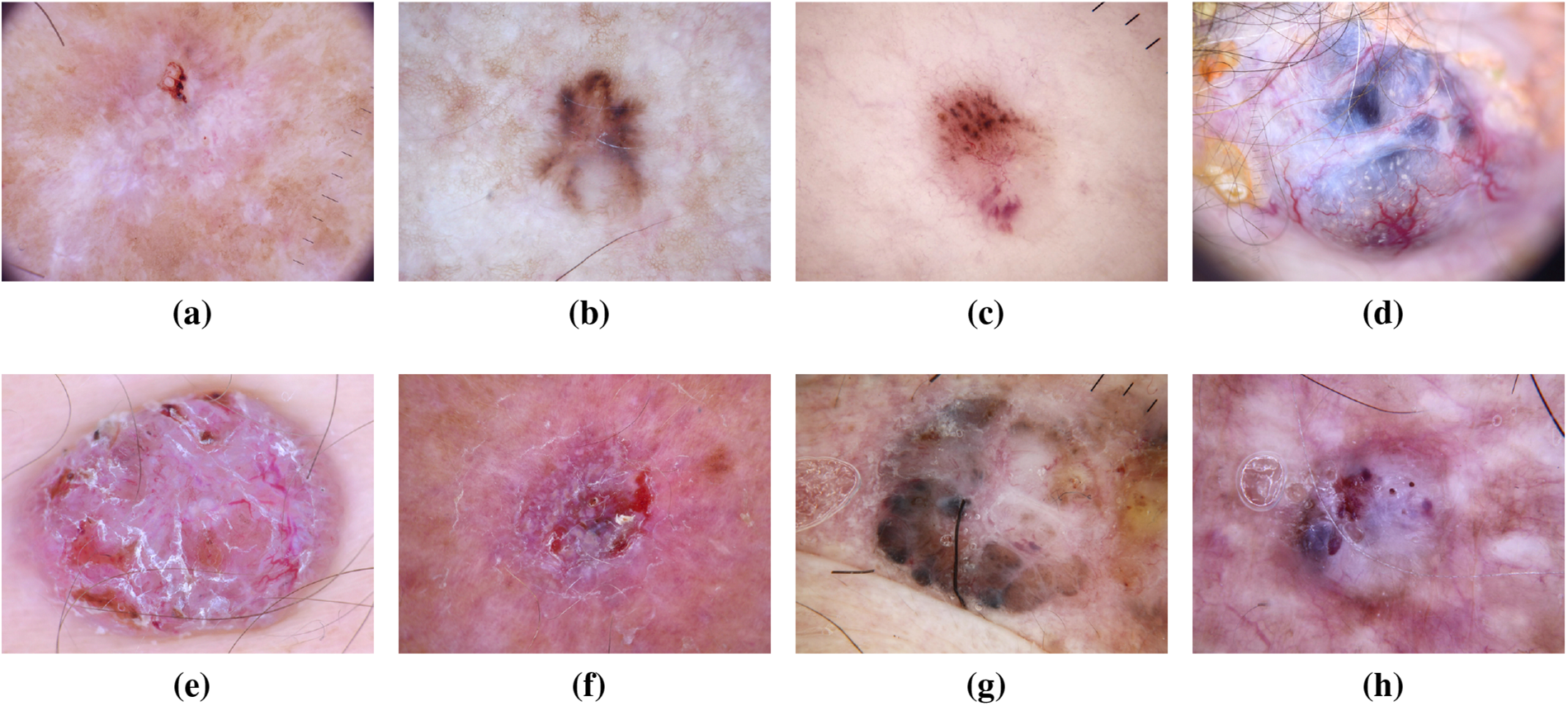
Figure 7: From (a) to (h): different states of Basal Cell Carcinoma images of ISIC dataset
SCC, a common skin cancer type, is more prevalent in individuals with light skin, although it can affect individuals with darker skin tones. It often presents as a red, firm lump, a scaly area, or a recurrent sore [33,34]. Frequent sun-exposed areas are susceptible to SCC development. Research leveraging the ISIC dataset has facilitated the exploration of unique patterns and features associated with SCC. The dataset’s contribution has led to advancements in early detection techniques and the development of targeted treatment approaches [35]. Recent investigations using the ISIC dataset have focused on unraveling the genomic landscape of SCC, providing insights into the molecular mechanisms driving its development and progression [36]. This deeper understanding has paved the way for personalized treatment strategies, potentially improving outcomes for individuals diagnosed with SCC. Additionally, machine learning applications trained on the ISIC dataset have demonstrated promising capabilities in distinguishing between different subtypes of SCC, aiding clinicians in refining their diagnostic assessments [37]. Furthermore, collaborative efforts within the dermatology research community, fueled by the ISIC dataset, have facilitated international studies on the epidemiology of SCC [38]. This collective approach has not only enhanced our understanding of risk factors associated with SCC but has also informed public health strategies for prevention and early intervention. The ongoing contributions of the ISIC dataset continue to play a crucial role in advancing SCC research, with implications for improved diagnostics, treatment outcomes, and public health policies. Fig. 8 has an image annotation of SSC.
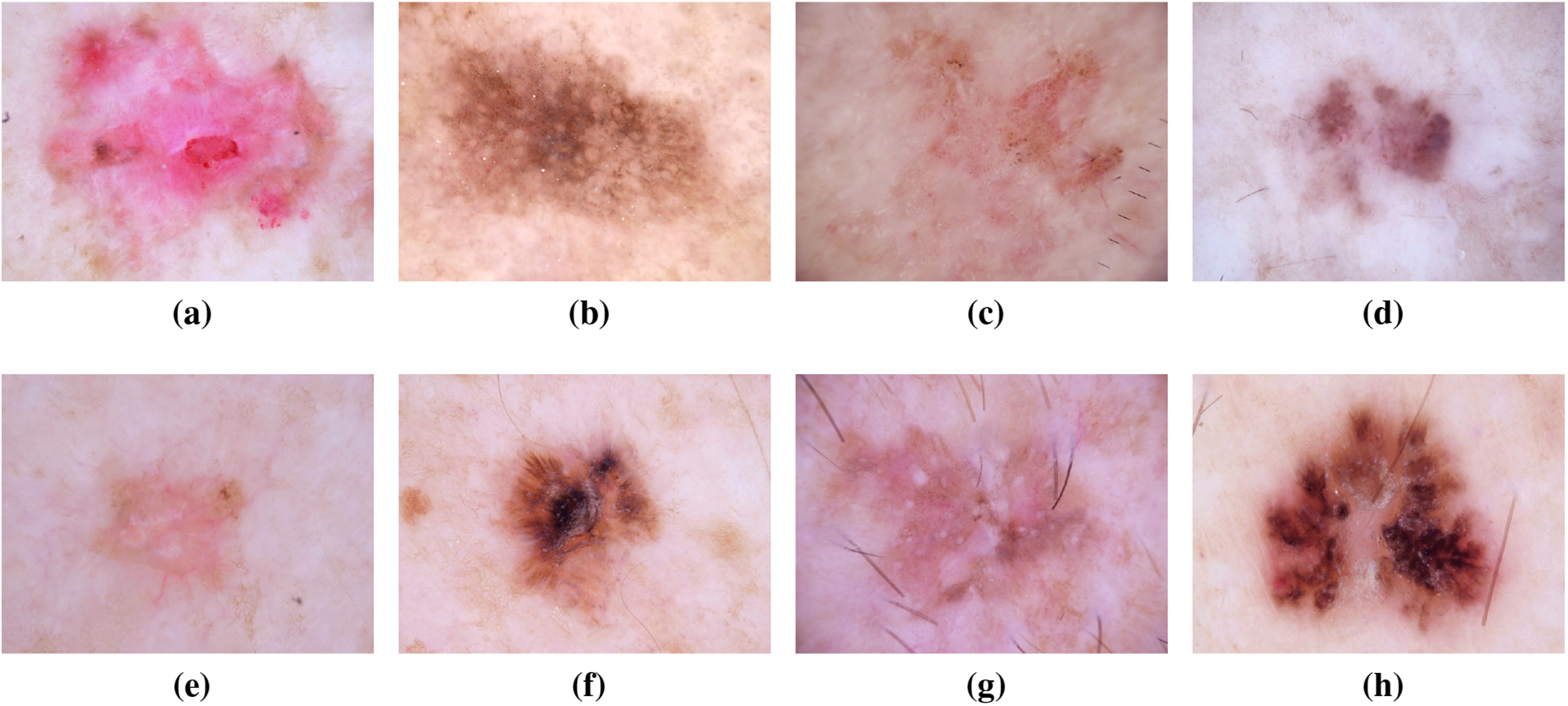
Figure 8: From (a) to (h): different stages of Squamous Cell Carcinoma images of ISIC dataset
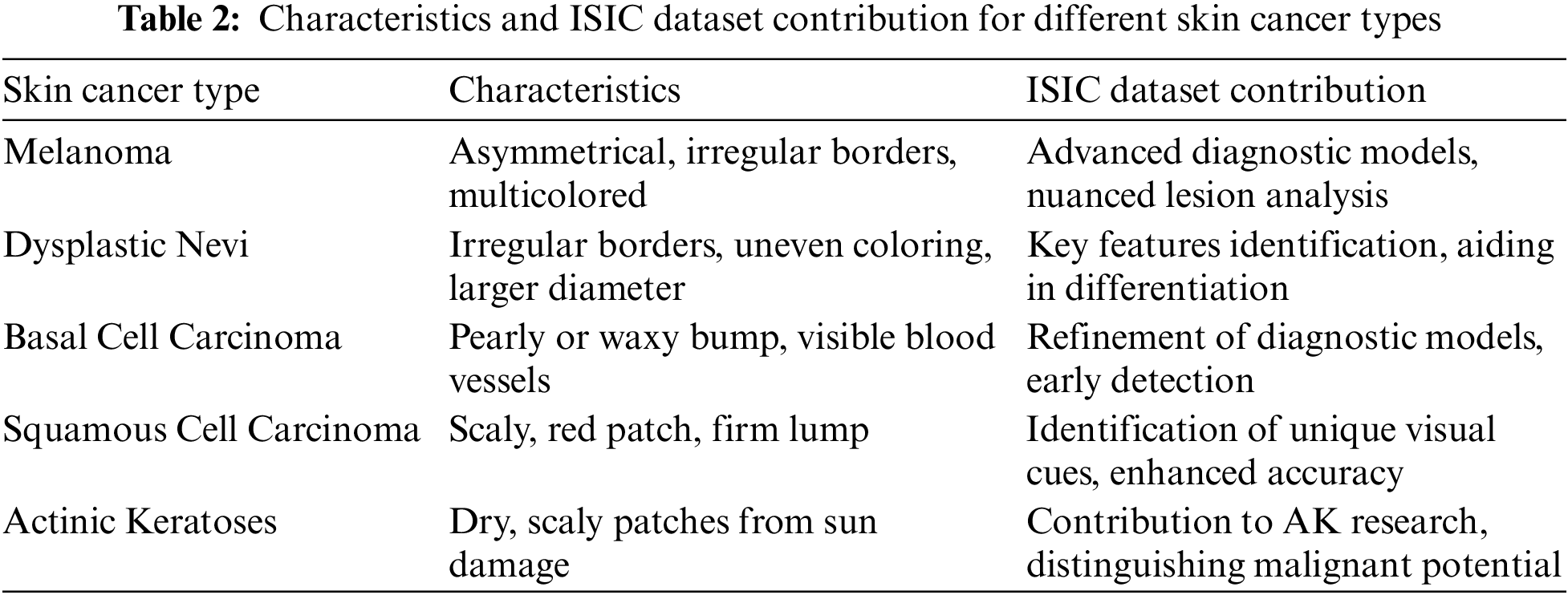
While not categorized as skin cancer, AKs are precursors with the potential to progress into squamous cell carcinoma. These scaly, dry skin lesions result from excessive sun exposure and typically appear on the head, neck, hands, and forearms. The ISIC dataset has been crucial in studying the evolution of AKs, aiding in distinguishing benign lesions from those with malignant potential. Insights from the dataset have informed timely interventions to prevent the progression of AKs into advanced stages of skin cancer [39]. Recent analyses leveraging the extensive data within the ISIC dataset have provided a deeper understanding of the genetic and molecular alterations associated with AKs. This knowledge has not only refined diagnostic criteria but has also contributed to the identification of biomarkers indicative of malignant transformation. Machine learning applications trained on the ISIC dataset have demonstrated promising capabilities in predicting the likelihood of AKs advancing to squamous cell carcinoma, allowing for more personalized and proactive patient management [40,41]. Fig. 9 has an image annotation of AKs. Moreover, collaborative efforts among dermatologists and researchers, facilitated by the shared ISIC dataset, have enabled the development of risk stratification models for individuals with AKs. These models consider a range of factors, including clinical features and genetic markers, to tailor surveillance and intervention strategies. The global exchange of insights within the ISIC community has played a pivotal role in shaping guidelines for the management of AKs, emphasizing the importance of preventive measures and early interventions to mitigate the risk of progression to invasive skin cancer [42]. Where Table 2 presents the characteristics and ISIC data contribution for different skin cancer types.
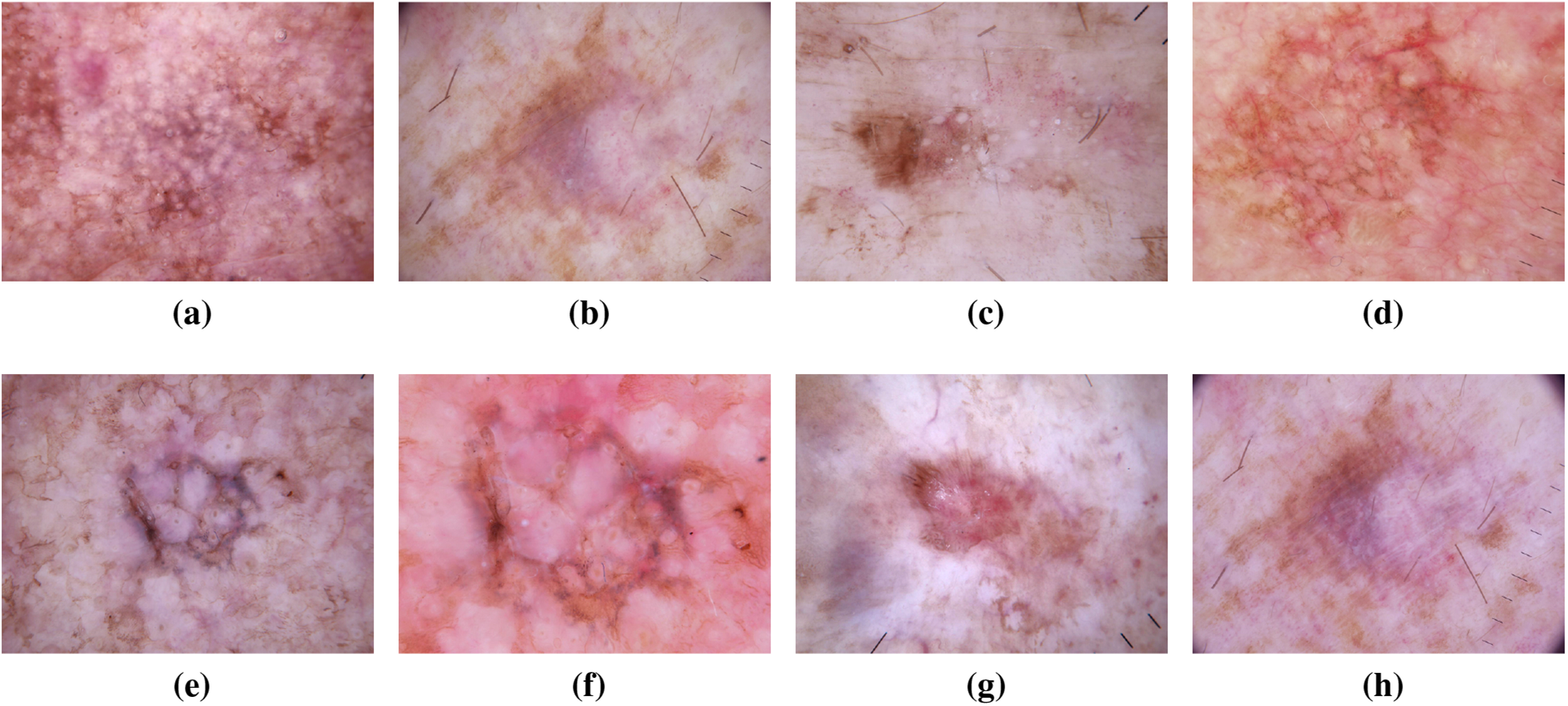
Figure 9: From (a) to (h): various states of Actinic Keratoses images of ISIC dataset
Skin cancer symptoms encompass changes in moles, the appearance of new lesions, or alterations in skin texture. Early detection is crucial, prompting regular self-examination and professional checks. Precautions include sun safety practices wearing protective clothing, using sunscreen, and avoiding excessive sun exposure [43]. Prevalence varies among skin cancer types. BCC is most common, linked to sun exposure. SCC follows, associated with sun exposure and tobacco use. Melanoma, the most serious type, requires vigilance for changes in moles. Dysplastic nevi, atypical moles, necessitate close monitoring: AKs, precursors to SCC, mandate sun protection [44]. While BCC has a higher patient prevalence due to sun exposure, early detection, and preventive measures are universally vital. Regular skin checks and adherence to sun safety mitigate risks across all skin cancer types. Individuals need to be proactive in monitoring their skin, seeking professional evaluation for any concerning changes, and adopting sun-safe behaviors to reduce the overall burden of skin cancer [45]. Public awareness campaigns play a pivotal role in educating individuals about skin cancer symptoms, the importance of early detection, and the significance of sun protection. Dermatologists emphasize the need for routine skin examinations, especially for those with fair skin or a history of sun exposure. With advancements in technology, mobile applications, and telemedicine services are also becoming valuable tools for promoting skin health and providing accessible avenues for skin cancer assessments. Regular check-ups with healthcare providers further enhance the collective effort in preventing, detecting, and managing skin cancer, ultimately contributing to improved outcomes and reduced morbidity [46].
Fig. 4 illustrates common types of skin cancers, emphasizing the diverse visual representations captured in the ISIC dataset that contribute to a deeper understanding of each skin cancer type. Fig. 10 presents the incidence of skin cancer across different regions over the past years.
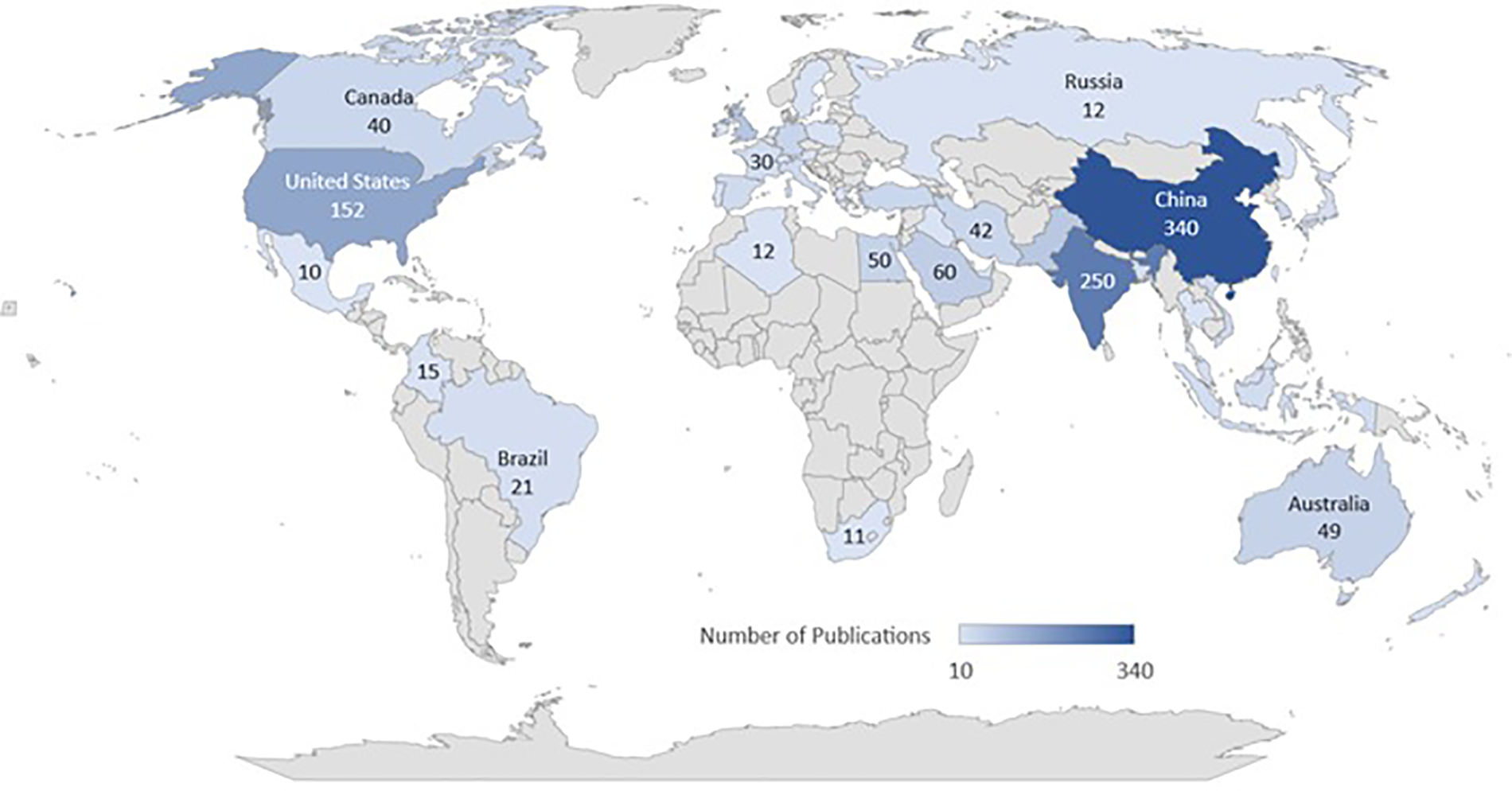
Figure 10: Worldwide publication on skin cancer images using ISIC dataset
Our research delved into the utilization of ISIC datasets for various research objectives, spanning the past 4 to 5 years. Due to the widespread adoption of ISIC datasets, offering a comprehensive list of studies is unfeasible. Nevertheless, we meticulously selected some of the most prominent and well-cited papers for our analysis. Our examination revealed a prevalent trend where recent research often incorporated multiple datasets, showcasing the versatile nature of ISIC datasets. The ISIC dataset has evolved significantly over the years, starting with ISIC 2016, comprising 900 training images and 379 test images, totaling 1279 images. Each image was sized at 512 × 512 pixels and included ground truth data indicating lesion malignancy. Subsequent years saw substantial expansions: ISIC 2017 contained 2600 images; ISIC 2018 introduced a dataset with 10,015 training images and 1512 test images, totaling 11,527 images at 600 × 450 pixels. The trend continued with ISIC 2019, offering 25,331 training images and 8238 test images, totaling 33,569 images at 1024 × 1024 pixels. In 2020, the largest ISIC dataset yet was released, comprising 33,126 training images and 10,982 test images, totaling 44,108 images with varying resolutions, commonly at 768 × 786 pixels.
Our analysis highlighted the predominant focus on tasks such as classification and segmentation within the studies deemed relevant. Binary classification emerged as a popular research area, particularly with the advent of ISIC 2018 and ISIC 2019 datasets. Moreover, the ISIC 2020 challenge specifically concentrated on melanoma detection, potentially leading to an influx of additional binary classification papers. While SCS tasks are not as prevalent as cancer diagnosis, ISIC datasets, particularly ISIC 2016 to 2018, provided valuable delineated segmentation masks. Additionally, other notable applications of ISIC datasets include studies on color constancy impact and data augmentation using generative adversarial networks. Table 3 presents the annotation of ISIC dataset in various publications.
3.4 Overview of SCC and SCS with ISIC Dataset
The ISIC dataset has emerged as a pivotal resource in advancing research on skin cancer detection and segmentation. Skin cancer, one of the most prevalent types of cancer, necessitates early and accurate diagnosis for effective treatment. The ISIC dataset addresses this imperative by providing a comprehensive collection of high-resolution dermoscopic images, encompassing various skin lesions, including malignant and benign cases [65]. Skin cancer classification involves the categorization of lesions into different classes, such as melanoma, basal cell carcinoma, and squamous cell carcinoma. Machine learning and deep learning techniques have been extensively applied to the ISIC dataset for automated classification, leveraging the rich visual information present in dermoscopic images [66]. CNNs and other deep learning architectures have demonstrated remarkable performance in distinguishing between different skin lesions with a high degree of accuracy [67]. In addition to SCC, SCS plays a crucial role in delineating the boundaries of skin lesions, aiding in a more detailed analysis of their characteristics. Segmentation algorithms applied to the ISIC dataset aim to precisely identify and outline the regions of interest within the images, facilitating a better understanding of lesion morphology and size [68]. Researchers often employ transfer learning, fine-tuning pre-trained models, and ensemble techniques to enhance the generalization and robustness of skin cancer classification and segmentation models. The ISIC dataset’s large-scale and diverse collection of images contributes to the development of robust models that can handle variations in skin types, lesion sizes, and imaging conditions [69].
As the field continues to evolve, collaborations like ISIC play a pivotal role in fostering advancements in skin cancer research. The integration of advanced computer vision techniques with the rich data provided by the ISIC dataset holds promising potential for improving early detection and aiding clinicians in making more informed decisions for optimal patient care [70,71]. Furthermore, the ISIC dataset supports benchmarking and comparison of different algorithms, fostering healthy competition and driving innovation in the field of dermatology [72]. The incorporation of clinical metadata, such as patient demographics and lesion histories, enhances the dataset’s utility for building models that not only classify and segment lesions but also consider the broader patient context [73].
The ISIC dataset’s global accessibility has facilitated collaborative efforts across institutions, enabling researchers worldwide to contribute to the collective knowledge in skin cancer research [74]. Open challenges and competitions based on the ISIC dataset serve as platforms for researchers to showcase novel approaches and methodologies, fostering a community-driven pursuit of improved diagnostic tools [75]. Ethical considerations and privacy concerns in handling medical image datasets, including the ISIC dataset, have prompted the development of secure and privacy-preserving methodologies [76]. Researchers are increasingly mindful of ensuring patient privacy and obtaining proper consent, laying the foundation for responsible and transparent use of medical data.
Continuous updates and expansions of the ISIC dataset, incorporating new cases and diverse populations, ensure that the models trained on this dataset remain relevant and applicable across different scenarios [77]. The evolving nature of skin cancer pathology demands a dynamic dataset, and the ISIC collaboration is responsive to this need [78]. In response to emerging challenges, such as data scarcity in specific subtypes or demographic groups, efforts are underway to enhance the representatives of the ISIC dataset. This includes targeted data collection initiatives and collaborations with diverse healthcare institutions to ensure a more comprehensive and inclusive dataset that can address disparities in skin cancer diagnosis and treatment [79]. The ongoing integration of multi-modal data, including not only dermoscopic images but also clinical and genetic information, further enriches the ISIC dataset. Fig. 11 presents the number of ISIC images used in the various publications for the skin cancer SCC and SCS.
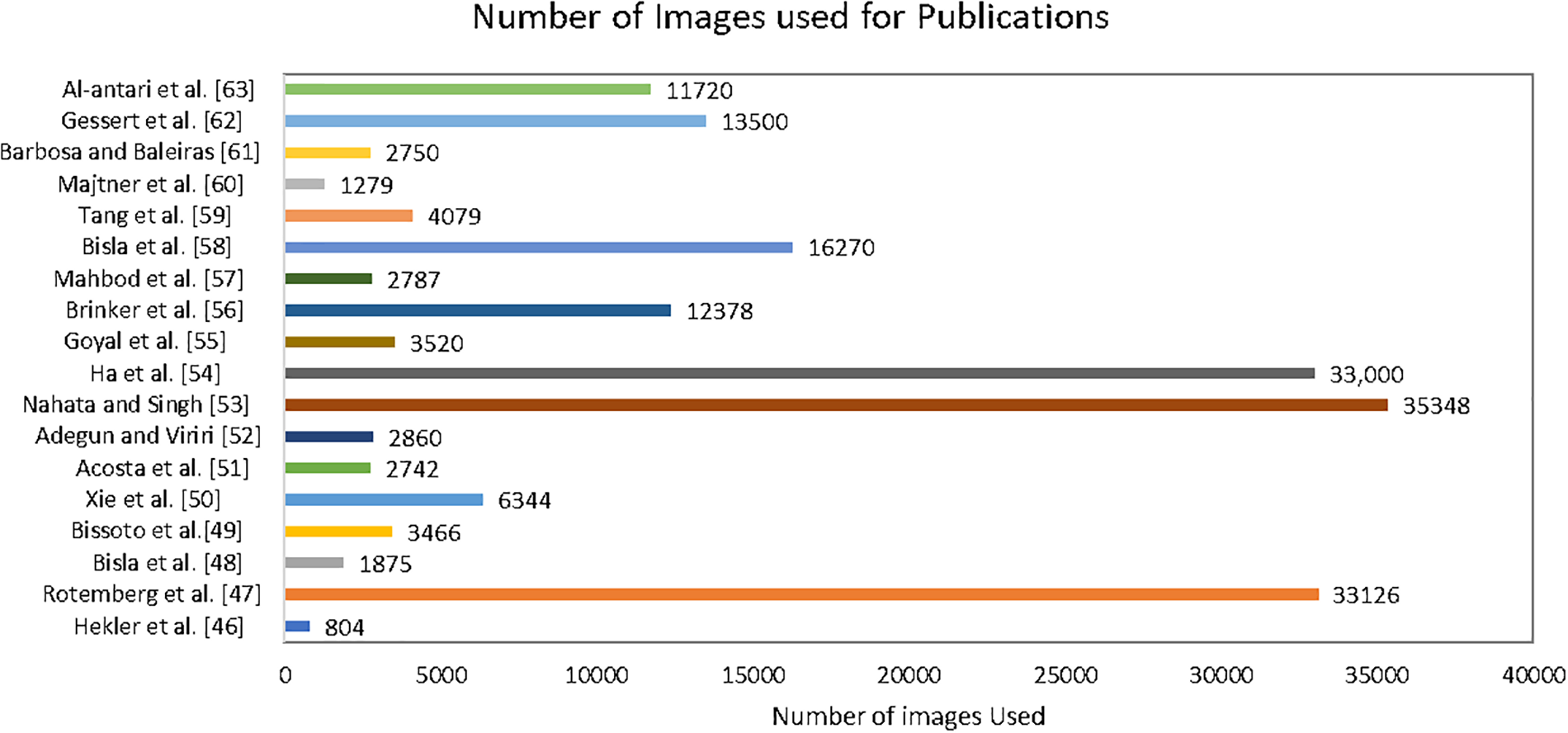
Figure 11: Number of images used in various publications
This holistic approach facilitates a more comprehensive understanding of skin cancer, enabling researchers to explore correlations between genetic markers, clinical parameters, and imaging features for a more nuanced diagnosis and prognosis [80]. As artificial intelligence applications in dermatology continue to evolve, there is a growing emphasis on the interpretability and explainability of models trained on the ISIC dataset. Transparent models can enhance trust among healthcare practitioners, fostering seamless integration of AI-driven tools into clinical workflows for more effective decision-making [81]. Moreover, the dynamic nature of the ISIC dataset not only fosters advancements in algorithmic precision but also encourages ongoing research into emerging skin cancer subtypes and their distinct diagnostic markers. This adaptability ensures that the dataset stays abreast of the ever-evolving landscape of dermatological knowledge, contributing to the refinement of diagnostic methodologies [82]. Additionally, the ISIC dataset’s open-access nature cultivates a global community of researchers, allowing for the seamless exchange of ideas, methodologies, and benchmarking standards. This collaborative ethos promotes a collective push towards more accurate, efficient, and universally applicable skin cancer diagnostic tools, ultimately benefiting patients worldwide [83]. Table 4 demonstrates the contribution of ISIC dataset in skin cancer types.
Through meticulous examination of various research papers, we have observed that researchers frequently utilize models such as AlexNet, VGG, ResNet, SqueezeMNet, among others, to achieve remarkable accuracy and F1-scores, ranging from 76.09% to 99.3%. Our analysis encompassed datasets spanning ISIC 2016, 2017, 2018, 2019, and 2020. However, it is imperative to highlight that our detailed review revealed a concerning trend associated with preprocessing techniques, particularly in data augmentation. We identified a recurrent issue of random data creation and duplication resulting from these techniques. This phenomenon has significantly contributed to mispredictions, overfitting, biases, and a lack of generalization in the outcomes. Such challenges underscore the necessity for a more careful consideration of preprocessing methods to ensure the integrity and reliability of the data and subsequent analysis in the field of skin cancer diagnosis.
3.5 CNNs for the SCC and SCS Using ISIC Dataset
CNNs play a crucial role in advancing the classification and segmentation of skin lesions, leveraging datasets like ISIC for improved diagnostic accuracy. CNNs directly learn from data, excelling in image recognition and analysis tasks related to skin cancer detection [101]. Recognized as one of the most proficient machine learning algorithms, CNNs demonstrate remarkable performance in various image processing tasks and computer vision applications, including skin lesion localization, segmentation, classification, and detection [102]. Typically comprising tens or hundreds of layers, each responsible for recognizing distinct aspects of skin lesion images, CNNs operate by applying convolutional filters during training. These filters start by detecting fundamental features such as color variations, edges, and textures, progressively becoming more sophisticated, and ultimately identifying specific lesion characteristics [103]. Hidden layers between the CNN’s input and output layers conduct operations that modify the data to learn features specific to the skin lesion dataset. The most commonly used layers include Convolution, activation (or ReLU), and pooling [104].
The Convolution layer is the fundamental building block, handling most of the computational workload. Through convolution, filters are applied to input skin lesion images, activating different lesion aspects. Mathematically, convolution can be expressed as:
where S(i, j) represents the output of the convolution operation at position (i, j) in the resulting image or feature map. And (I ∗ K)(i, j) This is the convolution operation between the input image I and the convolutional kernel K at the position (i, j) [105].
Activation functions, such as Rectified Linear Unit (ReLU), expedite training by introducing non-linearity and ensuring that only activated characteristics proceed to the subsequent layer. Mathematically, ReLU is defined as:
where pooling is another essential layer that reduces the network’s parameter count by performing nonlinear downsampling on the output [108]. Max pooling, for instance, can be expressed as:
These operations, including convolution, activation, and pooling, are repeated across multiple layers, where each layer learns to recognize diverse features of skin lesions. The final classification output is obtained from the top layer’s classification layer in the CNN architecture. Fig. 12 demonstrates the overall methodology of ISIC image classification and segmentation in detail.
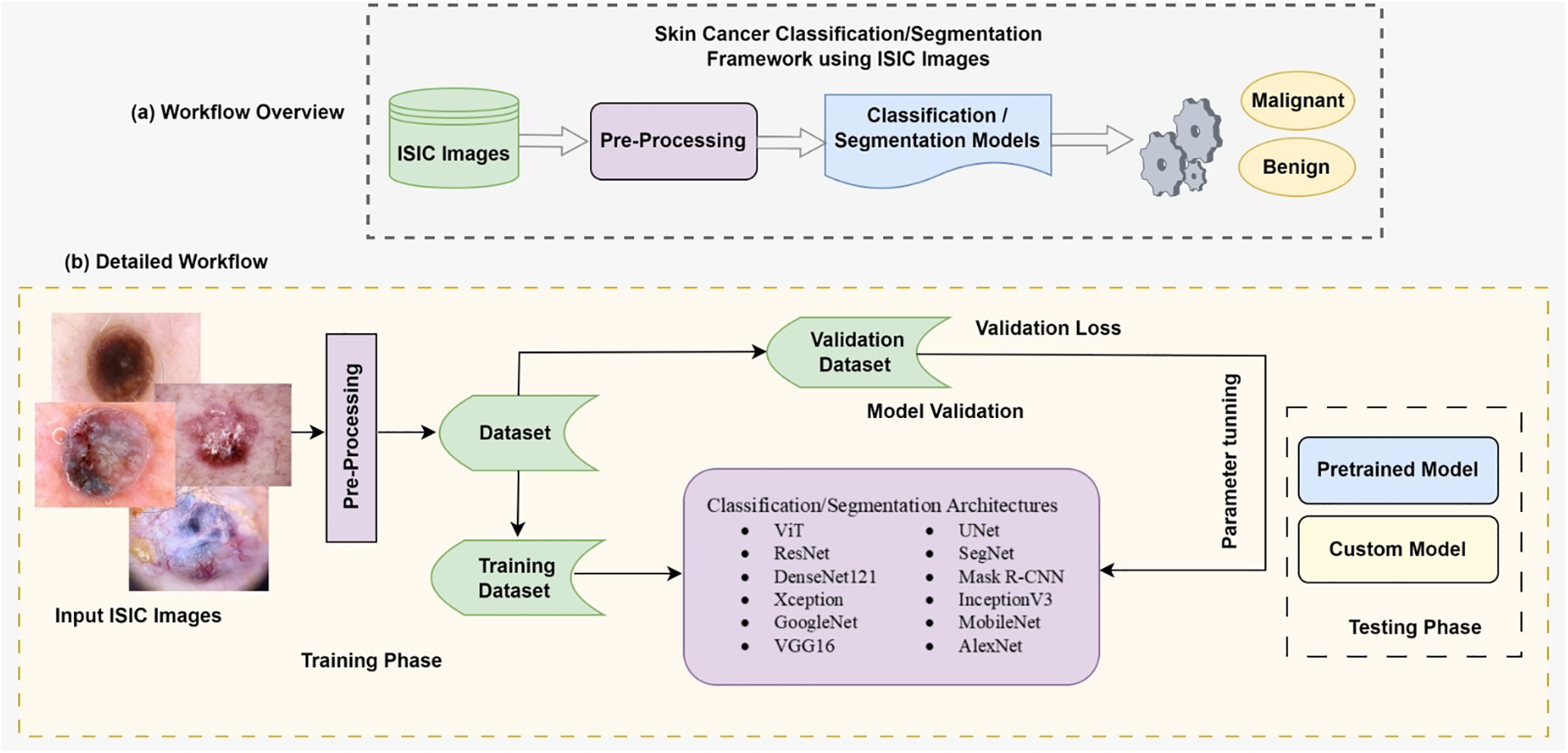
Figure 12: Overall methodology of ISIC images classification and segmentation
3.6 State-of-the-Art CNN Architectures for SCC and SCS Using ISIC Dataset
This section delves into the forefront of CNN architectures, demonstrating their mathematical and computational prowess in image classification and segmentation for skin cancer detection. These algorithms are utilized to enhance computational involvement for better results, specifically tailored to the ISIC dataset. Researchers have extensively explored these architectures to improve diagnostic accuracy. Key architectures employed in ISIC dataset studies include:
VGG is a convolutional neural network architecture developed by Karen Simonyan and Andrew Zisserman at Oxford University’s Visual Geometry Group [109]. The VGG-16 model, detailed in attained an impressive 92.7% top-5 test accuracy on the ImageNet dataset, which encompasses 14 million images across 1000 classes. Simonyan and Zisserman enhanced performance by substituting large-size kernel filters with multiple 3 × 3 kernel-sized filters, surpassing the capabilities of AlexNet [110]. VGG offers various configurations based on the number of convolution layers, with VGG-16 and VGG-19 being the most prevalent. The architecture of VGG-16, illustrated in Fig. 13, consists of thirteen convolution layers, a max-pooling layer, three fully connected layers, and an output layer [111].
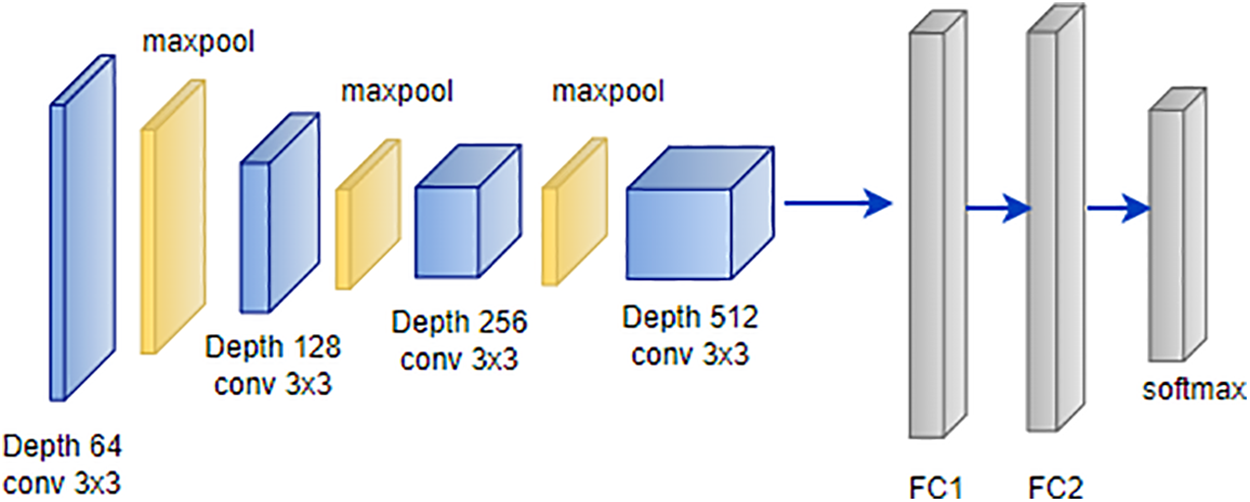
Figure 13: Architecture of VGG
This model’s architecture has demonstrated its efficacy in diverse image classification tasks, making it a valuable asset for researchers in the field [112]. The versatility of the VGG architecture extends to its application in skin cancer detection using the ISIC dataset. Researchers have harnessed the power of VGG-16 and VGG-19 configurations to address the intricacies of skin lesion classification and segmentation [113]. The rich diversity of skin images within the ISIC dataset aligns seamlessly with VGG’s ability to capture complex features. By adapting the pre-trained VGG models on ImageNet to the specific characteristics of skin cancer images from the ISIC dataset, studies have achieved remarkable results [114]. Fine-tuning VGG architectures with transfer learning techniques allows the models to grasp subtle patterns indicative of various skin conditions. The robust performance demonstrated by VGG in broader image classification tasks translates effectively to the nuanced domain of skin cancer detection within the ISIC dataset [115].
Eq. (4), inherent to VGG’s convolutional layers, encapsulates the mathematical foundation of its adaptability and effectiveness in processing skin images from the ISIC dataset. This synergy between VGG architecture and the ISIC dataset underscores the significance of leveraging state-of-the-art models for advancing the accuracy and reliability of skin cancer detection systems.
AlexNet, introduced by Hosny et al. [116], is a pioneering CNN architecture renowned for revolutionizing image classification tasks. Comprising five convolutional layers and three fully connected layers, as depicted in Fig. 6, AlexNet played a pivotal role in winning the ImageNet LSVRC in 2012 [117]. With a total of 60 million parameters, AlexNet effectively addressed overfitting concerns through the innovative integration of dropout layers, enhancing the model’s generalization capabilities:
where P(dropout) represents the probability of dropout, this equation signifies that during training, each neuron has a 50% chance of being dropped out, preventing reliance on specific neurons and promoting better generalization [118]. Beyond its success on ImageNet, AlexNet’s architecture has become a foundational blueprint for subsequent deep learning models, contributing significantly to the advancement of computer vision applications [119]. Trained on ImageNet LSVRC-2010, AlexNet demonstrated remarkable performance with top-1 and top-5 error rates of 37.5% and 17.0%, respectively, solidifying its position as a landmark in the evolution of deep learning architectures. The softmax activation function used in the final layer is defined as:
where
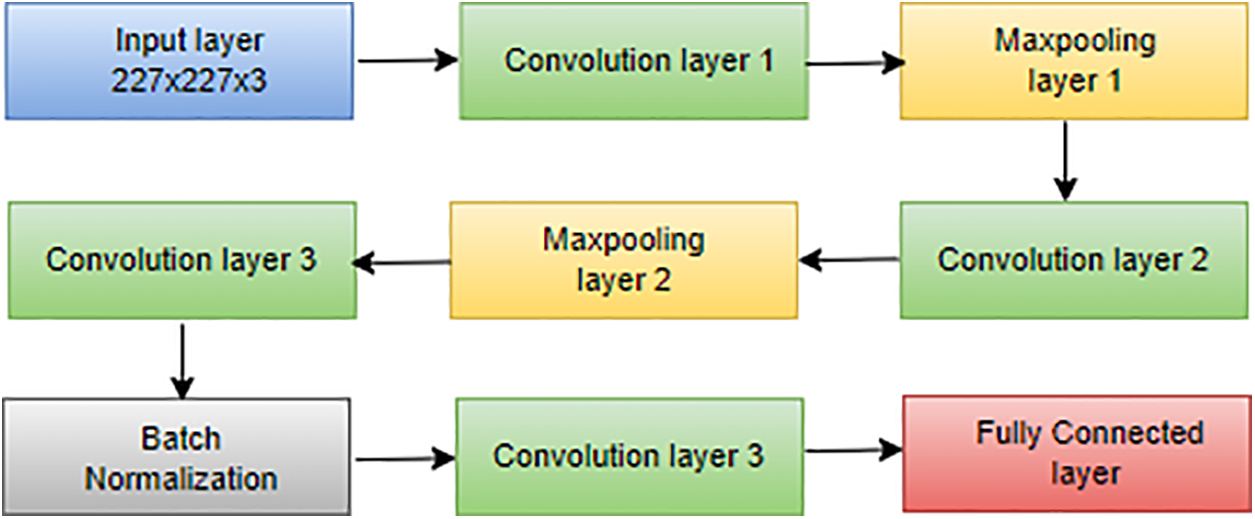
Figure 14: Architecture of AlexNet
AlexNet secured victory in the ImageNet 2012 competition, featuring an original eight-layer architecture. In the landscape of deep learning, the common strategy of adding more layers aims to boost performance and reduce error rates [121]. However, this approach introduces challenges like vanishing gradients, where the gradient approaches zero, and exploding gradients, where the gradient becomes excessively large. Addressing these challenges, He et al. [122] introduced skip connections, an innovative concept to alleviate problems associated with exploding and vanishing gradients. Skip connections involve bypassing certain levels between layers and directly linking layer activations to subsequent layers, forming what is known as residual blocks [123]. The ResNet architecture stands as a powerful tool for researchers aiming to construct and train deep neural networks effectively. The equations for ResNet skip connections can be expressed as follows, where
This novel approach serves as the foundation of the ResNet architecture, depicted in Fig. 15. ResNet’s skip connections enable the bypassing of problematic layers during training, effectively mitigating issues related to exploding and vanishing gradients [125]. This architectural innovation has proven instrumental in facilitating the training of deep neural networks, providing a practical solution to challenges encountered with increasing network depth [126].
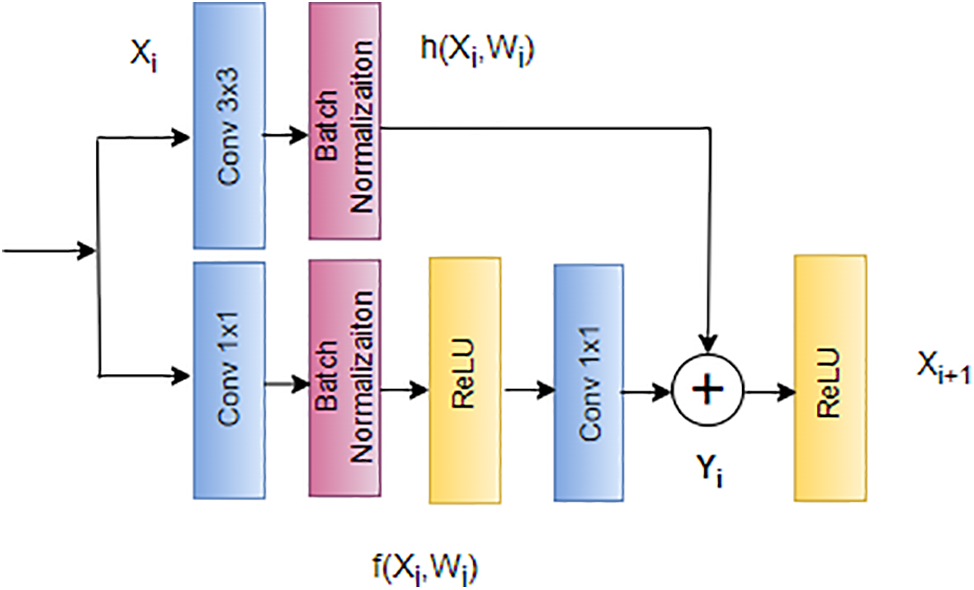
Figure 15: Architecture of ResNet
Howard et al. [127] introduced a lightweight network called MobileNet specifically designed for mobile applications. In MobileNet, the traditional 3 × 3 convolution operation found in standard CNNs is replaced with a combination of a 3 × 3 depthwise convolution and a 1 × 1 pointwise convolution operation. This strategic use of depthwise separable convolution, as opposed to the standard convolution operation, proves effective in reducing the overall number of training parameters [128]. The distinction between the standard convolution operation and the depthwise separable convolution employed in MobileNet is illustrated in Fig. 16.
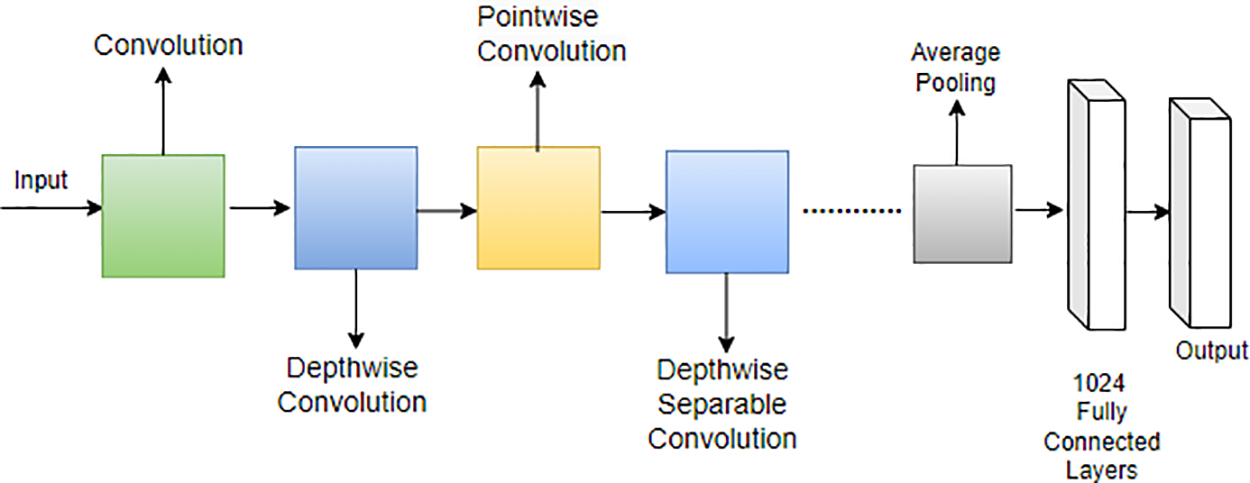
Figure 16: Architecture of MobileNet
This modification in the convolutional operation contributes to the efficiency of MobileNet, making it well-suited for deployment on mobile devices by significantly reducing computational demands and model size [129]. MobileNet represents a noteworthy advancement in tailoring neural network architectures for optimal performance on resource-constrained platforms [130]. Moreover, MobileNet’s architecture is characterized by the following equations for depthwise convolution and pointwise convolution:
Depthwise convolution:
Pointwise convolution:
In these equations, x represents the input, w denotes the depthwise convolutional kernel, y is the intermediate output after depthwise convolution, z is the final output after pointwise convolution, and b represents the bias term [131]. The depthwise convolution is followed by batch normalization and ReLU activation, contributing to the overall efficiency of MobileNet [132].
In the groundbreaking work by Huang et al., the DenseNet architecture, or densely connected convolutional network, was introduced, and its application to skin cancer classification and segmentation, specifically on the ISIC dataset, has demonstrated notable success [133,134]. Illustrated in Fig. 17, DenseNet’s unique connectivity pattern, where each layer is directly connected to every other layer, proves advantageous in capturing intricate features inherent in skin lesion images. For skin cancer classification, DenseNet leverages the comprehensive information flow between layers, utilizing feature maps from all preceding layers as inputs for each subsequent layer [135,99]. This connectivity scheme mitigates the vanishing gradient problem and enhances the model’s ability to discern nuanced patterns indicative of different skin conditions within the ISIC dataset [136,137]. In the domain of segmentation, DenseNet’s connectivity pattern contributes to robust feature propagation throughout the network, aiding in the precise delineation of lesion boundaries. The efficiency gains from reduced parameters make DenseNet well-suited for handling the complexities of skin cancer images in the ISIC dataset [138,139]. The connectivity equation for DenseNet, tailored to the ISIC dataset, is expressed as follows, where
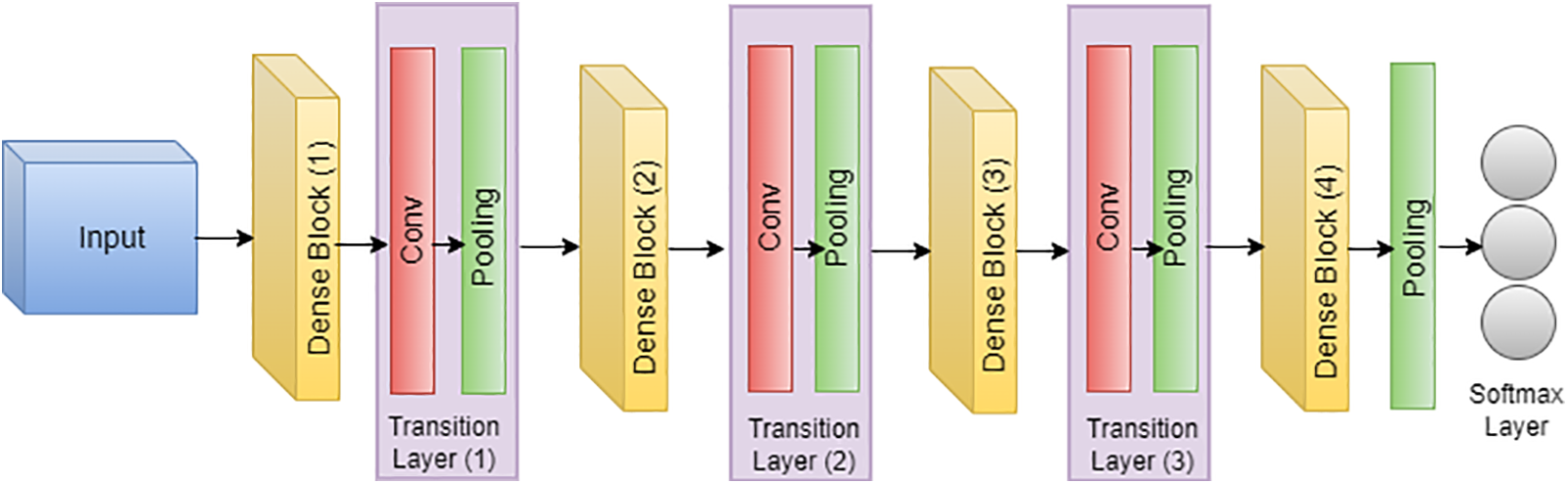
Figure 17: Architecture of DensNet
This adaptation underscores the efficacy of DenseNet in processing ISIC dataset images, emphasizing its role in advancing both skin cancer classification and segmentation tasks [140].
U-Net’s popularity in medical image segmentation, particularly for skin cancer segmentation on the ISIC dataset, can be attributed to its robust architecture [141,142]. The encoder-decoder structure, complemented by skip connections, facilitates the accurate localization of skin lesions. The model’s effectiveness lies in its ability to maintain a seamless flow of information between the encoder and decoder, enabling precise delineation of lesions in the challenging scenarios posed by the ISIC dataset [143,144]. The incorporation of skip connections proves pivotal in retaining fine-grained details during upsampling, a crucial factor contributing to U-Net’s remarkable success in skin cancer segmentation [145,146]. The following equation can succinctly represent the architecture of U-Net:
where
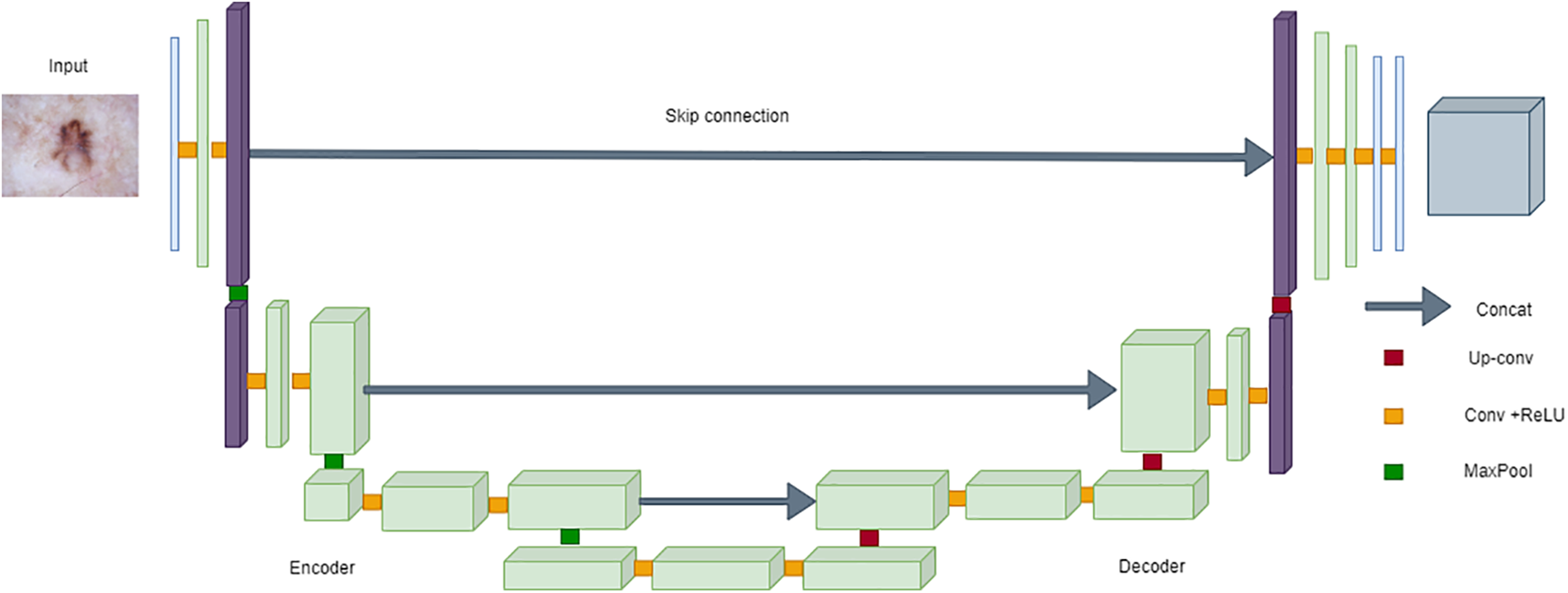
Figure 18: Architecture of UNet
SegNet is a convolutional neural network architecture tailored for pixel-wise image segmentation tasks, making it suitable for applications like skin lesion segmentation in the ISIC dataset [149,150]. Its design includes an encoder-decoder structure with symmetrically paired encoder and decoder layers. SegNet utilizes max-pooling indices from the encoder during the upsampling process, aiding in preserving spatial information crucial for accurate segmentation [151,152]. The model’s architecture enhances the localization of intricate features within skin lesions, contributing to its effectiveness in the challenging context of the ISIC dataset. SegNet’s performance is often attributed to its focus on retaining fine-grained details, demonstrating its utility in medical image segmentation, particularly for dermatological applications [153,154].
This equation represents the forward pass of SegNet, where X is the input image,
In the realm of SCC and SCS using the ISIC dataset, various models, including AlexNet, VGG, ResNet, and SqueezeMNet, have been extensively employed by researchers, yielding commendable accuracy and F1-scores ranging from 76.09% to 99.3%. Notably, in the domain of SCS, particularly utilizing the ISIC dataset across multiple iterations, significant advancements have been made with the adoption of specialized models such as InSiNet and U-Net. In recent investigations spanning ISIC 2018, ISIC 2019, and ISIC 2020 datasets, these models have demonstrated notable efficacy, achieving commendable accuracy rates. Specifically, across the respective datasets, segmentation accuracies of 94.59%, 91.89%, and 90.54% have been attained. These results underscore the continual refinement and adaptation of deep learning architectures to address the challenges inherent in SCS. Such accuracies not only reflect the robustness of the models but also signify their potential utility in clinical settings for aiding dermatologists in accurate diagnosis and treatment planning. Each model presents distinct advantages and disadvantages in the context of ISIC data utilization. AlexNet, renowned for its pioneering role in deep learning, offers a relatively straightforward architecture and efficient training, making it suitable for initial experimentation. However, its shallower depth may limit its ability to capture intricate features in complex skin lesion images. VGG, characterized by its deeper architecture, excels in feature extraction but suffers from increased computational complexity and memory requirements, potentially limiting its scalability. ResNet, with its residual connections, mitigates the vanishing gradient problem and facilitates the training of deeper networks, thereby enhancing feature representation. Nonetheless, its extensive depth may lead to overfitting, particularly with smaller datasets. SqueezeMNet, notable for its lightweight architecture, enables efficient inference on resource-constrained devices, but may sacrifice some accuracy compared to larger models. InSiNet incorporates attention mechanisms, allowing for dynamic focus on salient regions within the ISIC input image. This adaptability can enhance segmentation accuracy, particularly in cases with complex or heterogeneous cancer. U-Net, renowned for its ability to effectively capture fine-grained details in ISIC image segmentation tasks, features a symmetric architecture with contracting and expansive pathways. This design facilitates precise delineation of lesion boundaries, leading to accurate segmentation results. Despite their strengths, all these models are susceptible to the aforementioned issue of data duplication resulting from preprocessing techniques. This recurring challenge has posed significant obstacles, including mispredictions, overfitting, biases, and a lack of generalization, underscoring the critical need for meticulous consideration of preprocessing methods to uphold the integrity and reliability of both the data and subsequent analyses in skin cancer diagnosis.
After conducting an exhaustive review of existing literature, it has been discerned that the incorporation of preprocessing techniques to expand ISIC datasets presents a myriad of challenges, thereby exerting a discernible impact on the reliability and accuracy of the outcomes:
1. Random duplication in images: Preprocessing methodologies often engender the random duplication of images within the ISIC dataset. This inadvertent replication can lead to the recurrence of identical or highly similar images multiple times, thus precipitating erroneous predictions during both the training and evaluation phases of model development. Additionally, such random duplication exacerbates the problem of imbalanced data, where certain classes may be overrepresented while others are underrepresented, further complicating model training and evaluation.
2. Overfitting due to replicated data: The replication of data facilitated by preprocessing techniques may inadvertently induce overfitting of the model on the ISIC dataset. Overfitting, a phenomenon characterized by the model’s tendency to memorize the intricacies of the training data rather than generalize from it, can yield ostensibly superior performance metrics on the training dataset. However, such models often exhibit suboptimal generalization capabilities on unseen data, thereby impugning the reliability and robustness of the resultant outcomes.
3. Reduced diversity and generalizability: The process of replicating data through preprocessing interventions has the potential to attenuate the diversity inherent within the ISIC dataset. This reduction in diversity, precipitated by the introduction of identical or highly similar images, can impede the model’s capacity to generalize effectively to novel, unseen data instances. Consequently, this limitation culminates in a concomitant reduction in overall performance and reliability.
4. Inefficient resource utilization: The augmentation of ISIC datasets via preprocessing methodologies significantly amplifies the dataset’s size, thereby engendering inefficiencies in terms of memory and computational resource utilization during model training. The resultant elongated training times and augmented computational costs detrimentally affect the scalability and practical applicability of the model.
5. Introduction of biases: Preprocessing techniques may inadvertently introduce biases into the ISIC dataset, particularly in instances where the augmentation strategies are inadequately designed or implemented. The inadvertent introduction of biases can skew the model’s learning trajectory, precipitating biased predictions and ultimately compromising the fairness and reliability of the resultant outcomes.
Addressing these multifarious challenges necessitates a comprehensive understanding of the potential pitfalls associated with preprocessing techniques, as well as the judicious implementation of rigorous validation and quality control measures to safeguard the integrity and reliability of both the ISIC dataset and the ensuing model. Moreover, strategies to mitigate imbalanced data problems, such as class weighting or data resampling techniques, are imperative to ensure equitable representation of all classes and enhance the model’s performance and generalizability.
In future directions, addressing the challenge of random duplication in ISIC datasets resulting from the use of distinct augmentation techniques is paramount. Such duplications can lead to misleading outcomes and adversely affect real-time analysis, potentially increasing the incidence of true negative predictions and thereby promoting erroneous conclusions. To tackle this issue effectively, the integration of ViTs with their encoder-decoder architectures and attention mechanisms emerges as a promising approach. ViTs are renowned for their adeptness in feature extraction, while the ViT encoder-decoder framework offers the additional capability of generating diverse and realistic data samples. This integration presents a holistic solution for mitigating the impact of duplicated data on analysis outcomes. Moreover, by combining these techniques with ResNet, a well-established algorithm for feature extraction, there is an opportunity to bolster the detection of false positives in diagnosis as SCC and SCS. In the envisioned architecture, the encoder-decoder aspect of the ViT framework plays a crucial role. The encoder processes input images and extracts relevant features, while the decoder generates output predictions based on these features, thereby enabling both feature extraction and generation of diverse data samples. By amalgamating these methodologies, a more robust framework for ISIC image analysis can be established, leading to enhanced diagnostic accuracy and reliability in clinical settings.
In conclusion, this paper has highlighted the multifaceted challenges encountered in utilizing preprocessing techniques to expand ISIC datasets for skin cancer classification and segmentation. These challenges include random duplication in images, overfitting due to replicated data, reduced diversity and generalizability, inefficient resource utilization, and the introduction of biases. Collectively, these factors impede the reliability and accuracy of outcomes derived from machine learning models trained on such datasets. Addressing these challenges requires a comprehensive understanding of the pitfalls associated with preprocessing techniques, as well as the implementation of rigorous validation and quality control measures. Strategies to mitigate imbalanced data problems, such as class weighting or data resampling techniques, are essential to ensure equitable representation of all classes and enhance model performance and generalizability. Looking ahead, future directions outlined in this paper propose innovative approaches to overcome the challenge of random duplication in ISIC datasets. The integration of ViTs with its robust architecture offers an opportunity to enhance the detection of false positives in diagnosis, contributing to improved diagnostic accuracy and reliability in image datasets. This systematic review paper underscores the importance of addressing the challenges associated with preprocessing techniques in ISIC dataset utilization for skin cancer analysis. By delineating future directions and proposing innovative solutions, this paper contributes to advancing the field of skin cancer diagnostics, ultimately benefiting patient care and global healthcare initiatives. This study elucidates the pivotal role of mathematical and computational modeling in enhancing skin cancer detection methodologies, paving the way for more accurate and reliable diagnostic tools in dermatological image analysis research.
Acknowledgement: The authors gratefully acknowledge Pakistan Institute of Engineering and Applied Sciences for providing computational resources, including the NVIDIA GeForce GTX 1080 Ti 11 GB GPU, for the experimental work. This high-performance GPU played a crucial role in conducting comprehensive analyses and experiments, contributing significantly to the outcomes and findings presented in this research. The access to the NVIDIA GeForce GTX 1080 Ti GPU from the institute has been invaluable in advancing our understanding and enhancing the quality of the research outcomes.
Funding Statement: This research was conducted without external funding, and the authors acknowledge that no financial support was received for the development and publication of this paper.
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: A. Zameer, M. Hameed; data collection: M. Hameed; analysis and interpretation of results: A. Zameer, M. Hameed, M. Asif; draft manuscript preparation: M. Hameed, M. Asif. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data will be made available by the corresponding author upon reasonable request.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Ju A, Tang J, Chen S, Fu Y, Luo Y. Pyroptosis-related gene signatures can robustly diagnose skin cutaneous melanoma and predict the prognosis. Front Oncol. 2021;11:7. [Google Scholar]
2. WSRP Cancer Prevention Organisation, World Cancer Research Fund International, WCRF International. Available from: https://www.wcrf.org/. [Accessed 2023]. [Google Scholar]
3. Fontanillas P, Alipanahi B, Furlotte NA, Johnson M, Wilson CH, Pitts SJ, et al. Disease risk scores for skin cancers. Nat Commun. 2021;12:160. [Google Scholar] [PubMed]
4. Rosendahl C, Tschandl P, Cameron A, Kittler H. Diagnostic accuracy of dermatoscopy for melanocytic and nonmelanocytic pigmented lesions. J Am Acad Dermatol. 2011;64:1068–73. doi:10.1016/j.jaad.2010.03.039. [Google Scholar] [PubMed] [CrossRef]
5. Narasu J, Oza P, Hacihaliloglu I, Patel V. Medical transformer: gated axial attention for medical image segmentation. In: Medical image computing and computer assisted intervention–MICCAI 2021. Springer, Cham; 2021. vol. 12901. doi:10.1007/978-3-030-87193-2_4. [Google Scholar] [CrossRef]
6. Ramachandran S, Strisciuglio N, Vinekar A, John R, Azzopardi G. U-cosfire filters for vessel tortuosity quantification with application to automated diagnosis of retinopathy of prematurity. Neur Comput Appl. 2020;32:12453–68. doi:10.1007/s00521-019-04697-6. [Google Scholar] [CrossRef]
7. Xu F, Zhao H, Hu F, Shen M, Wu Y. A road segmentation model based on mixture of the convolutional neural network and the transformer network. Comput Model Eng Sci. 2023;135(2):1559–70. doi:10.32604/cmes.2022.023217. [Google Scholar] [CrossRef]
8. Cassidy B, Kendrick C, Brodzicki A, Jaworek-Korjakowska J, Yap M. Analysis of the ISIC image datasets: usage, benchmarks and recommendations. Med Image Anal. 2022;75:102305. doi:10.1016/j.media.2021.102305. [Google Scholar] [PubMed] [CrossRef]
9. Hardie R, Ali R, de Silva M, Kebede T. Skin lesion segmentation and classification for ISIC 2018 using traditional classifiers with hand-crafted features. arXiv Preprint arXiv:1807.07001. 2018. [Google Scholar]
10. Mahbod A, Schaefer G, Wang C, Ecker R, Ellinge I. Skin lesion classification using hybrid deep neural networks. In: ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2019; Brighton, UK. p. 1229–33. [Google Scholar]
11. Qureshi M, Khan S, Sharafat S, Quraishy M. Common cancers in Karachi, Pakistan: 2010–2019 cancer data from the dow cancer registry. Pak J Med Sci. 2020;36:1572–8. [Google Scholar] [PubMed]
12. Kaur R, GholamHosseini H, Sinha R, Lindén M. Automatic lesion segmentation using atrous convolutional deep neural networks in dermoscopic skin cancer images. BMC Med Imaging. 2022;22(1):103. doi:10.1186/s12880-022-00829-y. [Google Scholar] [PubMed] [CrossRef]
13. Gordon R. Skin cancer: an overview of epidemiology and risk factors. Semin Oncol Nurs. 2013;29:160. doi:10.1016/j.soncn.2013.06.002. [Google Scholar] [PubMed] [CrossRef]
14. Ahmed B, Qadir M, Melanoma S. Malignant melanoma: skin cancer-diagnosis, prevention, and treatment. Crit Rev in Eukaryotic Gene Expr. 2020;30(4). [Google Scholar]
15. Ashraf H, Waris A, Ghafoor M, Gilani S, Niazi I. Melanoma segmentation using deep learning with test-time augmentations and conditional random fields. Sci Rep. 2022;12:3948. doi:10.1038/s41598-022-07885-y. [Google Scholar] [PubMed] [CrossRef]
16. Nasiri S, Helsper J, Jung M, Fathi M. DePicT melanoma deep-CLASS: a deep convolutional neural networks approach to classify skin lesion images. BMC Bioinform. 2020;21:1–13. [Google Scholar]
17. Huang R, Liang J, Jiang F, Zhou F, Cheng N, Wang T, et al. MelanomaNet: an effective network for melanoma detection. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2019; Berlin, Germany. p. 1613–6. [Google Scholar]
18. Diab A, Fayez N, El-Seddek M. Accurate skin cancer diagnosis based on convolutional neural networks. Indones J Electr Eng Comput Sci. 2022;25:1429–41. [Google Scholar]
19. Saba T. Computer vision for microscopic skin cancer diagnosis using handcrafted and non-handcrafted features. Microsc Res Tech. 2021;84:1272–83. doi:10.1002/jemt.v84.6. [Google Scholar] [PubMed] [CrossRef]
20. Sung W, Chang C. Nevi, Dysplastic nevi, and melanoma: molecular and immune mechanisms involving the progression. Tzu-Chi Med J. 2022;34(1):1–7. doi:10.4103/tcmj.tcmj_158_20. [Google Scholar] [PubMed] [CrossRef]
21. Alsayyah A. Differentiating between early melanomas and melanocytic nevi: a state-of-the-art review. Pathol-Res Pract. 2023;249:154734. doi:10.1016/j.prp.2023.154734. [Google Scholar] [PubMed] [CrossRef]
22. Oh C, Jin A, Koh W. Trends of cutaneous basal cell carcinoma, squamous cell carcinoma, and melanoma among the Chinese, Malays, and Indians in Singapore from 1968–2016. JAAD Int. 2021;4:39–45. doi:10.1016/j.jdin.2021.05.006. [Google Scholar] [PubMed] [CrossRef]
23. van der Putten J, van der Sommen F. Efficient decoder reduction for a variety of encoder-decoder problems. IEEE Access. 2020;8:169444–55. doi:10.1109/Access.6287639. [Google Scholar] [CrossRef]
24. Zhang S, Huang S, Wu H, Yang Z, Chen Y. Intelligent data analytics for diagnosing melanoma skin lesions via deep learning in IoT system. Mob Inf Syst. 2021;2021:1–12. [Google Scholar]
25. Sprenger, Jeannine, J. Glycodendrimer mediation of Galectin-3 induced cancer cell aggregation. Montana State University-Bozeman; 2010. [Google Scholar]
26. Duncan J, Purnell J, Stratton M, Pavlidakey P, Huang C, Phillips C. Negative predictive value of biopsy margins of dysplastic nevi: a single-institution retrospective review. J Am Acad Dermatol. 2020;82:87–93. doi:10.1016/j.jaad.2019.07.037. [Google Scholar] [PubMed] [CrossRef]
27. Xu Y, Aylward J, Swanson A, Spiegelman V, Vanness E, Teng J, et al. Nonmelanoma skin cancers: basal cell and squamous cell carcinomas. Abeloff’s Clinic Oncol. 2020:1052–73. doi:10.1016/B978-0-323-47674-4.00067-0 [Google Scholar] [CrossRef]
28. Ciazynska M, Kaminska-Winciorek G, Lange D, Lewandowski B, Reich A, Slawinska M, et al. The incidence and clinical analysis of non-melanoma skin cancer. Sci Rep. 2021;11:4337. doi:10.1038/s41598-021-83502-8. [Google Scholar] [PubMed] [CrossRef]
29. Maurya A, Stanley R, Lama N, Nambisan A, Patel G, Saeed D, et al. Hybrid topological data analysis and deep learning for basal cell carcinoma diagnosis. J Imaging Inf Med. 2024, 1–15. [Google Scholar]
30. Datta S, Shaikh M, Srihari S, Gao M. Soft attention improves skin cancer classification performance; 2021, 13–23. [Google Scholar]
31. Fisher R, Rees J, Bertrand A. Classification of ten skin lesion classes: hierarchical KNN versus deep net. In: Medical image understanding and analysis. Springer, Cham; 2020. p. 86–98. doi:10.1007/978-3-030-39343-4_8. [Google Scholar] [CrossRef]
32. Gouda N, Amudha J. Skin cancer classification using ResNet. In: 2020 IEEE 5th International Conference on Computing Communication and Automation (ICCCA); 2020; Greater Noida, India. p. 536–41. doi:10.1109/ICCCA49541.2020.9250855 [Google Scholar] [CrossRef]
33. Stratigos A, Garbe C, Dessinioti C, Lebbe C, Bataille V, Bastholt L, et al. European interdisciplinary guideline on invasive squamous cell carcinoma of the skin: part 1. Epidemiology, diagnostics and prevention. Eur J Cancer. 2020;128:60–82. doi:10.1016/j.ejca.2020.01.007. [Google Scholar] [PubMed] [CrossRef]
34. Leiter U, Keim U, Garbe C. Epidemiology of skin cancer: update 2019. In: Sunlight, vitamin D and skin cancer; Springer, Cham; 2020. p. 123–39. doi:10.1007/978-3-030-46227-7_6. [Google Scholar] [CrossRef]
35. Yin W, Huang J, Chen J, Ji Y. A study on skin tumor classification based on dense convolutional networks with fused metadata. Front Oncol. 2022;12:989894.doi:10.3389/fonc.2022.989894. [Google Scholar] [PubMed] [CrossRef]
36. Huang H, Hsiao Y, Mukundan A, Tsao Y, Chang W, Wang H. Classification of skin cancer using novel hyperspectral imaging engineering via YOLOv5. J Clin Med. 2023;12:1134. doi:10.3390/jcm12031134. [Google Scholar] [PubMed] [CrossRef]
37. Olayah F, Senan E, Ahmed I, Awaji B. AI techniques of dermoscopy image analysis for the early detection of skin lesions based on combined CNN features. Diagnostics. 2023;13:1314. doi:10.3390/diagnostics13071314. [Google Scholar] [PubMed] [CrossRef]
38. Dildar M, Akram S, Irfan M, Khan H, Ramzan M, Mahmood A, et al. Skin cancer detection: a review using deep learning techniques. Int J Environ Res Public Health. 2021;18:5479. doi:10.3390/ijerph18105479. [Google Scholar] [PubMed] [CrossRef]
39. Guorgis G, Anderson C, Johan L, Magnus F. Actinic keratosis diagnosis and increased risk of developing skin cancer: a 10-year cohort study of 17,651 patients in Sweden. Acta Derm Venereol. 2020;100. [Google Scholar]
40. Gilchrest B. Actinic keratoses: reconciling the biology of field cancerization with treatment paradigms. J Investig Dermatol. 2021;141:727–31. doi:10.1016/j.jid.2020.09.002. [Google Scholar] [PubMed] [CrossRef]
41. Thomson J, Bewicke-Copley F, Anene C, Gulati A, Nagano A, Purdie K, et al. The genomic landscape of actinic keratosis. J Investig Dermatol. 2021;141:1664–74. doi:10.1016/j.jid.2020.12.024. [Google Scholar] [PubMed] [CrossRef]
42. Vimercati L, de Maria L, Caputi A, Cannone E, Mansi F, Cavone D, et al. Non-melanoma skin cancer in outdoor workers: a study on actinic keratosis in Italian navy personnel. Int J Environ Res Public Health. 2020;17:2321. doi:10.3390/ijerph17072321. [Google Scholar] [PubMed] [CrossRef]
43. Jones O, Ranmuthu C, Hall P, Funston G, Walter F. Recognising skin cancer in primary care. Adv Ther. 2020;37:603–16. doi:10.1007/s12325-019-01130-1. [Google Scholar] [PubMed] [CrossRef]
44. Al-Dmour N, Salahat M, Nair H, Kanwal N, Saleem M, Aziz N. Intelligence skin cancer detection using IoT with a fuzzy expert system. In: 2022 International Conference on Cyber Resilience (ICCR); 2022; Dubai, United Arab Emirates. p. 1–6. [Google Scholar]
45. Pfeifer G. Mechanisms of UV-induced mutations and skin cancer. Genome Instability Dis. 2020;1:99–113. doi:10.1007/s42764-020-00009-8. [Google Scholar] [CrossRef]
46. Sreedhar A, Aguilera-Aguirre L, Singh K. Mitochondria in skin health, aging, and disease. Cell Death Dis. 2020;11:444. doi:10.1038/s41419-020-2649-z. [Google Scholar] [PubMed] [CrossRef]
47. Hekler A, Kather J, Krieghoff-Henning E, Utikal J, Meier F, Gellrich F, et al. Effects of label noise on deep learning-based skin cancer classification. Front Med. 2020;7:177. doi:10.3389/fmed.2020.00177. [Google Scholar] [CrossRef]
48. Rotemberg V, Kurtansky N, Betz-Stablein B, Caffery L, Chousakos E, Codella N, et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci Data. 2021;8:34. doi:10.1038/s41597-021-00815-z. [Google Scholar] [CrossRef]
49. Bisla D, Choromanska A, Berman R, Stein J, Polsky D. Towards automated melanoma detection with deep learning: data purification and augmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops; 2019. p. 2720–8. [Google Scholar]
50. Bissoto A, Fornaciali M, Valle E, Avila S. (De) constructing bias on skin lesion datasets. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops; 2019. [Google Scholar]
51. Xie Y, Zhang J, Lu H, Shen C, Xia Y. SESV: accurate medical image segmentation by predicting and correcting errors. IEEE Trans Med Imaging. 2021;40:286–96. doi:10.1109/TMI.42. [Google Scholar] [PubMed] [CrossRef]
52. Jojoa Acosta M, Caballero Tovar L, Garcia-Zapirain M, Percybrooks W. Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Med Imaging. 2021;21:1–11. [Google Scholar]
53. Adegun A, Viriri S. Deep learning-based system for automatic melanoma detection. IEEE Access. 2020;8:7160–72. doi:10.1109/Access.6287639. [Google Scholar] [CrossRef]
54. Nahata H, Singh S. Deep learning solutions for skin cancer detection and diagnosis. In: Machine learning with health care perspective. Springer, Cham; 2020. doi:10.1007/978-3-030-40850-3_8. [Google Scholar] [CrossRef]
55. Ha Q, Liu B, Liu F. Identifying melanoma images using efficientnet ensemble: winning solution to the siim-isic melanoma classification challenge. arXiv preprint arXiv:2010.05351. 2020. [Google Scholar]
56. Goyal M, Oakley A, Bansal P, Dancey D, Yap M. Skin lesion segmentation in dermoscopic images with ensemble deep learning methods. IEEE Access. 2019;8:4171–81. [Google Scholar]
57. Brinker T, Hekler A, Enk A, Klode J, Hauschild A, Berking C, et al. A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur J Cancer. 2019;111:148–54. doi:10.1016/j.ejca.2019.02.005. [Google Scholar] [PubMed] [CrossRef]
58. Mahbod A, Schaefer G, Ellinger I, Ecker R, Pitiot A, Wang C. Fusing fine-tuned deep features for skin lesion classification. Comput Med Imaging Graph. 2019;71:19–29. doi:10.1016/j.compmedimag.2018.10.007. [Google Scholar] [PubMed] [CrossRef]
59. Bisla D, Choromanska A, Stein J, Polsky D, Berman R. Skin lesion segmentation and classification with deep learning system. arXiv Preprint arXiv:190206061. 2019. [Google Scholar]
60. Tang P, Liang Q, Yan X, Xiang S, Sun W, Zhang D, et al. Efficient skin lesion segmentation using separable-Unet with stochastic weight averaging. Comput Methods Programs Biomed. 2019;178:289–301. doi:10.1016/j.cmpb.2019.07.005. [Google Scholar] [PubMed] [CrossRef]
61. Majtner T, Yildirim Yayilgan S, Hardeberg J. Optimised deep learning features for improved melanoma detection. Multimed Tools Appl. 2019;78:11883–903. doi:10.1007/s11042-018-6734-6. [Google Scholar] [CrossRef]
62. Li Y, Shen L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors. 2018;18. [Google Scholar]
63. Gessert N, Nielsen M, Shaikh M, Werner R, Schlaefer A. Skin lesion classification using ensembles of multi-resolution efficientnets with meta data. MethodsX. 2020;7. doi:10.1016/j.mex.2020.100864. [Google Scholar] [PubMed] [CrossRef]
64. Al-Antari M, Rivera P, Al-Masni M, Valarezo E, Gi G, Kim T, et al. An automatic recognition of multi-class skin lesions via deep learning convolutional neural networks. In: ISIC2018: Skin Image Analysis Workshop and Challenge; 2018. [Google Scholar]
65. Wen D, Khan S, Xu A, Ibrahim H, Smith L, Caballero J, et al. Characteristics of publicly available skin cancer image datasets: a systematic review. Lancet Digital Health. 2022;4:e64–74. doi:10.1016/S2589-7500(21)00252-1. [Google Scholar] [PubMed] [CrossRef]
66. Javaid A, Sadiq M, Akram F. Skin cancer classification using image processing and machine learning. In: 2021 International Bhurban Conference on Applied Sciences and Technologies (IBCAST); 2021. p. 439–44. [Google Scholar]
67. Duggani K, Nath M. A technical review report on deep learning approach for skin cancer detection and segmentation. In: Data analytics and management: Proceedings of ICDAM; 2021. p. 87–99. [Google Scholar]
68. Yilmaz A, Kalebasi M, Samoylenko Y, Guvenilir M, Uvet H. Benchmarking of lightweight deep learning architectures for skin cancer classification using ISIC 2017 dataset. arXiv Preprint arXiv:2110.12270. 2021. [Google Scholar]
69. Hekler A, Utikal JS, Enk AH, Hauschild A, Weichenthal M, Maron RC, et al. Superior skin cancer classification by the combination of human and artificial intelligence. Eur J Cancer. 2019;120:114–21. doi:10.1016/j.ejca.2019.07.019. [Google Scholar] [PubMed] [CrossRef]
70. Magdy A, Hussein H, Abdel-Kader R, Abd El Salam K. Performance enhancement of skin cancer classification using computer vision. IEEE Access. 2023;11(72120–33). [Google Scholar]
71. Bhatt H, Shah V, Shah K, Shah R, Shah M. State-of-the-art machine learning techniques for melanoma skin cancer detection and classification: a comprehensive review. Intell Med. 2023;3:180–90. doi:10.1016/j.imed.2022.08.004. [Google Scholar] [CrossRef]
72. Razmjooy N, Ashourian M, Karimifard M, Estrela V, Loschi H, Do Nascimento D, et al. Computer-aided diagnosis of skin cancer: a review. Current Med Imaging. 2020;16:781–93. doi:10.2174/1573405616666200129095242. [Google Scholar] [CrossRef]
73. Alom M, Aspiras T, Taha T, Asari V. Skin cancer segmentation and classification with improved deep convolutional neural network. In: Medical imaging 2020: Imaging informatics for healthcare, research, and applications; 2020. p. 291–301. doi:10.1117/12.2550146. [Google Scholar] [CrossRef]
74. Manne R, Kantheti S, Kantheti S. Classification of skin cancer using deep learning, convolutional neural networks-opportunities and vulnerabilities–a systematic review. Int J Modern Trends Sci Technol. 2020;6:2455–3778. [Google Scholar]
75. Riaz L, Qadir H, Ali G, Ali M, Raza M, Jurcut A, et al. A comprehensive joint learning system to detect skin cancer. IEEE Access. 2023;11:79434–44. doi:10.1109/ACCESS.2023.3297644. [Google Scholar] [CrossRef]
76. Rezk E, Haggag M, Eltorki M, El-Dakhakhni W. A comprehensive review of artificial intelligence methods and applications in skin cancer diagnosis and treatment: emerging trends and challenges. Healthcare Analy. 2023; 100259. [Google Scholar]
77. Tahir M, Naeem A, Malik H, Tanveer J, Naqvi R, Lee S. MDSCC_Net: multi-classification deep learning models for diagnosing of skin cancer using dermoscopic images. Cancers. 2023;15:2179. doi:10.3390/cancers15072179. [Google Scholar] [PubMed] [CrossRef]
78. Tumpa P, Kabir M. An artificial neural network based detection and classification of melanoma skin cancer using hybrid texture features. Sens Int. 2021;2:100128. doi:10.1016/j.sintl.2021.100128. [Google Scholar] [CrossRef]
79. Li J. Skin cancer classification based on convolutional neural network. In: 2021 2nd International Conference on Artificial Intelligence and Computer Engineering (ICAICE); 2021. p. 532–40. [Google Scholar]
80. Dutta A, Kamrul Hasan M, Ahmad M. Skin lesion classification using convolutional neural network for melanoma recognition. In: Proceedings of International Joint Conference on Advances in Computational Intelligence: IJCACI 2020; 2021. p. 55–66. [Google Scholar]
81. Haggenmüller S, Maron R, Hekler A, Utikal J, Barata C, Barnhill R, et al. Skin cancer classification via convolutional neural networks: systematic review of studies involving human experts. Eur J Cancer. 2021;156:202–16. doi:10.1016/j.ejca.2021.06.049. [Google Scholar] [CrossRef]
82. Chen J, Zhu D, Hui B, Li R, Yue X, et al. Mu-Net: multi-path upsampling convolution network for medical image segmentation. Comput Model Eng Sci. 2022;131(1):73–95. doi:10.32604/cmes.2022.018565. [Google Scholar] [CrossRef]
83. Bian X, Pan H, Zhang K, Li P, Li J, Chen C. Skin lesion image classification method based on extension theory and deep learning. Multimed Tools Appl. 2022;81:16389–409. doi:10.1007/s11042-022-12376-3. [Google Scholar] [CrossRef]
84. Gajera H, Nayak D, Zaveri M. A comprehensive analysis of dermoscopy images for melanoma detection via deep CNN features. Biomed Signal Process Control. 2023;79:104186. doi:10.1016/j.bspc.2022.104186. [Google Scholar] [CrossRef]
85. Shinde R, Alam M, Hossain M, Md Imtiaz S, Kim J, Padwal A, et al. Squeeze-MNet: precise skin cancer detection model for low computing IoT devices using transfer learning. Cancers. 2022;15:12. doi:10.3390/cancers15010012. [Google Scholar] [PubMed] [CrossRef]
86. Alenezi F, Armghan A, Polat K. A multi-stage melanoma recognition framework with deep residual neural network and hyperparameter optimization-based decision support in dermoscopy images. Expert Syst Appl. 2023;215:119352. [Google Scholar]
87. Alenezi F, Armghan A, Polat K. Wavelet transform based deep residual neural network and ReLU based Extreme Learning Machine for skin lesion classification. Expert Syst Appl. 2023;213:11906. [Google Scholar]
88. Gouda W, Sama N, Al-Waakid G, Humayun M, Jhanjhi N. Detection of skin cancer based on skin lesion images using deep learning. Healthcare. 2022;10:1183. doi:10.3390/healthcare10071183. [Google Scholar] [PubMed] [CrossRef]
89. Bassel A, Abdulkareem A, Alyasseri Z, Sani N, Mohammed H. Automatic malignant and benign skin cancer classification using a hybrid deep learning approach. Diagnostics. 2022;12:2472. doi:10.3390/diagnostics12102472. [Google Scholar] [PubMed] [CrossRef]
90. Kousis I, Perikos I, Hatzilygeroudis I, Virvou M. Deep learning methods for accurate skin cancer recognition and mobile application. Electronics. 2022;11:1294. doi:10.3390/electronics11091294. [Google Scholar] [CrossRef]
91. Shorfuzzaman M. An explainable stacked ensemble of deep learning models for improved melanoma skin cancer detection. Multimed Syst. 2022;28:1309–23. doi:10.1007/s00530-021-00787-5. [Google Scholar] [CrossRef]
92. Abbas Q, Gul A. Detection and classification of malignant melanoma using deep features of NASNet. SN Comput Sci. 2022;4:21. doi:10.1007/s42979-022-01439-9. [Google Scholar] [CrossRef]
93. Reis H, Turk V, Khoshelham K, Kaya S. InSiNet: a deep convolutional approach to skin cancer detection and segmentation. Med Biol Eng Comput. 2022;60:643–62. doi:10.1007/s11517-021-02473-0. [Google Scholar] [PubMed] [CrossRef]
94. Ghosh P, Azam S, Quadir R, Karim A, Shamrat F, Bhowmik S, et al. SkinNet-16: a deep learning approach to identify benign and malignant skin lesions. Front Oncol. 2022;12:931141. doi:10.3389/fonc.2022.931141. [Google Scholar] [PubMed] [CrossRef]
95. Mazoure B, Mazoure A, Bédard J, Makarenkov V. DUNEScan: a web server for uncertainty estimation in skin cancer detection with deep neural networks. Sci Rep. 2022;12:179. doi:10.1038/s41598-021-03889-2. [Google Scholar] [PubMed] [CrossRef]
96. Rashid J, Ishfaq M, Ali G, Saeed M, Hussain M, Alkhalifah T, et al. Skin cancer disease detection using transfer learning technique. Appl Sci. 2022;12:5714. doi:10.3390/app12115714. [Google Scholar] [CrossRef]
97. Aljohani K, Turki T. Automatic classification of melanoma skin cancer with deep convolutional neural networks. AI. 2022;3:512–25. doi:10.3390/ai3020029. [Google Scholar] [CrossRef]
98. Demir A, Yilmaz F, Kose O. Early detection of skin cancer using deep learning architectures: ResNet-101 and inception-v3. In: Proceedings of the 2019 Medical Technologies Congress (TIPTEKNO); 2019; Izmir, Turkey. p. 1–4. doi:10.1109/TIPTEKNO47231.2019.8972045. [Google Scholar] [CrossRef]
99. Khan M, Sharif M, Akram T, Damasevicius R, Maskeliunas R. Skin lesion segmentation and multiclass classification using deep learning features and improved moth flame optimization. Diagnostics. 2021;11:811. doi:10.3390/diagnostics11050811. [Google Scholar] [PubMed] [CrossRef]
100. Abdar M, Samami M, Mahmoodabad S, Doan T, Mazoure B, Hashemifesharaki R, et al. Uncertainty quantification in skin cancer classification using three-way decision-based Bayesian deep learning. Comput Biol Med. 2021;135:104418. doi:10.1016/j.compbiomed.2021.104418. [Google Scholar] [PubMed] [CrossRef]
101. Inthiyaz S, Altahan B, Ahammad S, Rajesh V, Kalangi R, Smirani L, et al. Skin disease detection using deep learning. Adv Eng Softw. 2023;175:103361. doi:10.1016/j.advengsoft.2022.103361. [Google Scholar] [CrossRef]
102. Deshpande N, Gite S, Pradhan B, Assiri M. Explainable artificial intelligence–a new step towards the trust in medical diagnosis with AI frameworks: a review. Comput Model Eng Sci. 2022;133(3):843–72. doi:10.32604/cmes.2022.021225. [Google Scholar] [CrossRef]
103. Sharma R, Sharma J, Maheshwari R, Agarwal P. Thermogram adaptive efficient model for breast cancer detection using fractional derivative mask and hybrid feature set in the IoT environment. Comput Model Eng Sci. 2022; 130(2):923–947. doi:10.32604/cmes.2022.016065. [Google Scholar] [CrossRef]
104. Alosaimi W, Uddin M. Efficient data augmentation techniques for improved classification in limited data set of oral squamous cell Carcinoma. Comput Model Eng Sci. 2022;131(3):1387–401. doi:10.32604/cmes.2022.018433. [Google Scholar] [CrossRef]
105. Geng P, Lu J, Zhang Y, Ma S, Tang Z, Liu J. TC-Fuse: a transformers fusing CNNs network for medical image segmentation. Comput Model Eng Sci. 2023;137(2):2001–2023. doi:10.1016/j.est.2021.103009. [Google Scholar] [CrossRef]
106. Shao J, Chen S, Zhou J, Zhu H, Wang Z, Brown M. Application of U-Net and optimized clustering in medical image segmentation: a review. Comput Model Eng Sci. 2023;136(3):2173–2219. doi:10.32604/cmes.2023.025499. [Google Scholar] [CrossRef]
107. Adegun A, Viriri S. Deep learning techniques for skin lesion analysis and melanoma cancer detection: a survey of state-of-the-art. Artif Intell Rev. 2021;54(2):811–41. doi:10.1007/s10462-020-09865-y. [Google Scholar] [CrossRef]
108. Lu S, Yang J, Yang B, Yin Z, Liu M, Yin L, et al. Analysis and design of surgical instrument localization algorithm. Comput Model Eng Sci. 2023;137(1):669–685. doi:10.32604/cmes.2023.027417. [Google Scholar] [CrossRef]
109. Kumar G, Alqahtani H. Deep learning-based cancer detection-recent developments, trend and challenges. Comput Model Eng Sci. 2022;130(3):1271–1307. doi:10.32604/cmes.2022.018418. [Google Scholar] [CrossRef]
110. Qasim Gilani S, Syed T, Umair M, Marques O. Skin cancer classification using deep spiking neural network. J Digit Imaging. 2023;36:1137–47. doi:10.1007/s10278-023-00776-2. [Google Scholar] [PubMed] [CrossRef]
111. Tabrizchi H, Parvizpour S, Razmara J. An improved VGG model for skin cancer detection. Neur Process Lett. 2023;55:3715–32. doi:10.1007/s11063-022-10927-1. [Google Scholar] [CrossRef]
112. Ali M, Miah M, Haque J, Rahman M, Islam M. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach Learn App. 2021;5:100036. [Google Scholar]
113. Sharma A, Tiwari S, Aggarwal G, Goenka N, Kumar A, Chakrabarti P, et al. Dermatologist-level classification of skin cancer using cascaded ensembling of convolutional neural network and handcrafted features based deep neural network. IEEE Access. 2022;10:17920–32. doi:10.1109/ACCESS.2022.3149824. [Google Scholar] [CrossRef]
114. Raza R, Zulfiqar F, Tariq S, Anwar G, Sargano A, Habib Z. Melanoma classification from dermoscopy images using ensemble of convolutional neural networks. Mathematics. 2021;10:26. [Google Scholar]
115. Shaaban S, Atya H, Mohammed H, Sameh A, Raafat K, Magdy A. Skin cancer detection based on deep learning methods. In: The International Conference on Artificial Intelligence and Computer Vision; 2023. p. 58–67. [Google Scholar]
116. Hosny K, Kassem M, Fouad M. Classification of skin lesions into seven classes using transfer learning with AlexNet. J Digit Imaging. 2020;33:1325–34. doi:10.1007/s10278-020-00371-9. [Google Scholar] [PubMed] [CrossRef]
117. Medhat S, Abdel-Galil H, Aboutabl A, Saleh H. Skin cancer diagnosis using convolutional neural networks for smartphone images: a comparative study. J Radiat Res Appl Sci. 2022;15:262–7. [Google Scholar]
118. Abhvankar N, Pingulkar H, Chindarkar K, Siddavatam A. Detection of melanoma and non-melanoma type of skin cancer using CNN and RESNET. In: 2021 Asian Conference on Innovation in Technology (ASIANCON); 2021. p. 1–6. [Google Scholar]
119. Pavan M, Reddy U, Ghantasala C, Devi M, Nesarani A. Enhanced skin cancer classification with AlexNet and transfer learning. In: 2023 International Conference on Sustainable Communication Networks and Application (ICSCNA); 2023. p. 1020–4. [Google Scholar]
120. Saeed J, Zeebaree S. Skin lesion classification based on deep convolutional neural networks architectures. J Appl Sci Technol Trends. 2021;2:41–51. doi:10.38094/jastt20189. [Google Scholar] [CrossRef]
121. Bera S, Shrivastava V, Satapathy S. Advances in hyperspectral image classification based on convolutional neural networks: a review. Comput Model Eng Sci. 2022;133(2):219–250. doi:10.32604/cmes.2022.020601. [Google Scholar] [CrossRef]
122. Mohapatra S, Abhishek N, Bardhan D, Ghosh A, Mohanty S. Comparison of MobileNet and ResNet CNN architectures in the CNN-based skin cancer classifier model. Mach Learn Healthcare App. 2021:169–86. [Google Scholar]
123. Naeem A, Anees T, Fiza M, Naqvi R, Lee S. SCDNet: a Deep learning-based framework for the multiclassification of skin cancer using dermoscopy images. Sensors. 2022;22(22):5652. [Google Scholar] [PubMed]
124. Shah V, Autee P, Sonawane P. Detection of melanoma from skin lesion images using deep learning techniques. In: 2020 International Conference on Data Science and Engineering (ICDSE); 2020. p. 1–8. [Google Scholar]
125. Garcia S. Meta-learning for skin cancer detection using deep learning techniques. arXiv Preprint arXiv:210410775. 2021. [Google Scholar]
126. Islam M, Ali M, Ali M, Haque M, Das A, Hossain M, et al. Melanoma skin lesions classification using deep convolutional neural network with transfer learning. In: 2021 1st International Conference on Artificial Intelligence and Data Analytics (CAIDA); 2021. p. 48–53. [Google Scholar]
127. J. SM, P. M, Aravindan C, Appavu R. Classification of skin cancer from dermoscopic images using deep neural network architectures. Multimed Tools Appl. 2023;82:15763–15778. doi:10.1007/s11042-022-13847-3. [Google Scholar] [CrossRef]
128. Zeng Y, Zhu X. Skin cancer detection based on hybrid model by means of inception V3 and ResNet 50. In: 2022 3rd International Conference on Artificial Intelligence and Education (IC-ICAIE 2022); 2022. p. 275–82. [Google Scholar]
129. Huq A, Pervin M. Analysis of adversarial attacks on skin cancer recognition. In: 2020 International Conference on Data Science and Its Applications (ICoDSA); 2020. p. 1–4. [Google Scholar]
130. Bibi A, Khan M, Javed M, Tariq U, Kang B, Nam Y, et al. Skin lesion segmentation and classification using conventional and deep learning based framework. Comput Mater Contin. 2022;71:(2)2477–95. doi:10.32604/cmc.2022.018917. [Google Scholar] [CrossRef]
131. Sharmila B, Santhosh H, Parameshwara S, Swamy M, Baig W, Nanditha S. Optimizing deep learning networks for edge devices with an instance of skin cancer and corn leaf disease dataset. SN Comput Sci. 2023;4:793. doi:10.1007/s42979-023-02239-5. [Google Scholar] [CrossRef]
132. Tan E, Lim S, Lamont D, Epstein R, Lim D, Lin F. Development and validation of a deep learning model for improving detection of nonmelanoma skin cancers treated with Mohs micrographic surgery. JAAD Int. 2024;14:39–47. doi:10.1016/j.jdin.2023.10.007. [Google Scholar] [PubMed] [CrossRef]
133. Adegun A, Viriri S. FCN-based DenseNet framework for automated detection and classification of skin lesions in dermoscopy images. IEEE Access. 2020;8:150377–96. [Google Scholar]
134. Wei L, Ding K, Hu H. Automatic skin cancer detection in dermoscopy images based on ensemble lightweight deep learning network. IEEE Access. 2020;8:99633–47. [Google Scholar]
135. Abunadi I, Senan E. Deep learning and machine learning techniques of diagnosis dermoscopy images for early detection of skin diseases. Electronics. 2021;10:3158. doi:10.3390/electronics10243158. [Google Scholar] [CrossRef]
136. Balaha H, Hassan A, El-Gendy E, ZainEldin H, Saafan M. An aseptic approach towards skin lesion localization and grading using deep learning and Harris hawks optimization. Multimed Tools Appl. 2023: 1–29. [Google Scholar]
137. Gangwani D, Liang Q, Wang S, Zhu X. An empirical study of deep learning frameworks for melanoma cancer detection using transfer learning and data augmentation. In: 2021 IEEE International Conference on Big Knowledge (ICBK); 2021. p. 38–45. [Google Scholar]
138. Hossain M, Hossain M, Arefin M, Akhtar F, Blake J, et al. Combining state-of-the-art pre-trained deep learning models: a noble approach for skin cancer detection using max voting ensemble. Diagnostics. 2024;14:89. [Google Scholar]
139. Wang G, Yan P, Tang Q, Yang L, Chen J. Multiscale feature fusion for skin lesion classification. Biomed Res Int. 2023;2023:5146543. doi:10.1155/2023/5146543. [Google Scholar] [PubMed] [CrossRef]
140. Jeong H, Park C, Henao R, Kheterpal M. Deep learning in dermatology: a systematic review of current approaches, outcomes, and limitations. JID Innov. 2023;3:100150. doi:10.1016/j.xjidi.2022.100150. [Google Scholar] [PubMed] [CrossRef]
141. Nawaz M, Nazir T, Masood M, Ali F, Khan M, Tariq U, et al. Melanoma segmentation: a framework of improved DenseNet77 and UNET convolutional neural network. Int J Imaging Syst Technol. 2022;32:2137–53. doi:10.1002/ima.v32.6. [Google Scholar] [CrossRef]
142. Zafar K, Gilani S, Waris A, Ahmed A, Jamil M, Khan M, et al. Skin lesion segmentation from dermoscopic images using convolutional neural network. Sensors. 2020;20(6):1601. doi:10.3390/s20061601. [Google Scholar] [PubMed] [CrossRef]
143. Ramadan R, Aly S. CU-Net: a new improved multi-input color U-Net model for skin lesion semantic segmentation. IEEE Access. 2022;10:15539–64. doi:10.1109/ACCESS.2022.3148402. [Google Scholar] [CrossRef]
144. Farahani A, Mohseni H. Medical image segmentation using customized U-Net with adaptive activation functions. Neur Comput Appl. 2021;33:6307–23. doi:10.1007/s00521-020-05396-3. [Google Scholar] [CrossRef]
145. Chen P, Huang S, Yue Q. Skin lesion segmentation using recurrent attentional convolutional networks. IEEE Access. 2022;10:94007–18. doi:10.1109/ACCESS.2022.3204280. [Google Scholar] [CrossRef]
146. Liu L, Zhang X, Li Y, Xu Z. An improved multi-scale feature fusion for skin lesion segmentation. Appl Sci. 2023;13:8512. doi:10.3390/app13148512. [Google Scholar] [CrossRef]
147. Gu R, Wang L, Zhang L. DE-Net: a deep edge network with boundary information for automatic skin lesion segmentation. Neurocomputing. 2022;468:71–84. doi:10.1016/j.neucom.2021.10.017. [Google Scholar] [CrossRef]
148. Izadi S, Mirikharaji Z, Kawahara J, Hamarneh G. Generative adversarial networks to segment skin lesions. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); 2018. p. 881–4. [Google Scholar]
149. Arora G, Dubey A, Jaffery Z, Rocha A. Architecture of an effective convolutional deep neural network for segmentation of skin lesion in dermoscopic images. Expert Syst. 2023;40:e12689. doi:10.1111/exsy.v40.6. [Google Scholar] [CrossRef]
150. Adegun A, Viriri S. An enhanced deep learning framework for skin lesions segmentation. In: Computational Collective Intelligence: 11th International Conference; 2019; Hendaye, France, p. 414–25. [Google Scholar]
151. Ninh Q, Tran T, Tran T, Tran T, Pham V. Skin lesion segmentation based on modification of SegNet neural networks. In: 2019 6th NAFOSTED Conference on Information and Computer Science (NICS); 2019. p. 575–8. [Google Scholar]
152. Tao S, Jiang Y, Cao S, Wu C, Ma Z. Attention-guided network with densely connected convolution for skin lesion segmentation. Sensors. 2021;21:3462. doi:10.3390/s21103462. [Google Scholar] [PubMed] [CrossRef]
153. Zhang Y, Zhou T, Wang S, Wu Y, Gu P, Chen D. SamDSK: combining segment anything model with domain-specific knowledge for semi-supervised learning in medical image segmentation. arXiv Preprint arXiv:230813759. 2023. [Google Scholar]
154. Sirmacek B, Kivits M. Semantic segmentation of skin lesions using a small data set. arXiv Preprint arXiv:191010534. 2019. [Google Scholar]
155. Garg S, Balkrishan J. Skin lesion segmentation in dermoscopy imagery. Int Arab J Inf Technol. 2022;19:29–37. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF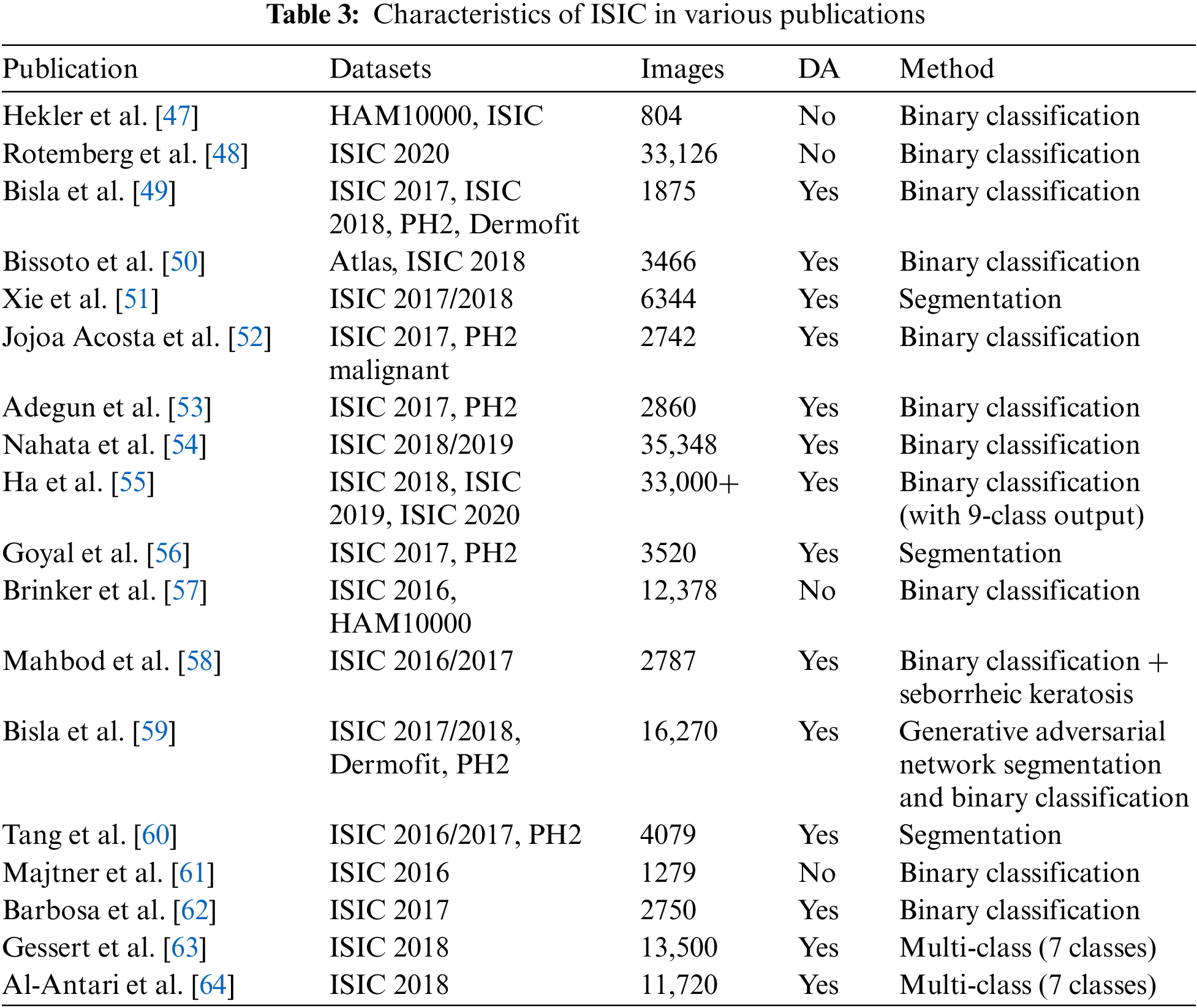
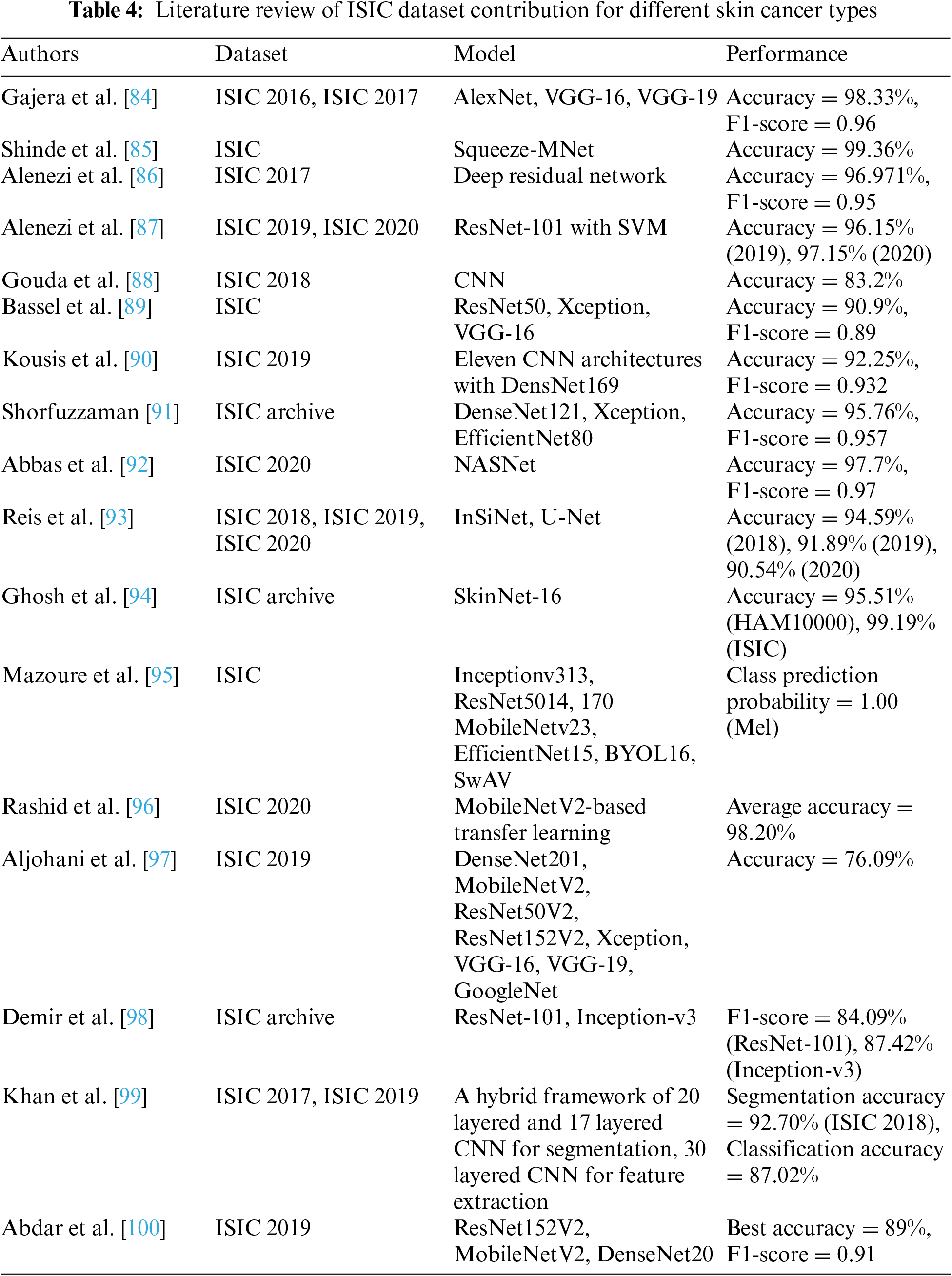
 Downloads
Downloads
 Citation Tools
Citation Tools