 Open Access
Open Access
ARTICLE
DCFNet: An Effective Dual-Branch Cross-Attention Fusion Network for Medical Image Segmentation
1 School of Computer Science and Engineering, Central South University, Changsha, 410083, China
2 School of Humanities, Central South University, Changsha, 410083, China
3 Xiangya Hospital Central South University, Changsha, 410008, China
4 Changsha Aier Eye Hospital, Changsha, 410015, China
* Corresponding Author: Yalong Xiao. Email:
(This article belongs to the Special Issue: Intelligent Medical Decision Support Systems: Methods and Applications)
Computer Modeling in Engineering & Sciences 2024, 140(1), 1103-1128. https://doi.org/10.32604/cmes.2024.048453
Received 08 December 2023; Accepted 16 January 2024; Issue published 16 April 2024
Abstract
Automatic segmentation of medical images provides a reliable scientific basis for disease diagnosis and analysis. Notably, most existing methods that combine the strengths of convolutional neural networks (CNNs) and Transformers have made significant progress. However, there are some limitations in the current integration of CNN and Transformer technology in two key aspects. Firstly, most methods either overlook or fail to fully incorporate the complementary nature between local and global features. Secondly, the significance of integrating the multi-scale encoder features from the dual-branch network to enhance the decoding features is often disregarded in methods that combine CNN and Transformer. To address this issue, we present a groundbreaking dual-branch cross-attention fusion network (DCFNet), which efficiently combines the power of Swin Transformer and CNN to generate complementary global and local features. We then designed the Feature Cross-Fusion (FCF) module to efficiently fuse local and global features. In the FCF, the utilization of the Channel-wise Cross-fusion Transformer (CCT) serves the purpose of aggregating multi-scale features, and the Feature Fusion Module (FFM) is employed to effectively aggregate dual-branch prominent feature regions from the spatial perspective. Furthermore, within the decoding phase of the dual-branch network, our proposed Channel Attention Block (CAB) aims to emphasize the significance of the channel features between the up-sampled features and the features generated by the FCF module to enhance the details of the decoding. Experimental results demonstrate that DCFNet exhibits enhanced accuracy in segmentation performance. Compared to other state-of-the-art (SOTA) methods, our segmentation framework exhibits a superior level of competitiveness. DCFNet’s accurate segmentation of medical images can greatly assist medical professionals in making crucial diagnoses of lesion areas in advance.Keywords
Medical image segmentation is a significant and complex research area [1] that plays a crucial role in the quantitative analysis of medical images. It encompasses various challenges commonly encountered in medical applications, such as segmenting polyps, glands, and breast tumors. This process holds immense importance in medical image-assisted diagnosis as it enables the extraction of meaningful features from medical images, aiding doctors in making more accurate diagnoses. However, medical image segmentation continues to face substantial challenges due to factors like blurred edges, similar morphologies, low contrast, noise interference, texture heterogeneity, and uncertainty in segmentation regions, as shown in Fig. 1. Additionally, the heterogeneity of tumor cancer further adds to these challenges [2]. Consequently, the development of automated methods that are both precise and robust for medical image segmentation has long presented a significant obstacle for medical image analysts [3]. The topic of image segmentation has gained significant attention due to the advancements in deep learning [4] technology.

Figure 1: The example images of similar morphology, blurred edges, low contrast, and noise interference in polyp, gland, and breast ultrasound
In recent years, the convolutional neural network (CNN) [5] has become the dominant framework for various models due to its exceptional ability to represent features. The CNN model incorporates a symmetric encoder-decoder structure, and its success can largely be attributed to the skip connection, which enhances the feature details in the decoder. Several variant models, such as UNet++ [6] and MultiresUNet [7], have made significant advancements based on the CNN. Despite these achievements, CNN variants still have limitations. The inherent locality of convolution operations prevents them from explicitly capturing long-range dependencies. Additionally, there is a significant disparity in feature representation between the encoder and decoder, thus compromising the consistency of the feature representation.
The utilization of the Transformer in natural language processing was initially introduced by Dosovitskiy et al. [8,9]. Since then, its application in computer vision tasks has garnered significant achievements. The Transformer-based models, such as the UNEt TRansformers (UNETR [10]) model proposed by Hatamizadeh, have been successfully employed in computer vision tasks. These models utilize the self-attention mechanism to effectively capture the global context feature of medical images. However, the drawback of Transformers lies in their neglect of local detail features, which consequently results in high computational costs. To address this issue and enhance both computational efficiency and image segmentation performance, a novel approach called the shifted window-based hierarchical vision Transformer (Swin Transformer [11]) has been proposed by Liu et al. This approach leverages both the neural network’s inductive bias and the self-attention mechanism in the Transformer to bolster the network structure.
In light of the above, it can be observed that there is a natural complementarity between the Transformer and CNN methodologies. Various methods, such as TransUNet [12], CTC-Net [13], and CASF-Net [14], have been devised to merge the capabilities of CNN and Transformer to fully harness their strengths. Notably, TransUNet [12] employs a CNN to extract low-level features, followed by the utilization of a Transformer to simulate global interactions. However, it is important to acknowledge that this implementation merely represents a sequential integration of convolution and Transformer mechanisms. It failed to effectively produce complementary features. CTC-Net [13] leverages the dual-branch encoders of CNN and Swin Transformer to generate complementary features. In a similar vein, CASF-Net [14] integrates global and local features and strategically maximizes the advantages offered by both CNN and Transformer, leading to feature enhancement. Despite the remarkable results achieved by these models, they overlook the importance of merging the features from dual-branch multi-scale encoders to improve the decoding of intricate details.
Based on the analysis conducted, there is a need for further investigation into the seamless integration of CNN and Swin Transformer, capitalizing on the respective strengths of both models. To address this, we introduce a groundbreaking network called DCFNet, which leverages a dual-branch cross-attention feature fusion architecture. The backbone of DCFNet comprises two branches: CNN and Swin Transformer. The CNN branch is responsible for capturing local features, while the Swin Transformer branch captures global context features. To enhance the details of the decoder’s features, it is crucial to effectively aggregate the encoder features across multiple scales. In the feature cross-fusion (FCF) module, reference is made to the Channel-wise Cross-fusion Transformer (CCT) mechanism [15] proposed by Wang et al. Subsequently, the feature fusion module (FFM) is suggested to merge dual branch feature maps to generate complementary features. Furthermore, channel attention blocks (CAB) are incorporated to emphasize channels that contribute significantly to the features and suppress low-contribution feature channels, thereby guiding the decoding process. The ultimate predicted segmentation outcome is obtained through the summation of the CNN branch and the Swin Transformer branch. The primary contributions of this paper can be summarized as follows:
• We introduce an innovative network called DCFNet, which incorporates a dual-branch cross-attention fusion approach. Initially, we utilize the CNN and Swin Transformer dual-branch backbone networks to extract both local and global context features from the input image. Subsequently, we employ the Feature Cross-Fusion (FCF) module to merge the context features obtained from the dual-branch encoders, thereby enhancing the decoder feature details. Lastly, to further enhance the feature details of the dual-branch decoders, we introduce the Channel Attention Block (CAB) to guide the decoding process.
• We propose the FCF module to integrate the local and global context features of the dual encoders. To achieve this integration, we leverage the Channel-wise Cross-fusion Transformer (CCT) module within the FCF module, allowing for multi-scale encoder feature fusion. Additionally, we introduce the FFM sub-mechanism within the FCF module for feature fusion. By utilizing FFM, dual-branch feature maps can focus on the related feature regions from a spatial perspective to produce fused features. Moreover, the establishment of long-range dependencies among the dual-branch feature maps and the salient feature map, generated by the spatial attention block (SAB) mechanism, contributes to the enhancement of significant feature information aggregation.
• We developed the Channel Attention Block (CAB) mechanism, which prioritizes the relevant channel features between the up-sampled decoder feature map and the enhanced feature map created by the FCF module. This approach suppresses any extraneous channel features and maximizes the integration of contributing channel features to enhance decoding feature details.
• Extensive experimentation on four segmentation tasks demonstrates that the proposed DCFNet in this study outperforms the most state-of-the-art models.
Convolutional neural networks (CNNs) have become the dominant segmentation frameworks in the medical image field, with Fully convolutional networks (FCNs) being particularly prominent [5]. Among the FCNs, UNet [16] has garnered noteworthy results in segmenting medical images. Subsequently, several CNN-based models, including UNet++ [6], V-Net [17], ResUNet++ [18], Attention U-Net [19], TransUNet [12], MultiResUNet [7], and UCTransNet [15], have been proposed. These models have been specially designed and have demonstrated significant segmentation performance in biomedical images. In conclusion, the application of CNN-based methods has led to significant advancements in this field.
2.2 Vision Transformer Based Methods
Recently, there has been an increasing prevalence in the application of a Transformer-based [9,20] architecture in computer vision. This can be attributed to the effectiveness of the multi-head self-attention mechanism in modeling the interaction between sequential tokens, which is derived from the origins of Transformers in natural language processing tasks. In the context of computer vision, the Vision Transformer (ViT) has achieved state-of-the-art performance in ImageNet classification tasks by utilizing the Transformer to model full-size images with global self-attention. Another notable development is the Swin Transformer [11], which has also produced state-of-the-art results. In the field of medical imaging, the Swin-Unet [21] and TransUNet [12] models have successfully incorporated Transformer architectures to enhance medical image segmentation performance. Additionally, the Gated Axial-Attention model (MedT [22]) has been proposed to address the challenge of limited data samples in medical images. Furthermore, the hybrid Transformer architecture (UTNet [23]) integrates self-attention into the convolutional neural network (CNN) to create a more powerful model. Lastly, DS-TransUNet [24] utilizes dual-scale Swin Transformer encoders to extract semantic features from different perspectives and incorporates a Transformer-based feature fusion module to fuse feature information from different scales.
2.3 Combining CNN and Transformer Methods
Several scholarly works [12,13,25] have made attempts to enhance the performance of medical image segmentation by combining the advantages of CNN and Transformer. For instance, TransUNet [12] employs a sequence of Transformer and CNN encoders to capture semantic feature information. CASF-Net [14] utilizes a cross-fusion module to merge features from dual CNN and Transformer branches, thereby enhancing the quality of image semantic segmentation. Additionally, CTC-Net [13] introduces a feature fusion module that combines CNN and Swin Transformer branches, resulting in improved complementary feature information. To further enhance the spatial context semantic information between Transformer decoding features and complementary features, skip concatenation is implemented. HiFormer [26] consists of a hierarchical CNN-Transformer feature extractor module. The outputs of the first and last layers are fed through a Double-Level Fusion (DLF) feature fusion module. However, the encoder feature extraction may ignore the importance of multi-scale feature information. H2Former [27] combines CNN and Transformer with multi-scale channel attention, but using same-layer skip connections may not be the best way to improve decoder performance.
The simplified frameworks of our DCFNet and various combinational methods of Transformer and CNN are depicted in Fig. 2. To provide a clear illustration of the different model structures, we present the overall architecture. Fig. 2a showcases the approach employed by TransUNet [12], where the encoder initially utilizes CNN to extract local semantic features. Subsequently, the Transformer encoder is employed to model the global context, and finally, the feature information is decoded using CNN. However, this approach fails to fully integrate the advantages of both CNN and Transformer. On the other hand, as shown in Fig. 2b, TransFuse [25] consists of a CNN encoder, dual CNN decoders, a Transformer encoder, and a feature fusion module. The dual encoders extract spatial features and interact with long-range context. The first decoder combines the Transformer and upsampling features, while the second decoder generates the final fused feature output. As depicted in Fig. 2c, CTC-Net [13] comprises a Swin Transformer encoder, a CNN encoder, a Swin Transformer decoder, and a feature fusion module. The CNN encoder is responsible for capturing local contextual features, while the Swin Transformer encoder focuses on modeling global contextual feature information. The feature fusion module generates complementary feature maps. However, CTC-Net does not utilize the CNN to decode features to further enhance the feature details of decoding. To address this issue and minimize the aggregation differences between the CNN and Swin Transformer, we propose a novel DCFNet dual-branch cross-attention feature fusion network. In Fig. 2d, DCFNet employs the CNN and Swin Transformer branches to capture global and local context features, respectively. The multi-scale feature aggregation mechanism integrates the multi-scale features of the encoder to generate enhanced features. The dual-stream feature fusion module combines the enhanced features from both branches to produce complementary features that enhance the feature details of the dual-branch decoder. The detailed methods of each component will be presented in the Methods section.

Figure 2: Comparisons of simplified frameworks. (a) TransUNet comprises a CNN encoder, a CNN decoder, and a Transformer block. (b) TransFuse incorporates a CNN encoder, dual CNN decoders, a Transformer encoder, and a feature fusion module. (c) CTC-Net integrates a CNN encoder, a Swin Transformer encoder, a Swin Transformer decoder, and an effective feature fusion module. (d) Our DCFNet incorporates a Swin Transformer branch, a CNN branch, a multi-scale feature aggregation, and a dual-stream feature fusion module
Within this section, we present an overview of our DCFNet framework and the principles of each component. The motivation behind the dual-branch network is derived from the advantageous features exhibited by both CNN and Swin Transformer. Specifically, when dealing with biomedical image segmentation, the CNN architecture may encounter difficulties in accurately segmenting small or thin objects due to the loss of pertinent features. Conversely, the Swin Transformer proves beneficial for establishing global context interactions in scenarios involving long and narrow objects, thereby improving object segmentation performance. By effectively combining local and global features, we can generate enhanced complementary features that further augment the quality of biomedical image segmentation. Subsequently, we will proceed to introduce the architecture of DCFNet, along with the operational principles of each component, encompassing the dual-branch network architecture, dual-branch feature cross-fusion module, channel-wise cross-fusion Transformer, and channel attention block mechanism.
The DCFNet is comprised of dual parallel branches (a Convolutional Neural Network (CNN) and a Swin Transformer), which serve as feature extractors. This architecture is illustrated in Fig. 3. Given a medical image with a spatial resolution of

Figure 3: Illustration of the proposed DCFNet. The generated feature maps of each stage of the dual-branch of CNN and Swin Transformer are streamed separately into the CCT mechanism for multi-scale feature channel-wise cross-fusion in the FCF module. Then the feature maps from the dual-CCT are streamed into the FFM mechanism for feature cross-attention fusion. In addition, the up-sampled feature maps of the decoder and the feature maps generated by the FFM mechanism are fed into the CAB mechanism for channel enhancement and suppression. Finally, the predicted mask is generated from the CNN decoder and Swin Transformer decoder
3.2 Global and Local Feature Extraction
The DCFNet, incorporating both the CNN and Swin Transformer branches, facilitates the extraction of distinctive features in medical images from diverse vantage perspectives. The CNN branch adeptly captures localized features, with an emphasis on intricate feature particulars. In contrast, the Swin Transformer branch excels in capturing global features, prioritizing the acquisition of interdependencies among spatially long-range components.
In the implementation, the encoder of the CNN branch utilizes the ResNet34 [28] architecture, as shown in Fig. 3. The ResNet34 model enables the representation of multi-scale features with fine-grained, the feature information tends to not disappear as the depth increases. On the other hand, the Swin Transformer branch combines the benefits of self-attention mechanisms found in the Transformer with efficient computational resource utilization. Both the Swin Transformer and CNN dual-branch architectures can be independently applied. However, the optimal approach involves combining the strengths of both CNN and Swin Transformer to produce complementary feature representations. This combination ultimately enhances the representation of feature details in the dual-branch decoder. For CNN and Swing Transformer branches, Assuming
where W-MSA denotes the window-based multi-head self-attention, while SW-MSA denotes the shifted window-based multi-head self-attention. LN and MLP denote the Layer normalization, Multi-Layer Perceptron.
3.3 Multi-Scale Channel-Wise Cross-Fusion Transformer
The multi-scale channel-wise cross-fusion Transformer (CCT) involves three main steps: multi-scale feature embedding, multi-head channel-wise cross attention, and multi-layer perceptron (MLP). To illustrate this process, we will focus on the CNN branch architecture (note that the Swin Transformer branch architecture is similar to this). Refer to Fig. 4 in the paper (CCT) for a visual representation. In this architecture, we are given feature maps, denoted as

Figure 4: Illustration of the channel-wise cross-fusion Transformer (CCT) module. The CCT can learn the dependencies between the different input feature layers in a Transformer cross-fusion way
Firstly, tokenization is performed to reshape the feature tensor
where
where
3.4 Feature Cross-Attention Fusion Module
The dual-branch network structures enable the extraction of distinctive features from multiple perspectives in medical images. Specifically, the CNN architecture is adept at capturing local features, while the Swin Transformer architecture focuses on modeling global context interaction. The confusion of these dual-branch feature representations is of utmost importance in enhancing the accuracy of medical image segmentation. To facilitate the generation of mutually complementary feature information, we have devised a dual-branch feature fusion module (FFM), illustrated in Fig. 5a. Overview, The FFM mechanism consists of two steps. First is to send the
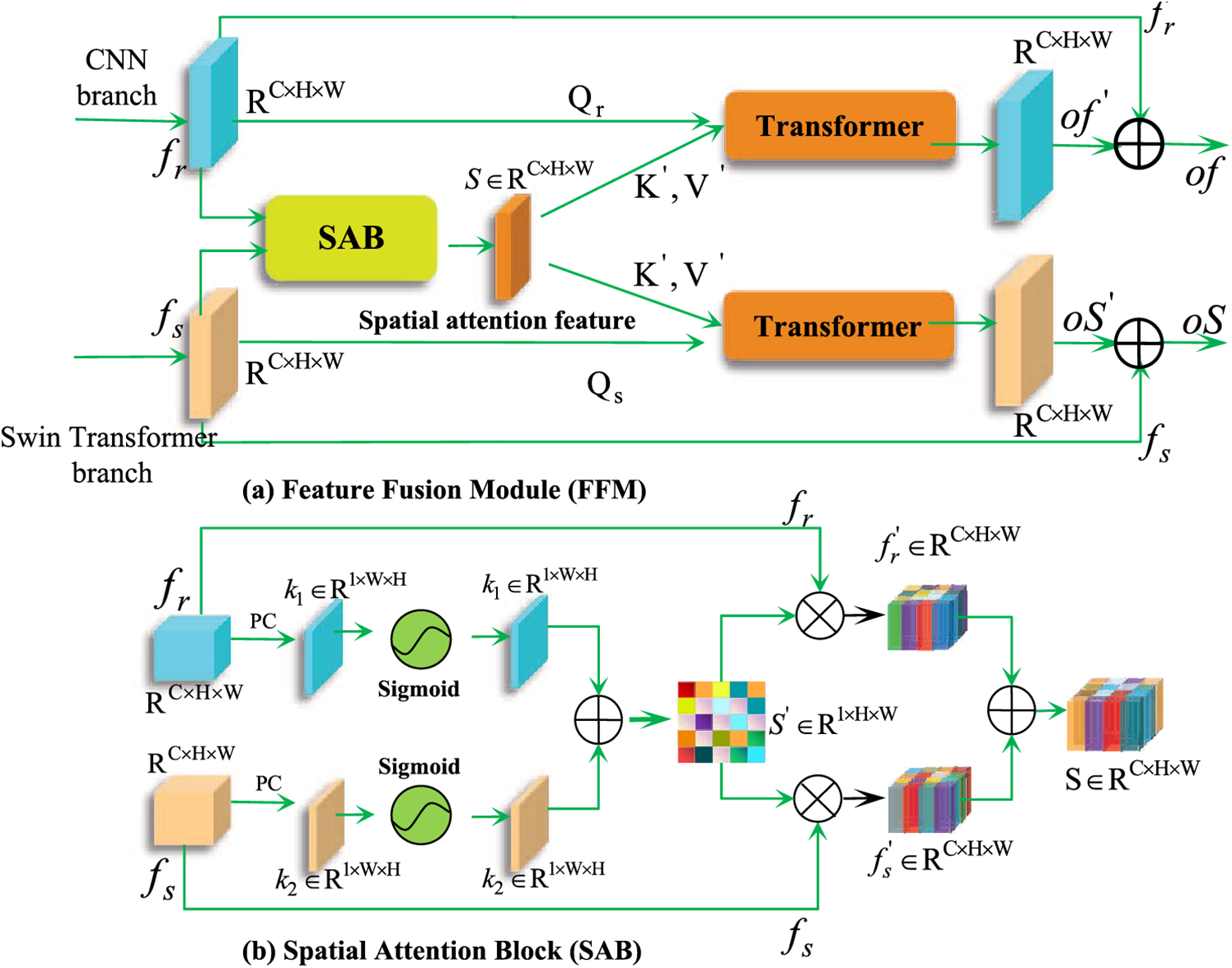
Figure 5: Illustration of the feature fusion module (FFM) module and the sub-mechanism spatial attention block (SAB) module. By utilizing FFM, dual-branch feature maps can focus on the related feature regions from a spatial perspective to produce fused features
To further augment the significant regions within the dual-branch feature maps while concurrently suppressing the irrelevant regions, we have also devised the spatial attention block (SAB). This can be observed in Fig. 5b. In the initial step, the pixel compression technique is employed to compress
where
In the subsequent stage, to enhance the concentration of the branching feature maps on the significant regions of the spatial attention feature
The incorporation of the self-attention component is of utmost importance when it comes to integrating multi-scale features. The underlying attention function is primarily located in the dot product operation of scaling, which is defined in the Eq. (10). The relevant vectors are as:
Nonetheless, Eq. (10) has a drawback in terms of the computational complexity required for the materialization of the softmax logits and the attention maps. Specifically, this process incurs a spatial complexity of
In light of this, Eq. (11) is regarded as the self-attention mechanism for the Transformer module in FFM. By incorporating Value (
where
The proper integration of channel feature information between the up-sampled features and the FCF-enhanced features plays a critical role in enhancing the segmentation result of DCFNet. Employing the channel attention block (CAB) mechanism allows for the enhancement of channel features by highlighting their significant contributions within the up-sampled feature maps of the decoder and the FCF module-enhanced feature maps, while simultaneously suppressing low-contribution channel features. This approach is particularly advantageous in enhancing the feature details of the decoder. To achieve this, we have devised the CAB. To illustrate, we take the Swin Transformer branch as a representative example (the CNN branch follows a similar pattern), as depicted in Fig. 6. Given the up-sampled feature map

Figure 6: Illustration of the channel attention block (CAB) module, which serves for channel enhancement and suppression
where
In this section, we present the datasets used in our experiment and provide the corresponding experimental details. We then compare our method to the current state-of-the-art methods. Additionally, we conduct ablation experiments to establish the rationality of our network design and the effectiveness of each component.
We use BUS [29], Gland Segmentation (GlaS) [30], KvasirCapsule-SEG [31] and KvasirSessile-SEG [32] datasets to evaluate our method DCFNet.
The BUS dataset, consisting of 163 breast ultrasound images taken from numerous female patients at the UDIAT Diagnostic Center, is detailed in [29]. The average pixel size was 760
The Gland Segmentation (GlaS) dataset [30] originates from the 2015 challenge concerning gland segmentation in histology images, offering images of Haematoxylin and Eosin (H and E) stained slides. The dataset comprises 165 images dispersed between 85 for training and 80 images for testing.
4.1.3 KvasirCapsule-SEG Dataset
KvasirCapsule-SEG [31], an enhanced subset of Kvasir includes polyp images along with their segmentation ground truth and bounding box information. The polyp class in Kvasir-Capsule comprises only 55 images that the author annotated with the assistance of a specialist gastroenterologist. Additionally, the dataset features an increased number of polyp images with improved annotations for better accuracy. The polyp class in Kvasir-Capsule comprises only 55 images that the author annotated with the assistance of a specialist gastroenterologist. 55 images are divided into 44 images for training purposes and 11 for testing.
4.1.4 KvasirSessile-SEG Dataset
The Kvasir-SEG comprises 1000 polyp images and their corresponding ground truth data from the Kvasir Dataset. This dataset includes 196 polyps that measure less than 10 mm and have been classified as Paris class 1 sessile or Paris class IIa. With the assistance of expert gastroenterologists, this dataset was meticulously selected. It is a subset of Kvasir-SEG, referred to as Kvasir-Sessile [32], which is particularly challenging for segmentation. Additionally, the images have been divided into 136 for training and 60 for testing purposes.
The DCFNet model was constructed utilizing the PyTorch framework and experimented on a single NVIDIA RTX A5000 24G card. The maximum number of iterations is 2000, while the optimizer is Adam with a learning rate of
The DCFNet’s model parameters can be found in Table 1. The Depth (encoder) and Depth (decoder) refer to the respective depths of the Swin Transformer encoder and decoder. Similarly, the Num-heads (encoder) and Num-heads (decoder) indicate the number of attention heads in the Swin Transformer encoder and decoder. Furthermore, the Num-heads (FFM) and Num-heads (CCT) denote the number of attention heads in the FFM and CCT components.

4.3 Comparison with State-of-the-Art Methods
In this subsection, a series of experiments were conducted to evaluate the performance of DCFNet in comparison to state-of-the-art (SOTA) methods across four medical image segmentation tasks. Additionally, the experimental results are presented and visual examples are provided to assess the learning and generalization abilities of DCFNet. The GT denotes GroundTruth, a term utilized to represent the real labels.
4.3.1 Comparative Result Analysis on the BUS Dataset
The DCFNet is evaluated alongside ten state-of-the-art segmentation general models in the BUS dataset. It is worth noting that segmenting ultrasound images of breast tumors poses a considerable challenge due to their indistinct boundaries and poor contrast. Additionally, the DCFNet model is compared with six different breast tumor segmentation tasks to provide a more comprehensive assessment of its effectiveness. The quantitative findings from our analysis of the BUS dataset [29] can be found in Table 2. The utilization of red bold highlights the best performance, while blue bold highlights suboptimal performance. DCFNet demonstrates superior results in terms of Dice and IoU metrics, surpassing previous state-of-the-art models in both general and ultrasound breast tumor segmentation tasks. Specifically, compared with sub-optimal general methods: TransUNet, UCtransNet, DCSAU-Net, and CANet in general medical image segmentation task, DCFNet achieves Dice (IoU) with an improvement range from 4.643% (3.069%) to 4.754% (4.384%). Although UCTransNet achieved the best performance in terms of F1 score, DCFNet was only slightly lower by 0.86%. In the task of ultrasound segmentation, DCFNet demonstrated significant enhancements in Dice (IoU) performance compared to the suboptimal models MGCC and M2SNet, with improvements of 3.164% (4.716%) and 3.289% (4.533%), respectively. Furthermore, DCFNet showcased improvements of 1.972% and 1.085% in F1 performance. In the realm of general segmentation tasks and specifically in the domain of ultrasound segmentation tasks, DCFNet has emerged as the most superior model in terms of the ASD metric, attaining an impressive value of 24.515%. Furthermore, DCFNet has demonstrated commendable performance across all segmentation metrics, showcasing a well-balanced and comprehensive capability. The aforementioned results demonstrate the potential of DCFNet, our model that combines dual-branch features, and its superior segmentation proficiency compared to other state-of-the-art models.
The DCFNet and other partially superior methods were visualized in Fig. 7. The areas where the DCFNet outperforms the other methods are highlighted by red boxes or red arrows. The visualization of the segmentation results effectively showcases the advantages and strong learning capability of our proposed method.

Figure 7: The qualitative comparative results on representative images of BUS dataset. The red boxes or red arrow highlight regions where DCFNet performs better than the other methods
4.3.2 Comparative Result Analysis on the GlaS Dataset
The quantitative comparative analysis of the GlaS [30] dataset, as presented in Table 3, demonstrates the segmentation indicators of DCFNet and other state-of-the-art (SOTA) methods. The experimental results highlight the best performance, indicated by red bold, and the suboptimal performance, indicated by blue bold. Performance comparison and analysis with other suboptimal SOTA models: TransUNet, UCTransNet, M2SNet and HiFormer, our method DCFNet achieves 3.089% (4.494%), 1.733% (2.723%), 2.932% (4.795%) and 1.614% (2.599%) improvement in Dice(IoU) performance, respectively. Furthermore, DCFNet showcased improvements of 2.180%, 1.221%, 1.755%, and 1.132% in F1 performance. The DCFNet’s ASD metric achieved an optimal value of 10.626%, whereas the implementation of HiFormer was suboptimal. Compared to other state-of-the-art methods, our suggested DCFNet attains the most optimal balance between Dice, IoU, F1, and ASD indicators.

The segmentation maps of partially superior methods are visualized in Fig. 8. The areas where DCFNet outperforms other methods are highlighted with red boxes or a red arrow. Our DCFNet produces segmentation results that closely match the ground truth in comparison to other state-of-the-art models. These analyses provide evidence that DCFNet can achieve more precise segmentation while maintaining accurate shape.

Figure 8: The qualitative comparative results on representative images of the GlaS dataset. The red boxes or red arrow highlight regions where DCFNet performs better than the other methods
4.3.3 Comparative Result Analysis on the KvasirCapsule-SEG and KvasirSessile-SEG
Table 4 shows the quantitative comparative analysis of the KvasirCapsule dataset. In the KvasirCapsule [31] dataset, compared with the previous state-of-the-art (SOTA) methods, our proposal method achieves 96.402%, 93.081%, 95.771%, and 4.429% in (Dice, IoU, F1 and ASD) metrics. Specifically, our results surpass the partial suboptimal models HiFormer, CANet, and the DCSAU-Net by 0.030% (0.046%), 0.053% (0.078%) and 0.163% (0.254%) in Dice (IoU), respectively. Despite the slightly lower F1 score of DCFNet compared to DCSAU-Net by 0.068%, DCFNet demonstrates superior performance with an ASD value of 4.429%. It is worth noting that DCFNet manages to strike a balance across all performance metrics. The numerical results indicate that DCFNet can attain superior segmentation performance even with limited polyp datasets. As shown in Fig. 9, even though the segmentation results of DCFNet, UCTransNet, HiFormer, and M2SNet are very similar to ground truth, the segmentation maps generated by DCFNet demonstrates smoother edges and a closer resemblance to the actual segmented image.


Figure 9: The qualitative comparative results on representative images of KvasirCapsule-SEG. The red boxes or red arrow highlight regions where DCFNet performs better than the other methods
For the KvasirSessile dataset [32], the segmentation performance of the different methods varies significantly. Table 5 shows the quantitative comparative analysis, and the proposed DCFNet surpasses the suboptimal state-of-the-art methods M2SNet, HiFormer, and TransUNet in metrics of Dice, IoU, and F1. Numerically, DCFNet achieves 74.024% (63.762%) on these Dice (IoU) metrics, respectively, which are 6.654% (6.103%), 6.506% (7.143%) and 6.231% (6.864%) better than sub-optimal M2SNet, HiFormer and TransUNet while DCFNet has demonstrated a notable enhancement in F1 scores, with improvements ranging from 4.742% to 5.269%. Moreover, DCFNet has achieved a commendable ASD score of 29.210%, exhibiting a noteworthy reduction of 5.242% when compared to the sub-optimal performing model, HiFormer. In addition, in Fig. 10, we present a visualization of the generated mask image of the partially performer superior models. Although the KvasirSessile dataset presents a significant challenge, DCFNet achieves more precise segmentation results by accurately isolating the position of polyps from normal regions. In summary, the qualitative segmentation performance and numerical results show the excellent segmentation ability and the successful architecture of DCFNet.


Figure 10: The qualitative comparative results on representative images of KvasirSessile-SEG. The red boxes or red arrow highlight regions where DCFNet performs better than the other methods
To demonstrate the soundness of our DCFNet and the comprehensibility of each component, we conducted ablation studies on BUS, GlAS, and KvasirCapsule-SEG. As KvasirSessile-SEG and KvasirCapsule-SEG are both part of the polyp segmentation task, we opted for KvasirCapsule-SEG. Through a comprehensive ablation study, we evaluated the effectiveness of each component in DCFNet by removing them one by one. Within the study, “CCT” represents the Channel-wise Cross-fusion Transformer, “FFM” denotes the designed feature fusion module, “CAB” denotes the channel attention block between decoders, and “w/o” denotes the word “without” for simplicity. DCFNet represents the complete framework.
5.1.1 Effectiveness on CCT Component
To investigate the performance of the aggregated multi-scale features of the CCT module on segmentation, we remove the CCT mechanism from the complete DCFNet. As shown in Table 6, without the CCT component, the performance of DCFNet w/o CCT is significantly lower than the complete DCFNet in three datasets. Specifically, without the CCT component, the performance reduction of Dice (IoU) is 4.588% (6.862%), 0.776% (1.162%), and 0.023% (0.007%) on the BUS, GLAS, and KvasirCapsule-SEG datasets, respectively. In the BUS and GLaS datasets, the absence of the CCT component results in F1 (%) lags behind the full DCFNet of 2.112% and 0.789%, and the ASD (%) performance higher than 4.301% and 0.771%, respectively. On the KvasirCapsule dataset, lacking the CCT component, the performance metrics for F1 (ASD) despite achieving optimal performance of 95.789(%) and 4.417(%). However, the DCFNet of incorporating CCT achieves balance when assessing various performance metrics on the three datasets. The results presented in this study indicate that the comprehensive DCFNet with CCT significantly improves segmentation capabilities, thus confirming the effectiveness of CCT. Furthermore, Fig. 11 illustrates that the absence of CCT results in incorrect predictions for the BUS and GlaS datasets. The results, both numerical and visual, offer compelling evidence that the integration of multi-scale features through the CCT fused encoder successfully enhances the performance of segmentation in the decoder.


Figure 11: Segmentation results for different components applied to the BUS, GlaS, and KvasirCapsule-SEG datasets. The red boxes or red arrow highlight regions where DCFNet performs better than the other methods
5.1.2 Effectiveness on FFM Component
To assess the effectiveness of the FFM components integrated into the CNN and Swin Transformer branches, we eliminate the FFM mechanism in the entire DCFNet. Connecting the enhanced features generated by the CCT component directly to the CAB component results in the model “DCFNet w/o FFM”. The results presented in Table 6 and Fig. 11 reveal that the performance of DCFNet is inferior in datasets when the FFM component is absent, in comparison to the complete DCFNet model. A decline in performance can be observed, indicated by a decrease in Dice (IoU) scores from 78.613% (68.091%) and 92.109% (85.980%) to 76.202% (65.866%) and 91.284% (84.870%) for BUS and GlaS, respectively. Except for the BUS dataset, the F1 metric displays inferior performance compared to the full DCFNet on the GlaS and KvasirCapsule datasets, with values of 89.345% and 95.702%, respectively. Moreover, the ASD metric of the complete DCFNet model incorporating FFM exhibits lower values in comparison to the model w/o FFM across all three datasets. The presented numerical results provide empirical support for the notion that the FFM mechanism enhances the segmentation capability of the model by effectively integrating spatially distinct regions with notable features.
5.1.3 Effectiveness on CAB Component
To gain a more comprehensive understanding of the detailed information about the enhanced decoder feature of the CAB mechanism. we replaced the CAB module with the cascade block, which led to the creation of a new model referred to as “DCFNet w/o CAB”. The results, as presented in Table 6, demonstrate a significant decrease in the Dice (IoU) metric across three datasets when comparing DCFNet w/o CAB to the complete DCFNet. According to the findings, the performance of Dice, IoU and F1 score on the BUS dataset has shown a decrease of 7.191%, 8.280%, and 5.519%. These results suggest that by incorporating the CAB component, DCFNet can potentially achieve improved performance.
Moreover, it is evident from Fig. 11, which visualizes the absence of CAB, that there is an increase in mispredicted normal tissue and missed tumor identification. The findings from both numerical and visual segmentation analyses substantiate the fact that the CAB mechanism enhances the feature channels that make significant contributions while suppressing those with low contributions. As a result, the detailed information of the decoder feature is effectively enhanced.
In addition, it is noteworthy that despite the absence of the CCT or FFM element, the Dice, IoU, and F1 scores achieved by “DCFNet w/o FFM” or “DCFNet w/o CCT” surpass the majority of segmentation models listed in Tables 2–4. This observation implies that our network demonstrates a resilient segmentation performance, even in the absence of specific components.
5.2 Ablation Study on Dual-Branch Structure
Within this subsection, we conduct ablation experiments on the distinct contributions of both the CNN and Swin Transformer branches to validate the overall performance resulting from their combined efforts. We specifically refer to the single branch network of the Swin Transformer as “Model 1”. To evaluate the synergistic potential of CNN in conjunction with the Swin Transformer branch network, we amalgamate the CNN branch and Model 1, resulting in the creation of “Model 2”. Subsequently, we designate the collection of all the feature fusion components (CCT + FFM + CAB) as “CFC”. Similarly, the comprehensive DCFNet is obtained by merging the CFC with Model 2.
The obtained quantitative results of ablation experiments conducted on dual-branch network architectures are presented in Table 7. The results indicate that the performance of the Swin Transformer’s single branch Model 1 is inferior to both Model 2 and the complete DCFNet. Specifically, in the BUS and GlaS datasets, Model 2 demonstrates a notable improvement of 4.4% (4.497%) and 4.446% (6.924%) in metrics of Dice (IoU) performance, in comparison to the Model 1 network. Surprisingly, the absence of the CFC mechanism in Model 2 yields a Dice (IoU) performance that surpasses that of the majority of models presented in Tables 2 and 3 within the BUS and GlaS datasets. This result can be attributed to the successful synergy between the CNN and Swin Transformer branches, underscoring the enhanced performance achieved through their combination.

Furthermore, the comprehensive DCFNet has surpassed Model 1 and Model 2 in terms of performance. Specifically, in the BUS, GlaS, and KvasirCapsule datasets, DCFNet achieves a Dice (IoU) performance of 78.613% (68.091%), 92.109% (85.980%), and 96.402% (93.081%), respectively. These results are significantly higher than those of Model 2, with improvements of 4.046% (5.264%), 0.402% (0.7%), and 0.406% (0.726%) respectively. These findings provide strong evidence for the effectiveness of the CFC mechanism fusion dual-branch networks.
Building upon the favorable outcomes presented in the aforementioned empirical findings, we proceed to delve into the merits of our model, its inherent constraints, and the future work for its enhancement.
The advantages of our work are reflected in the multi-scale feature aggregation of the method and the efficient fusion of local and global features. In the introduction of a dual-branch backbone network, it is recommended to incorporate both local and global features. Additionally, in the encoder, the introduction of a CCT fusion encoder can produce multi-scale features that are more conducive to enhanced decoding features. Finally, we designed the FFM and CAB mechanisms to fully fuse the dual-branch network and highlight significant spatial and channel features while suppressing irrelevant features as much as possible. The experiment’s results also confirmed this point. Meanwhile, our DCFNet model may need to be optimized and tuned appropriately for different medical image types. To achieve the best segmentation performance. In comparison to CASFNet [14] and CTC-Net [13] models, we have implemented the Swing Transformer due to its higher computational efficiency, instead of the Transformer. This results in a reduction of computational burden. Furthermore, our model can integrate the multi-scale features of the encoder, surpassing the limitation of solely incorporating dual-scale feature fusion at the same layer. Similarly, H2Former [27] performs simple cascading with the encoder at the same level, ignoring the importance of aggregating multi-scale, feature-enhanced decoders.
Against our model, our model may have the following limitations. Table 1 showcases our architecture’s implementation of the Transformer in three distinct stages. These stages encompass the encoding and decoding processes of the Swing Transformer branch, as well as the parameters associated with the dual-branch CCT and FFM mechanisms. Notably, the CCT effectively integrates features from the encoder pyramid, while also incorporating the fusion capabilities of the FFM. Despite its impressive segmentation performance, it does come at the cost of sacrificing certain model parameters.
Despite the relatively competitive segmentation performance of our model, our approach continues to face difficulties when applied to ultrasonic tumor images and polyp segmentation images. These challenges arise from the presence of low-contrast images and images with complex structures and similar textures. A clear illustration of this can be observed in Figs. 7 and 10, where although our method demonstrates highly similar segmentation results, there remains a notable discrepancy between the region boundary and the actual label.
After analyzing the limitations of the previous work, we have identified several areas where our model can be further improved. These areas of improvement can be summarized as follows: Firstly, our future work will focus on enhancing the balance between model inference efficiency and achieving a high-performance segmentation effect. Secondly, if a lightweight model is obtained through model compression, based on Swing Transformer and CNN, while ensuring the performance of model segmentation, it has the potential to achieve clinical real-time segmentation in the future by striking a balance between computational efficiency and performance. Finally, our model has only been validated on 2D images, lacking validation in the segmentation of 3D medical images. This represents a direction that necessitates further exploration in our future endeavors.
This article presents an innovative approach called the Dual-Branch Feature Cross-Fusion Network (DCFNet) for medical image segmentation. DCFNet effectively combines the strengths of Convolutional Neural Network (CNN) and Swin Transformer dual-branch network features. To integrate the complementary global and local features, we propose the Feature Cross-Fusion (FCF) module at the dual-branch encoder stage and the Channel Attention Block (CAB) at the dual-branch decoder stage. Specifically, we utilize the Channel-wise Cross-fusion Transformer (CCT) module in the FCF to aggregate multi-scale encoder features. Additionally, we introduce the Feature Fusion Module (FFM) sub-mechanism to merge the dual-encoder features and generate complementary features that enhance the dual-branch decoder’s capabilities. During the decoder stage, we employ CAB to capture channel features that significantly contribute to improving feature details. Through experiments conducted on four publicly available medical image datasets, we provide compelling evidence that our proposed DCFNet outperforms several state-of-the-art networks in medical image segmentation. Furthermore, we demonstrate the effectiveness of our network through ablation experiments conducted separately for each component and network structure. Our study establishes that the proposed DCFNet is a promising method for automatically segmenting medical imaging lesions.
Acknowledgement: This work is supported by the High Performance Computing Center of Central South University. We would like to express our gratitude to the editors and reviewers for their valuable suggestions, which greatly improved this article.
Funding Statement: This work is supported by the National Key R&D Program of China (2018AAA0102100), the National Natural Science Foundation of China (No. 62376287), the International Science and Technology Innovation Joint Base of Machine Vision and Medical Image Processing in Hunan Province (2021CB1013), the Key Research and Development Program of Hunan Province (2022SK2054), the Natural Science Foundation of Hunan Province (No. 2022JJ30762, 2023JJ70016), the 111 Project under Grant (No. B18059).
Author Contributions: The authors confirm contribution to the paper as follows: Study conception and design: Chengzhang Zhu and Renmao Zhang; Data curation: Rong Hu and Xuanchu Duan; Analysis and interpretation of results: Chengzhang Zhu, Renmao Zhang, and Yalong Xiao; Funding acquisition: Chengzhang Zhu, Yalong Xiao, and Beiji Zou; Methodology: Chengzhang Zhu and Renmao Zhang; Software: Chengzhang Zhu, Renmao Zhang, and Yalong Xiao; Supervision: Beiji Zou; Validation: Chengzhang Zhu, Renmao Zhang, and Yalong Xiao; Visualization: Yalong Xiao; Writing-original draft: Xian Chai; Writing-review and editing: Zhangzheng Yang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data will be made available on request.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Liu, X., Song, L., Liu, S., Zhang, Y. (2021). A review of deep-learning-based medical image segmentation methods. Sustainability, 13(3), 1224. [Google Scholar]
2. Xi, J., Yuan, X., Wang, M., Li, A., Li, X. et al. (2020). Inferring subgroup-specific driver genes from heterogeneous cancer samples via subspace learning with subgroup indication. Bioinformatics, 36(6), 1855–1863. [Google Scholar] [PubMed]
3. Rueckert, D., Glocker, B., Kainz, B. (2016). Learning clinically useful information from images: Past, present and future. https://doi.org/10.1016/j.media.2016.06.009 [Google Scholar] [CrossRef]
4. Hesamian, M. H., Jia, W., He, X., Kennedy, P. J. (2019). Deep learning techniques for medical image segmentation: Achievements and challenges. Journal of Digital Imaging, 32(4), 582–596. [Google Scholar] [PubMed]
5. Long, J., Shelhamer, E., Darrell, T. (2017). Fully convolutional networks for semantic segmentation, vol. 39. Amsterdam: Elsevier. [Google Scholar]
6. Zhou, Z., Rahman Siddiquee, M. M., Tajbakhsh, N., Liang, J. (2018). UNet++: A nested U-Net architecture for medical image segmentation. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp. 3–11. Granada, Spain, Springer. [Google Scholar]
7. Ibtehaz, N., Rahman, M. S. (2020). MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Networks, 121, 74–87. [Google Scholar] [PubMed]
8. Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X. et al. (2021). An image is worth 16x16 words: Transformers for image recognition at scale. https://doi.org/10.48550/arXiv.2010.11929 [Google Scholar] [CrossRef]
9. Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L. et al. (2017). Attention is all you need. Advances in Neural Information Processing Systems, 30. [Google Scholar]
10. Hatamizadeh, A., Tang, Y., Nath, V., Yang, D., Myronenko, A. et al. (2022). UNetR: Transformers for 3D medical image segmentation. Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, IEEE. [Google Scholar]
11. Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y. et al. (2021). Swin transformer: Hierarchical vision transformer using shifted windows. Proceedings of the IEEE/CVF International Conference on Computer Vision,Montreal, QC, Canada, IEEE. [Google Scholar]
12. Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E. et al. (2021). TransUNet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306. [Google Scholar]
13. Yuan, F., Zhang, Z., Fang, Z. (2023). An effective CNN and transformer complementary network for medical image segmentation. Pattern Recognition, 136, 109228. [Google Scholar]
14. Zheng, J., Liu, H., Feng, Y., Xu, J., Zhao, L. (2023). CasF-Net: Cross-attention and cross-scale fusion network for medical image segmentation. Computer Methods and Programs in Biomedicine, 229, 107307. [Google Scholar] [PubMed]
15. Wang, H., Cao, P., Wang, J., Zaiane, O. R. (2022). UCTransNet: Rethinking the skip connections in U-Net from a channel-wise perspective with transformer. Proceedings of the AAAI Conference on Artificial Intelligence, vol. 36. Arlington, Virginia, USA. [Google Scholar]
16. Ronneberger, O., Fischer, P., Brox, T. (2015). U-Net: Convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, Springer. [Google Scholar]
17. Milletari, F., Navab, N., Ahmadi, S. A. (2016). V-Net: Fully convolutional neural networks for volumetric medical image segmentation. 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, IEEE. [Google Scholar]
18. Jha, D., Smedsrud, P. H., Riegler, M. A., Johansen, D., De Lange, T. et al. (2019). ResUNet++: An advanced architecture for medical image segmentation. 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, IEEE. [Google Scholar]
19. Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich, M. et al. (2018). Attention U-Net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999. [Google Scholar]
20. Zheng, S., Lu, J., Zhao, H., Zhu, X., Luo, Z. et al. (2021). Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 6881–6890. [Google Scholar]
21. Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X. et al. (2022). Swin-UNet: UNet-like pure transformer for medical image segmentation. Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, pp. 36–46. Tel-Aviv, Israel, Springer. [Google Scholar]
22. Valanarasu, J. M. J., Oza, P., Hacihaliloglu, I., Patel, V. M. (2021). Medical transformer: Gated axial-attention for medical image segmentation. Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, pp. 36–46. Strasbourg, France, Springer. [Google Scholar]
23. Gao, Y., Zhou, M., Metaxas, D. N. (2021). UTNet: A hybrid transformer architecture for medical image segmentation. Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, pp. 61–71. Strasbourg, France, Springer. [Google Scholar]
24. Lin, A., Chen, B., Xu, J., Zhang, Z., Lu, G. et al. (2022). DS-TransUNet: Dual swin transformer U-Net for medical image segmentation. IEEE Transactions on Instrumentation and Measurement, 71, 1–15. [Google Scholar]
25. Zhang, Y., Liu, H., Hu, Q. (2021). TransFuse: Fusing transformers and CNNs for medical image segmentation. Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, Strasbourg, France, Springer. [Google Scholar]
26. Heidari, M., Kazerouni, A., Kadarvish, M. S., Azad, R., Aghdam, E. K. et al. (2023). HiFormer: Hierarchical multi-scale representations using transformers for medical image segmentation. 2023 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), pp. 6202–6212. Waikoloa, HI, USA. [Google Scholar]
27. He, A., Wang, K., Li, T., Du, C., Xia, S. et al. (2023). H2Former: An efficient hierarchical hybrid transformer for medical image segmentation. IEEE Transactions on Medical Imaging, 42(9), 2763–2775. [Google Scholar] [PubMed]
28. He, K., Zhang, X., Ren, S., Sun, J. (2016). Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, Nevada, USA, IEEE. [Google Scholar]
29. Yap, M. H., Pons, G., Marti, J., Ganau, S., Sentis, M. et al. (2017). Automated breast ultrasound lesions detection using convolutional neural networks. IEEE Journal of Biomedical and Health Informatics, 22(4), 1218–1226. [Google Scholar] [PubMed]
30. Sirinukunwattana, K., Pluim, J. P., Chen, H., Qi, X., Heng, P. A. et al. (2017). Gland segmentation in colon histology images: The GlaS challenge contest. Medical Image Analysis, 35, 489–502. [Google Scholar] [PubMed]
31. Jha, D., Tomar, N. K., Ali, S., Riegler, M. A., Johansen, H. D. et al. (2021). NanoNet: Real-time polyp segmentation in video capsule endoscopy and colonoscopy. 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, IEEE. [Google Scholar]
32. Jha, D., Smedsrud, P. H., Johansen, D., de Lange, T., Johansen, H. D. et al. (2021). A comprehensive study on colorectal polyp segmentation with ResUNet++, conditional random field and test-time augmentation. IEEE Journal of Biomedical and Health Informatics, 25(6), 2029–2040. [Google Scholar] [PubMed]
33. Wang, H., Xie, S., Lin, L., Iwamoto, Y., Han, X. H. et al. (2022). Mixed transformer U-Net for medical image segmentation. CASSP 2022–2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 2390–2394. Singapore, Singapore. [Google Scholar]
34. Xu, Q., Ma, Z., Na, H., Duan, W. (2023). DCSAU-Net: A deeper and more compact split-attention U-Net for medical image segmentation. Computers in Biology and Medicine, 154, 106626. [Google Scholar] [PubMed]
35. Xie, X., Zhang, W., Pan, X., Xie, L., Shao, F. et al. (2023). CANet: Context aware network with dual-stream pyramid for medical image segmentation. Biomedical Signal Processing and Control, 81, 104437. [Google Scholar]
36. Wang, Y., Deng, Z., Hu, X., Zhu, L., Yang, X. et al. (2018). Deep attentional features for prostate segmentation in ultrasound. Medical Image Computing and Computer Assisted Intervention–MICCAI 2018, pp. 523–530. Granada, Spain, Springer. [Google Scholar]
37. Badrinarayanan, V., Kendall, A., Cipolla, R. (2017). SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 39(12), 2481–2495. [Google Scholar] [PubMed]
38. Iqbal, A., Sharif, M. (2022). MDA-Net: Multiscale dual attention-based network for breast lesion segmentation using ultrasound images. Journal of King Saud University-Computer and Information Sciences, 34(9), 7283–7299. [Google Scholar]
39. Tang, F., Wang, L., Ning, C., Xian, M., Ding, J. (2023). CMU-Net: A strong ConvMixer-based medical ultrasound image segmentation network. 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), pp. 1–5. Cartagena, Colombia, IEEE. [Google Scholar]
40. Tang, F., Ding, J., Wang, L., Xian, M., Ning, C. (2023). Multi-level global context cross consistency model for semi-supervised ultrasound image segmentation with diffusion model. arXiv preprint arXiv:2305.09447. [Google Scholar]
41. Zhao, X., Jia, H., Pang, Y., Lv, L., Tian, F. et al. (2023). M2SNet: Multi-scale in multi-scale subtraction network for medical image segmentation. arXiv preprint arXiv:2303.10894. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools