 Open Access
Open Access
ARTICLE
Transparent and Accurate COVID-19 Diagnosis: Integrating Explainable AI with Advanced Deep Learning in CT Imaging
1 Department of Information Systems, College of Computer and Information Sciences, King Saud University, Riyadh, 11543, Saudi Arabia
2 Department of Computer Engineering, College of Computer and Information Sciences, King Saud University, Riyadh, 11543, Saudi Arabia
3 Science Technology and Innovation Department, King Saud University, Riyadh, 11543, Saudi Arabia
* Corresponding Author: Mohammad Mehedi Hassan. Email:
(This article belongs to the Special Issue: Intelligent Medical Decision Support Systems: Methods and Applications)
Computer Modeling in Engineering & Sciences 2024, 139(3), 3101-3123. https://doi.org/10.32604/cmes.2024.047940
Received 23 November 2023; Accepted 17 January 2024; Issue published 11 March 2024
Abstract
In the current landscape of the COVID-19 pandemic, the utilization of deep learning in medical imaging, especially in chest computed tomography (CT) scan analysis for virus detection, has become increasingly significant. Despite its potential, deep learning’s “black box” nature has been a major impediment to its broader acceptance in clinical environments, where transparency in decision-making is imperative. To bridge this gap, our research integrates Explainable AI (XAI) techniques, specifically the Local Interpretable Model-Agnostic Explanations (LIME) method, with advanced deep learning models. This integration forms a sophisticated and transparent framework for COVID-19 identification, enhancing the capability of standard Convolutional Neural Network (CNN) models through transfer learning and data augmentation. Our approach leverages the refined DenseNet201 architecture for superior feature extraction and employs data augmentation strategies to foster robust model generalization. The pivotal element of our methodology is the use of LIME, which demystifies the AI decision-making process, providing clinicians with clear, interpretable insights into the AI’s reasoning. This unique combination of an optimized Deep Neural Network (DNN) with LIME not only elevates the precision in detecting COVID-19 cases but also equips healthcare professionals with a deeper understanding of the diagnostic process. Our method, validated on the SARS-COV-2 CT-Scan dataset, demonstrates exceptional diagnostic accuracy, with performance metrics that reinforce its potential for seamless integration into modern healthcare systems. This innovative approach marks a significant advancement in creating explainable and trustworthy AI tools for medical decision-making in the ongoing battle against COVID-19.Keywords
The COVID-19 pandemic has emerged as a defining global health crisis of the 21st century, with a staggering toll of over six million lives lost by 2023 [1]. In confronting this challenge, the medical community initially relied on Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR) tests for detection, a method praised for its specificity in detecting the SARS-CoV-2 virus [2,3]. However, the RT-PCR test has several limitations. These include its sensitivity to sample quality and timing, with studies indicating a significant rate of false negatives, especially when samples are collected in the early or late stages of infection. Additionally, RT-PCR tests can have lengthy turnaround times, which is a critical factor in pandemic management. Moreover, the demand for RT-PCR testing can lead to resource constraints, impacting test availability and accessibility [4,5].
In the wake of these challenges, medical imaging has assumed a critical role as a supplementary diagnostic tool, especially when combined with clinical assessments, epidemiological information, and laboratory results [6]. Among the imaging modalities, chest computed tomography (CT) scans have been paramount in the rapid identification and isolation of infected individuals, offering greater detail in soft tissue contrast than X-rays, which are more accessible but less precise [7,8].
The surge in infection rates has underscored the necessity for automated analysis of CT images to accelerate the detection process. The manual interpretation of these detailed scans is a time-intensive task, emphasizing the imperative for automation in the diagnostic evaluation of COVID-19 [9]. Recent developments in deep learning (DL) for COVID-19 detection via chest CT images have shown promise, with a range of pre-trained models, including VGG19, RESNet50, and DenseNet169, delivering substantial accuracy improvements [3,10,11]. The integration of transfer learning has further refined these models, enabling approaches such as DenseNet201 to excel in classifying COVID-19 cases [12–16].
Yet, the opaque nature of DL models’ decision-making processes presents a significant barrier, which the field of Explainable AI (XAI) seeks to address by rendering the workings of AI transparent and intelligible to all stakeholders [17,18]. This is achieved through the implementation of various methodologies, including but not limited to model simplification [19–21], rule extraction [17,22–24], and the deployment of sophisticated visualization techniques [25–27]. Model simplification involves reducing the complexity of ML/DL models to make their internal mechanisms and decision-making processes more understandable. Rule extraction identifies and clarifies the rules and patterns classifiers use for predictions, illuminating their logic. Additionally, advanced visualization employs graphical methods to clarify data, features, and decision processes in these models, improving user understanding. Together, these methods demystify ML/DL classifiers, building trust and reliability in their applications. However, the application of XAI to the high-dimensional data of CT scans often necessitates more complex modeling approaches than those suitable for X-ray images.
In this context, our research contributes a comprehensive framework for COVID-19 classification from CT scans that synthesizes cutting-edge DL techniques with the latest in XAI. We implement feature extraction and data augmentation strategies with pre-trained deep convolutional neural network (CNN) models, leading to a high-performing, generalizable deep neural network (DNN) classification model. Our design incorporates a carefully curated selection of fully connected, dropout, and batch normalization layers to enhance the post-feature extraction classification efficacy.
We extend the capabilities of our framework by integrating the local interpretable model-agnostic explanations (LIME) technique, which explicates the decision-making process for AI-driven classifications. This innovative approach promises not only to set a new standard for COVID-19 detection from CT scans but also to advance the medical diagnostic field toward a future where AI decision-making is both interpretable and reliable. The key contributions of our work include:
• The use of DenseNet201 architecture for extracting features indicative of COVID-19 from CT scan images can eliminate the vanishing gradient problem, improve feature reuse and feature propagation, and reduce the number of parameters. Moreover, the combination of extracted features by DenseNet201 with the average global pooling layer reduces the overfitting problem and unnecessary use of memory.
• The employment of data augmentation to diversify the training data enhances the model’s ability to generalize and reduces overfitting.
• The application of XAI methods to ensure transparent classification processes, particularly through the use of LIME to clarify distinguishing features of COVID-19 in CT scans.
• The validation of our approach on a publicly available, extensive dataset of CT scans for SARS-CoV-2 detection [28].
• The advancement of AI in medical diagnostics toward a model that combines accuracy with transparency and trustworthiness.
Through this multifaceted approach, we aim to establish a new benchmark for the detection and classification of COVID-19 from CT images and to push the field toward transparent, explainable, and reliable AI in medical diagnostics.
The rest of this paper is organized as follows. Section 2 presents the work done in the field of deep learning techniques and XAI methods for chest CT images. Section 3 describes background knowledge about CNN and DenseNet. The proposed technique is described in Section 4. The experimental results and discussions are given in Section 5. The concluding remarks are drawn in Section 6.
Extensive research has been conducted to detect COVID-19 through radiological imaging, with chest CT scans demonstrating lower false positive rates compared to other techniques such as X-rays. In their pioneering work, Farjana et al. [3] harnessed transfer learning with pre-trained models such as VGG19, RESNet50, and DenseNet169 to diagnose COVID-19 from CT scans, overcoming challenges of limited data due to privacy constraints. The DenseNet169 model was distinguished by its exceptional performance, achieving 98.5% accuracy in binary classification, outshining its counterparts.
Joshi et al. [10] explored a transfer learning framework using the VGG16 model, concentrating on lung CT scans reflective of the virus’s primary impact. They employed preprocessing methods like CLAHE and data augmentation to enhance the dataset, culminating in a high degree of accuracy and precision, each marked at 95.
In response to the critical need for prompt diagnosis, Gupta et al. [29] developed a deep learning-based system for automated COVID-19 detection from CT scans. Leveraging a sizable dataset, their innovative approach utilized both established and new models, with the DarkNet19 model attaining an impressive 98.91% accuracy through rigorous validation.
Perumal et al. [30] introduced the DenSplitnet model, which utilized Dense blocks in conjunction with Self-Supervised Learning for pre-training. The model’s unique dual-pathway approach at the classification stage significantly enhanced generalizability, achieving notable accuracy and precision metrics on a dedicated dataset.
Ibrahim et al. [31] unveiled the COV-CAF framework, which integrates conventional and deep learning techniques to categorize COVID-19 severity from 3D chest CT volumes. Their two-phased methodology, combining fuzzy clustering for segmentation and a hybrid of automatic and manual feature extraction, demonstrated its potential by achieving high accuracy and sensitivity on diverse datasets.
Gaur et al. [32] proposed a system utilizing empirical wavelet transformation for CT image preprocessing, which showcased high classification accuracy and an impressive AUC score, signaling its clinical efficacy.
Soares et al. [28] constructed an explainable Deep Learning model (xDNN) that reported an accuracy of 88.6%. In a similar vein, Lu et al. [33] developed a CGENet model rooted in graph theory, which achieved an accuracy of 97.78%. Concurrently, Basu et al. [34] employed a feature selection strategy, attaining an identical peak accuracy and a substantial precision-recall curve score.
Jaiswal et al. [12] examined various deep learning architectures for COVID-19 patient classification, identifying DenseNet201-based deep transfer learning as the most accurate, achieving a 96.25% accuracy rate. Basu et al. [34] introduced a composite framework for COVID-19 detection from CT scans, which incorporated deep learning with optimization techniques, achieving high accuracy rates with their novel method.
Recent efforts have focused on employing explainable AI to demystify COVID-19 detection. Boutorh et al. [24] utilized CNNs for image analysis and machine learning algorithms for symptom data, employing LIME for greater transparency, with the CNN model exhibiting 96% accuracy and F1-score.
Ye et al. [22] proposed an explainable classifier that assists radiologists by using LIME and Shapley values to assess the contribution of individual superpixels in the diagnosis, enabling both local and global interpretability.
Chadaga et al. [19] put forward a decision support system that integrates machine learning and deep learning to diagnose COVID-19 using an ensemble of clinical and blood marker data, with a multi-level stacked model showcasing exceptional accuracy and interpretability.
Mahmoudi et al. [23] introduced a method to increase the interpretability of DL algorithms in image classification, with a focus on X-ray and CT scans for COVID-19 diagnosis, while Mercaldo et al. [17] proposed a transfer learning model that classifies CT scans across three categories, enhancing the diagnostic process. Rostani et al. [35] applied an explainable model for COVID-19 diagnosis, with their model achieving significant accuracy and specificity, guided by the XAI technique of random forest.
While these advancements signify progress, the current XAI methods still struggle with capturing the multifaceted three-dimensional interactions in CT images.
The application of machine learning algorithms has been revolutionary across numerous sectors, particularly in healthcare for disease prognosis, diagnostics, image analysis, object detection, signal processing, and natural language processing. This study employs a sophisticated deep learning model known as DenseNet201, rooted in the Convolutional Neural Network (CNN) architecture. This background section aims to concisely describe the CNN and DenseNet frameworks to provide a foundational understanding for the subsequent discussions.
3.1 Convolutional Neural Network
The Convolutional Neural Network (CNN), often inspired by biological visual systems, is a multilayered neural architecture designed for processing data with a grid-like topology, such as images. In CNNs, initial layers are tasked with feature extraction identifying edges, textures, and other elementary patterns. These are then composed into higher-order features in subsequent layers [36,37]. To manage the high dimensionality of these features, pooling operations are employed, typically following the convolution layers. The resulting feature maps are then relayed into a fully connected neural network that classifies the images by employing back-propagation algorithms during training. The intricate layers of processing that include convolutional, pooling, and fully connected nodes, endow CNNs with the ability to achieve high accuracy and robust performance, particularly in image processing and computer vision applications.
One challenge in feature mapping within convolutional layers is the exact localization of features; minute shifts in the input image can lead to entirely different feature maps. This sensitivity is often mitigated through downsampling strategies, which while reducing resolution, retain essential structural elements, thereby enhancing classification robustness. Downsampling is commonly achieved by modifying the convolution stride or implementing pooling layersamax or average post-convolution. The application of nonlinear functions, such as the Rectified Linear Unit (ReLU), precedes pooling, adding to the network’s complexity and capacity for feature distinction. A standard CNN pipeline thus consists of sequential layers: input, convolution, nonlinearity application, and pooling. Network fine-tuning, an essential phase, optimizes the model’s hyperparameters for improved performance. The operations within a typical convolutional layer are mathematically represented as:
where
The max pooling operation for the
where
Here
3.2 Dense Convolutional Network (DenseNet)
DenseNet, a paradigm shift in deep learning network design, advances the foundational work of its precursor, ResNet, by enhancing feature propagation and reuse. Pioneered by Huang et al. [38], DenseNet’s architecture is lauded for its innovative connectivity and feature concatenation, which are its defining characteristics.
• Layer Connectivity: DenseNet’s hallmark is its exhaustive connectivity, where each layer receives concatenated inputs from all previous layers, formalized as:
where
• Feature Integration: Utilizing concatenation rather than summation, DenseNet fortifies feature propagation within the network, thereby enhancing feature diversity and network capacity.
3.2.2 Mathematical Formulation
Mirroring the Taylor series expansion, DenseNet can be conceptualized as a series expansion of features, where the network depth equates to higher-order terms in the series. This is expressed mathematically as a multi-layer composite function:
where each
3.2.3 Structural Implementation
Dense Blocks: The ‘dense block’ is the cornerstone of DenseNet, where each layer within the block is a convolutional block that enriches the feature set for subsequent layers. This is captured by:
Transition Layers: To control the model’s parameter growth, transition layers compress the channel dimension, often through convolution and pooling operations:
1. The architecture initiates with a convolutional layer, which is succeeded by a max-pooling operation, reminiscent of the initial layers observed in ResNet.
2. It is organized into four dense blocks, each potentially comprising a predetermined count of convolutional layers. A configuration analogous to that of ResNet-18 might feature four layers per block.
3. Interspersed with transition layers, the network effectively manages the feature maps’ dimensionality throughout the depth of the model.
4. The architecture culminates with a global pooling layer, which seamlessly transitions into a fully connected layer, producing the final classification output.
The proposed methodology is to develop and train a deep learning framework, tailored to classify COVID-19 infection from chest CT-scan imagery. Subsequent to the classification process, an XAI technique is employed to elucidate the diagnostic rationale by highlighting the pertinent features within the image that underpin its categorization. This dual-step process not only provides a binary classification but also augments the interpretability of the model, offering a transparent window into the decision-making process that is vital for clinical validation and trust. The schematic architecture of the proposed methodology is depicted in Fig. 1. A detailed description of the proposed deep learning model and XAI method are given in the subsequent subsections.

Figure 1: Schematic view of the proposed methodology
4.1 Proposed Deep Learning Model
Our proposed methodology integrates the use of a pre-trained Densenet-201 neural network model with XAI techniques to enhance the classification of COVID-19 from chest CT scans. The choice of DenseNet201 as the primary architecture for our deep learning model was driven by several factors. DenseNet201 is known for its efficiency in feature propagation and reuse, which is crucial for medical image analysis where subtle features can be significant for accurate diagnosis. This architecture, with its densely connected layers, ensures minimal information loss, an essential factor for the detailed and nuanced features present in CT scans. Additionally, DenseNet201 has a lower parameter count compared to other complex models like ResNet or VGG, which reduces the risk of overfittingaa vital consideration given the limited size of medical imaging datasets. Moreover, DenseNet201’s architecture, with its inherent feature reuse capability, allows for deeper network construction without a proportional increase in computational complexity. This characteristic is particularly beneficial for detecting COVID-19 in CT scans, where the identification of intricate patterns associated with the disease requires deep and complex network architectures.
As illustrated in Fig. 1, the process begins with the collection and pre-processing of CT images, which includes resizing, normalization, and formatting to meet the input specifications of the Densenet-201 model.
The dataset is partitioned into training and testing sets with an 80:20 ratio. Feature extraction is performed via the Densenet-201 model, yielding a feature set shaped into a 4 × 4 × 1920 matrix, which is then reduced in dimensionality through 2D Global Average Pooling. To further enhance the model’s predictive power, the training data undergoes augmentation to introduce a richer variety of patterns.
Following this, we fine-tune and adapt the pre-trained network on the COVID-19 dataset by adjusting the parameters of the last
where
New layers are incorporated to replace the final classification layers, aligning the network’s output to the task at hand, described as:
with
Hyperparameters are meticulously tuned, and regularization strategies such as dropout are implemented to mitigate overfitting, formulated as:
where
In this paper, we apply a combination of different fully connected layers, dropout layers, and batch normalization to achieve better classification performance for COVID-19 CT-scan data. In other words, we have designed and developed four different DenseNet201 architectures by varying the layers in a post-processing deep learning structure. These four distinct deep learning approaches are referred to as Model-1, Model-2, Model-3 and Model-4. The corresponding structure of each model is shown in Figs. 2–5. These configurations represent variations in the architecture intended to optimize the model’s performance by adjusting the depth and complexity of the network.

Figure 2: Model-1 DenseNet201 architecture

Figure 3: Model-2 DenseNet201 architecture
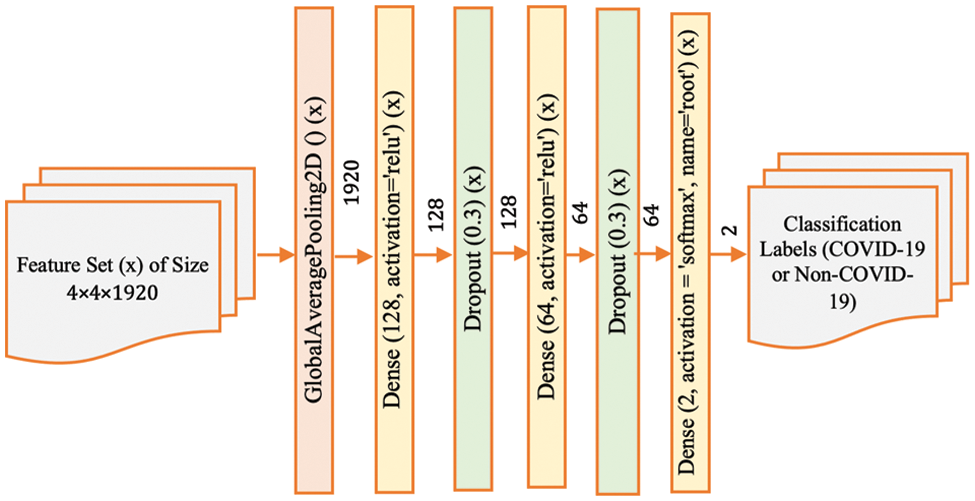
Figure 4: Model-3 DenseNet201 architecture
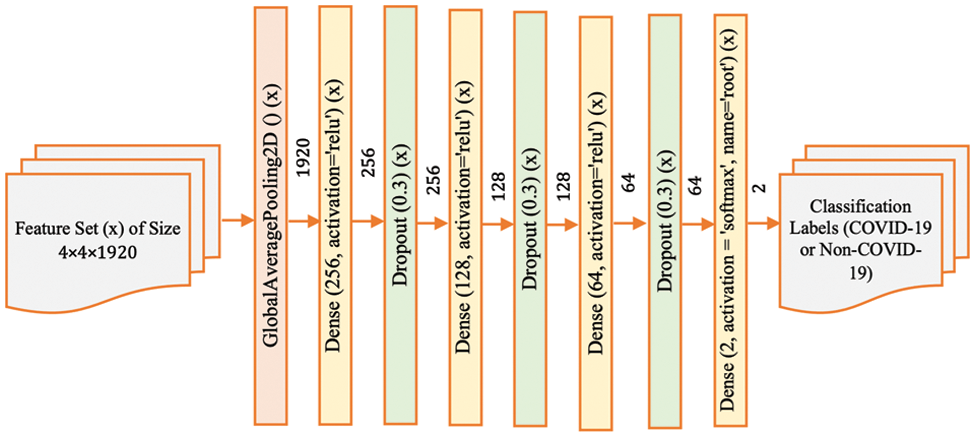
Figure 5: Model-4 DenseNet201 architecture
Model-1 (Fig. 2) presents the base architecture, where the feature set extracted from CT scan images is passed through a global average pooling layer, followed by a sequence of dense layers with activation functions and dropout layers to prevent overfitting. The final output layer uses a softmax activation function to produce the classification labels for COVID-19 or non-COVID-19.
Model-2 (Fig. 3) modifies the base architecture by adjusting the neuron count in the dense layers, potentially enhancing the model’s ability to capture more complex patterns in the data. This change aims to balance the model’s capacity with the available training data, seeking to improve generalization without significantly increasing the risk of overfitting.
Model-3 (Fig. 4) further adjusts the architecture by introducing additional dropout layers. These layers are strategically placed to introduce more robustness to the network, enabling it to maintain performance on unseen data by reducing the reliance on any single feature during training. This helps to ensure that the network’s predictions are based on a broader set of features, which can be particularly useful when dealing with medical images where certain features might be more subtle or nuanced.
Finally, Model-4 (Fig. 5) represents the most complex variant, with multiple dense and dropout layers. This configuration is designed to explore the network’s capacity to discern intricate and potentially abstract features within the CT scans that are indicative of COVID-19. By incorporating additional layers, the network has the potential to model more complex relationships within the data. However, this also increases the model’s parameters, requiring careful tuning to avoid overfitting, especially if the amount of training data is limited.
Each configuration’s contribution lies in its potential to identify the most effective architecture for the specific task of COVID-19 classification from CT images. By evaluating the performance of each model variation, we can determine the optimal balance between model complexity and predictive power. This systematic approach allows for an empirical assessment of how changes in the neural network’s structure affect its ability to diagnose COVID-19 accurately. The resulting insights can not only inform the development of more effective diagnostic tools for COVID-19 but also contribute to the broader field of medical image analysis, where model interpretability and reliability are of paramount importance.
4.2 Explainable Artificial Intelligence
Explainable Artificial Intelligence (XAI), a subset of artificial intelligence that focuses on clarifying the reasoning behind decisions made by AI models, has gained prominence. Take, for instance, a scenario where our developed AI system identifies a CT-Scan image as non-COVID. XAI would provide the rationale behind the system’s diagnosis as non-COVID. To illustrate the significance of interpretability/explainability, consider the following scenario.
Scenario: Imagine a deep learning algorithm correctly identifies an image as a cat. It is beneficial to understand the factors influencing this decision. The justification might be the presence of characteristics like fur, whiskers, and claws, which led to its classification as a cat. Such insights can enhance the confidence of users in the autonomous system’s decisions.
Various interpretive tools are available to elucidate the rationale behind a classifierâs judgments. In our research, we employ the well-known Local Interpretable Model-agnostic Explanations (LIME) technique. Here, we concisely expound on the LIME algorithm, as detailed in a referenced study.
Within LIME, the classification of the ith image, denoted as
Here, G represents the class of potential interpretable models.
In the LIME framework, Eq. (12) explains the decision
Contrary to many XAI techniques such as Grad-Cam and Tylor decomposition [21] that provide overall understanding, LIME offers specific interpretability by providing explanations for particular forecasts. The emphasis on local attention is especially well-suited for medical imaging, as the analysis of individual instances, rather than overall patterns, is frequently more therapeutically significant. LIME’s capacity to identify the exact areas inside CT scans that influence the model’s judgments enables doctors to comprehend the diagnostic reasoning on an individual basis. This approach has an advantage over other strategies that may only provide a general measure of the value of elements without specifically identifying their spatial relevance within the image.
5 Experimental Setup and Findings
In our research, we have utilized the SARS-CoV-2 CT scan dataset compiled by Soares et al. [28], which encompasses a total of 2481 2D CT images. This dataset is bifurcated into two subsets: one containing 1252 CT scans from patients diagnosed with COVID-19, and the other comprising 1229 CT scans from individuals not infected by the virus. We sourced our data from a cohort of 120 patients based in Sao Paulo, Brazil, with the COVID-19 positive group including 32 males and 28 females, and the uninfected group consisting of an equal number of males and females, 30 each. Access to the dataset is available at www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset. The two classes of the dataset can be shown in Fig. 6.
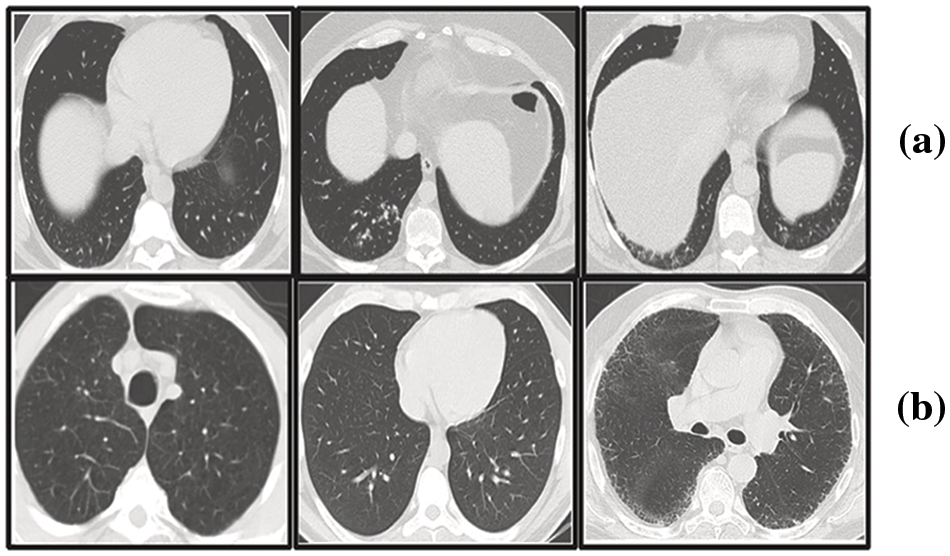
Figure 6: Representative sections from the SARS-COV-2 2D-CT-scan collection featuring both COVID-positive and COVID-negative cases
The proposed COVID-19 XAI framework in this study is implemented on a Google Colab environment with Keras, TensorFlow, and some other required libraries built with Python 3.6. The resources offered to the active session are hardware accelerator T4 GPU, 78.2 GB disk, and 12.7 GB system RAM. The dataset of CT images is divided randomly into 80% for the training set and 20% for the test set. From the training set, 10% randomly is selected for the validation set. The distribution of classes within the training, validation, and test sets is given in Table 1.

In our study, we modified the size of CT images to 128 × 128 pixels, diverging from the default 244 × 244 pixel input size of the pre-trained model. This resizing is critical for preserving the intricate details in the CT images without distortion, thereby enhancing both the efficiency and performance of the model. Our training set underwent a series of augmentation processes that retained the integrity of the original CT scans. These processes included random rotations within a 20-degree range, horizontal shifts up to a 0.2 ratio, vertical shifts up to a 0.2 ratio, and random horizontal flips.
For the classification phase, the models were trained over 100 epochs with a batch size of 32. The ImageDataGenerator class from the Keras library facilitated the augmentation of our training dataset. This tool generated a diverse set of augmented images for each epoch through random transformations applied to the training images. With a training set of 1785 images and a batch size of 32, we achieved 56 unique images per epoch. The entire training process for each model spanned approximately one hour. An adaptive learning rate was employed, initially set at 0.003 and decreasing by a factor of 0.7 when validation accuracy plateaued beyond a predefined patience number, set at 10. The minimum learning rate was capped at 0.0000001. The âadamâ optimizer was used for training the models. Regarding the training of the XAI model interpretation, the average duration was less than one minute.
Note that the study’s dataset, collected by Soares et al. [28] may not capture the global diversity in COVID-19 cases. Factors like age, gender, ethnicity, and underlying health conditions, which affect COVID-19’s radiographic presentation, could lead to variations in the model’s performance across different demographics. Furthermore, the dataset’s image quality, including variations in resolution and contrast of the 2D CT images, might impact the model’s generalizability and accuracy due to differences in imaging equipment and protocols.
To assess the performance of the COVID-19 detection part of the framework, we utilized the following performance metrics:
• Accuracy is the proportion of true results among the total number of cases examined.
• Precision measures the proportion of true positive identifications among all positive identifications made by the model.
• Recall (also known as sensitivity) assesses the proportion of actual positives that were correctly identified.
• F1-score is the harmonic mean of precision and recall, providing a single score that balances both concerns.
5.4 Experimental Results Analysis and Discussion
This section delineates and analyzes the performance outcomes of the proposed four models, specifically regarding precision, recall, F1-score, and accuracy. The trajectory of model accuracy throughout the training process is illustrated in Fig. 7.
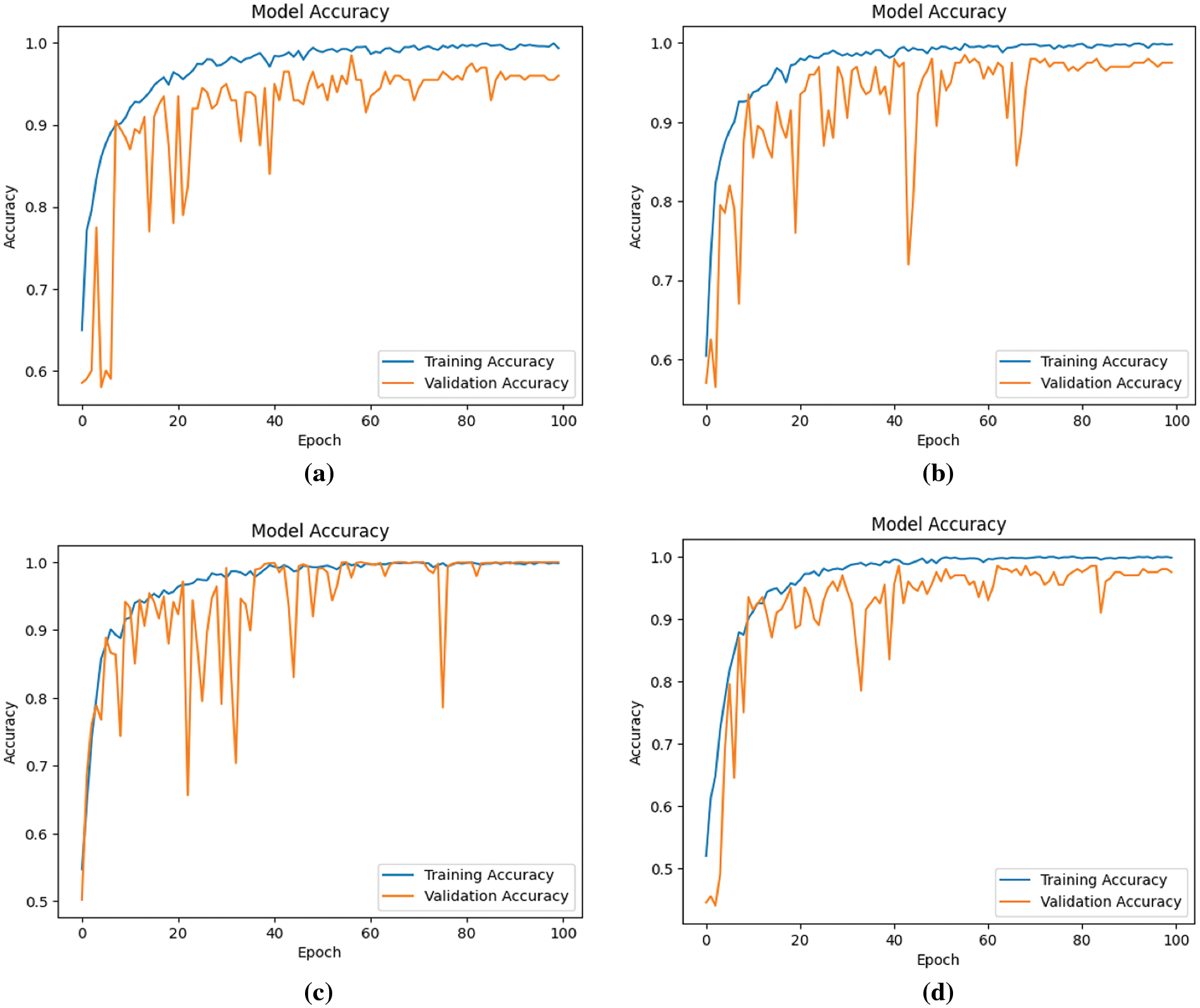
Figure 7: Accuracy of four models during the learning process (a) Model-1 (b) Model-2 (c) Model-3 (d) Model-4
Observations from Fig. 7 indicate an overall ascent in both training and validation accuracy across the models, notwithstanding minor fluctuations in validation accuracy. This enhancement in model performance, as reflected by an increasing number of epochs, suggests an improvement in learning efficacy. Concurrently, Fig. 8 displays the models’ loss during the learning progression.
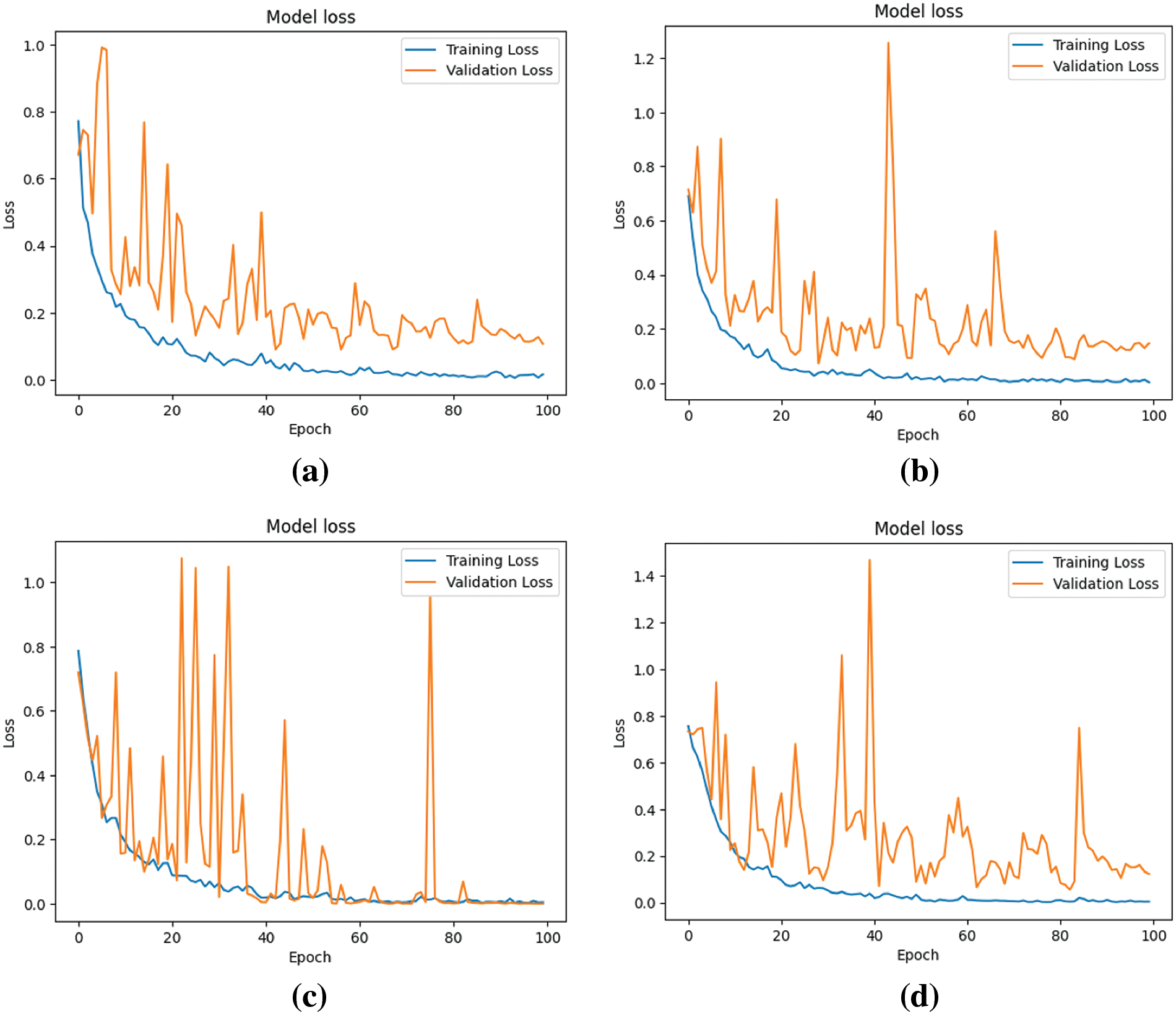
Figure 8: Loss of four models during the learning process (a) Model-1 (b) Model-2 (c) Model-3 (d) Model-4
Fig. 8 elucidates the models’ loss or cost, representing the errors encountered during learning. A trend is discerned where, across epochs, there is a decline in loss, implying enhanced model performance, despite nominal variances in validation loss.
The analysis of Figs. 7 and 8 indicates that Model 3 demonstrates a consistent alignment between the training and validation curves. This consistency suggests a substantial reduction in the overfitting issue during Model 3’s learning process, signifying an improvement in performance in comparison to the other models.
After the models’ training, the test dataset is employed to derive classification metrics. Fig. 9 delineates the confusion matrices for the four models in question.
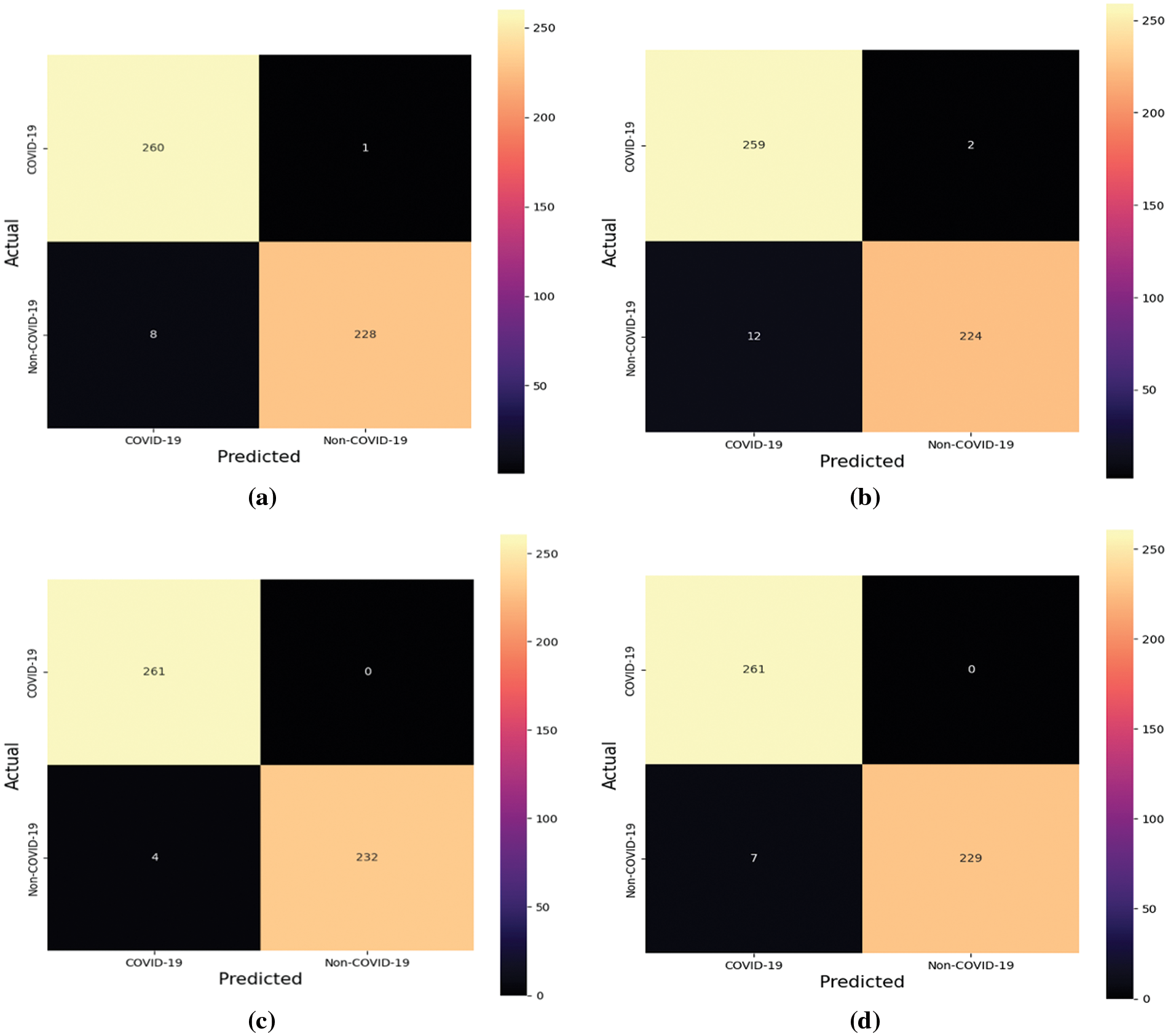
Figure 9: Confusion matrices of the four models (a) Model-1 (b) Model-2 (c) Model-3 (d) Model-4
Expounding on the data from the confusion matrices, additional evaluative metricsaprecision, recall, F1-score, and accuracyaare enumerated in Tables 2–5. These metrics provide a quantitative foundation for assessing the classification powers of the models post-training.




Table 2 portrays the performance of Model-1, which demonstrates high precision and recall, particularly for the COVID-19 class, leading to a noteworthy overall accuracy of 98.19%. The near-perfect recall indicates the model’s sensitivity in correctly identifying COVID-19 cases. The weighted average scores closely mirror the high accuracy, underscoring the model’s balanced performance across both classes.
Table 3 details the evaluation of Model-2, where we observe a slight decline in precision for the COVID-19 class compared to Model-1. Nevertheless, the model maintains high recall and achieves an aggregate accuracy of 97.18%. This suggests that while Model-2 is slightly less precise, it continues to identify COVID-positive cases reliably.
Table 4 delineates the results for Model-3, which excels with an exceptional recall of 1.0000 for COVID-19, indicating that all COVID-positive cases in the test set were correctly classified. Coupled with perfect precision for non-COVID-19 and an overall accuracy of 99.20%, Model-3 stands out as the superior model among the four, showcasing an exemplary balance between sensitivity and specificity.
Table 5 provides insights into Model-4, which, akin to Model-3, achieves a recall of 1.0000 for COVID-19. However, it has a slightly lower precision for the COVID-19 class than Model-3. With an overall accuracy of 98.59%, Model-4’s performance is robust, yet marginally less optimal than Model-3.
The precision metric across the models indicates the proportion of true positives against all positive calls made by the model, while recall illustrates the model’s ability to find all relevant cases within the dataset. The F1-score provides a harmonic mean of precision and recall, offering a single measure of the model’s accuracy per class. The support denotes the number of actual occurrences of the class in the specified dataset.
In summation, these tables collectively highlight the strengths and limitations of each model, with Model-3 demonstrating the most promising results. The meticulous evaluation through these metrics is vital, as it ensures that the models are not only accurate but also equitable in their predictions, an essential consideration in the clinical deployment of AI tools. However, our methodology, while robust, relies on a pre-trained Densenet-201 neural network model, which might limit its ability to capture novel features specific to COVID-19 that were not present in the data used for its initial training. Furthermore, our model variations, designed to optimize performance, require extensive computational resources for training and evaluation, which may not be readily available in all clinical settings.
5.5 Performance Analysis with Other State-of-the-Art Depp Learning Models
We have also compared the proposed deep learning model with other state-of-the-art deep learning methods on the same SARS-CoV-2 CT scan dataset [28]. The results are presented in Table 6.

The proposed Model-3, as seen in Table 6, demonstrates superior performance compared to the other models presented in the table, achieving an accuracy of 0.9920. This implies that Model-3 exhibits a high level of accuracy in identifying cases of COVID-19, and its predictions hold significance and use.
Upon doing a comparative analysis between the suggested Model-3 and other models, namely VGG-19, ResNET-50, and different iterations of DenseNet, it becomes evident that Model-3 exhibits a higher level of performance. Model-3 consistently exhibits superior performance across all criteria, even when compared to more sophisticated models such as COV-CAF and xDNN. This finding suggests that the performance of Model-3 is notable in accurately detecting the proper condition while also mitigating the occurrence of false positives and false negatives. Consequently, the model exhibits a well-balanced and dependable nature, making it suitable for clinical diagnostics.
The exemplary performance in terms of precision and recall indicates that Model-3 exhibits a high level of proficiency in accurately detecting genuine instances of COVID-19 while minimizing errors. This attribute holds significant importance in a medical setting, where the consequences of false negatives can be quite detrimental. In a similar vein, the optimal F1-score demonstrates an optimal equilibrium between precision and recall, making it particularly suitable for medical diagnostics. In this context, it is imperative to avoid overlooking genuine cases (high recall) while simultaneously preventing an excessive number of false alarms (high precision).
In summary, Table 6 presents a comprehensive overview of the efficacy of the proposed Model-3 in the diagnostic process of COVID-19 using CT scans. This highlights the potential of Model-3 as a dependable instrument within the realm of medical imaging.
In our paper, we emphasize the importance of ethical considerations in deploying AI within healthcare, with a strong focus on patient privacy and data security. We detail the measures taken to ensure the confidentiality and security of patient data, acknowledging this as a crucial aspect of our AI system’s design and operation. Our approach is guided by ethical principles such as transparency, ensuring clear communication about the AI system’s decision-making processes, fairness, aiming to prevent any form of bias, and accountability, defining clear responsibilities for AI-driven outcomes.
5.6 Explanation of the Decision Made by Deep Learning Classifier Using LIME
In the medical domain, physicians and professionals often seek to understand why a patient is diagnosed as COVID-19 positive, in addition to simply classifying the patient as suffering from the virus. We applied an XAI approach to justify the classifications achieved using CT-Scan images. Our goal was to elucidate the decisions made by our automated computational method for each patient. For this purpose, we employed the LIME approach to explain the classification outcomes generated by the deep neural network used in our study.
The CT-scan images presented in Fig. 10 illustrate the original input CT-scan image alongside the regions of interest that led to a COVID-19 classification. The first column in Fig. 10 shows the actual CT-scan images inputted into our automated deep learning system. A typical radiologist would look for the following imaging characteristics in COVID-19 cases through CT scans:
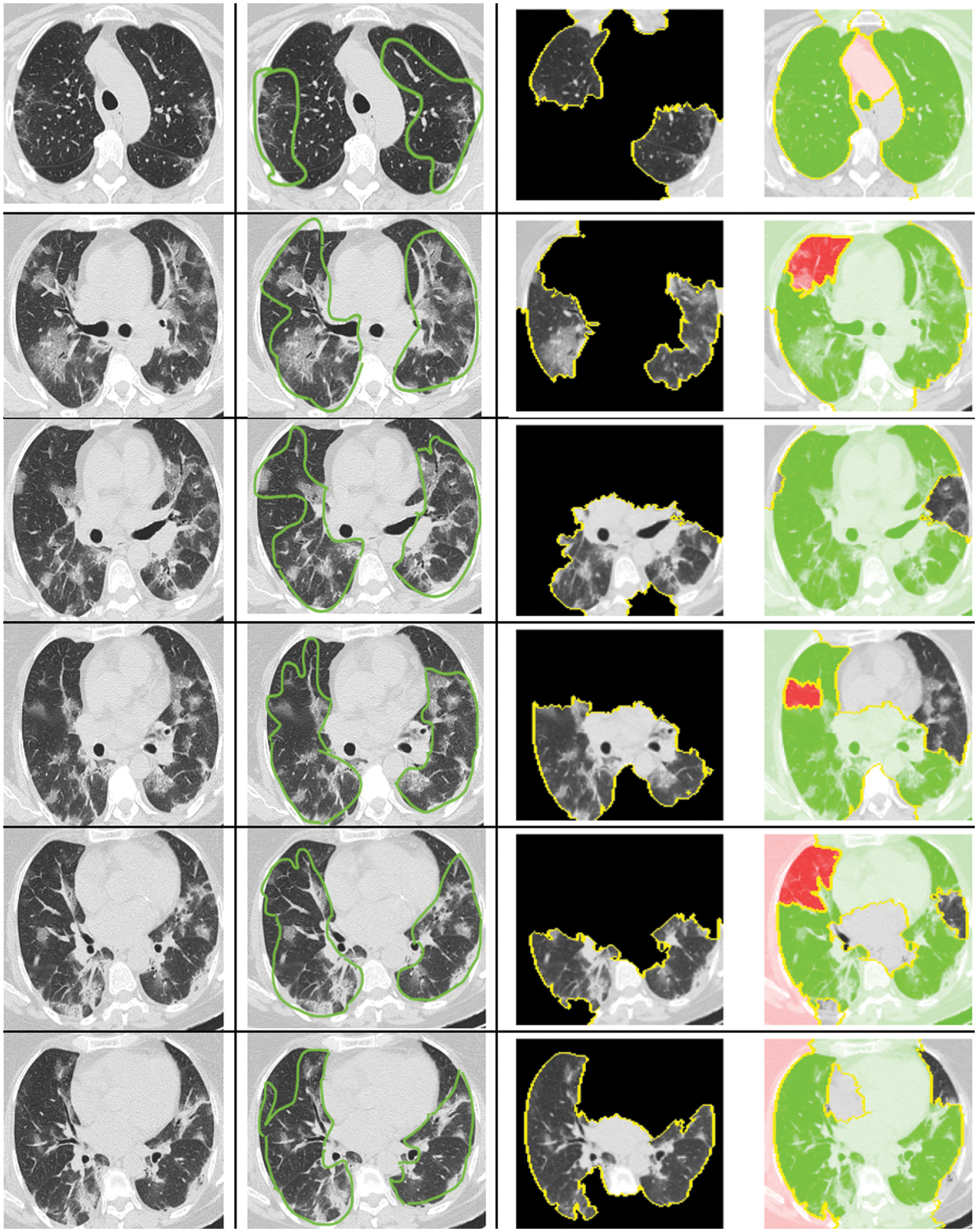
Figure 10: Explanation for the decision of classifying as COVID or non-COVID using LIME
• Ground-Glass Opacities (GGOs): These appear as hazy, opaque areas that do not obscure the underlying lung structures and are a common early finding in COVID-19.
• Bilateral and Peripheral Distribution: COVID-19 often affects both lungs, with abnormalities more commonly found along the outer edges.
• Multifocal Involvement: Multiple lung areas are often involved, indicated by several patches of ground-glass opacities.
• Consolidation: In severe cases, ground-glass opacities may progress to consolidation, where the lung tissue becomes more solid.
• Crazy-Paving Pattern: This is identified by ground-glass opacities with superimposed interlobular septal thickening and intralobular lines.
• Vascular Enlargement: This is seen as enlarged blood vessels in the CT-scan image, part of the inflammatory response.
• Absence of Features Typically Seen in Other Respiratory Diseases: Such as the lack of pleural effusions, lymphadenopathy, and cavitation.
As shown in column 2 of Fig. 10, the highlighted regions satisfy the majority of these characteristics. These annotations were made by a professional radiologist without prior knowledge of the patients’ COVID-19 status. Intriguingly, our computational approach accurately diagnosed these patients, and the XAI identified the correct region in the CT-scan image that a radiologist would find relevant.
The XAI process segments the raw CT-scan image into several partitions worthy of investigation. It then produces an image highlighting only the region of interest, as seen in column 3 of Fig. 10. This region contributes significantly to identifying a patient as COVID-19 positive using our automated approach. The identified region of interest aligns with what professional radiologists would consider a significant indication of COVID-19. Notably, the LIME approach emphasized the regions showing GGOs, leading to the classification of the patient as COVID-19 positive. Furthermore, the regions of interest annotated by the radiologist were also deemed important by the XAI approach.
What stands out in this analysis is that the LIME methodology appears to produce results that largely align with the regions marked by the radiologist, suggesting a high degree of accuracy. This alignment is crucial, as it not only validates the model’s diagnostic capabilities but also enhances radiologists’ trust in the AI system, providing a clear and interpretable rationale for its decisions. Such interpretability is essential in a clinical setting, where understanding the basis for AI-driven conclusions can directly inform patient care decisions and strategies.
Our model’s interpretability through LIME provides clinicians with understandable visual cues that delineate the AI’s reasoning, which is particularly beneficial in several key areas. Clinicians can corroborate AI-generated diagnoses with their expertise, enhancing diagnostic confidence, particularly in ambiguous cases. The visual cues provided by our model can assist in precise treatment planning by revealing the disease’s extent and guiding patient management strategies. Moreover, these explanations facilitate better communication with patients about their condition and the reasoning behind treatment recommendations, potentially improving compliance. Furthermore, the model serves as an educational tool, aiding in the professional development of less experienced radiologists by illuminating key radiographic features of COVID-19. Lastly, by highlighting critical areas in the scans, our model helps radiologists efficiently prioritize their review process, which is especially beneficial in high-volume clinical settings, reducing cognitive load and aiding in risk stratification for more personalized patient care.
This study presented an integrated framework that combined advanced deep learning techniques with Explainable Artificial Intelligence to identify COVID-19 from CT scan images. Leveraging LIME, our model demonstrated an ability to produce interpretable results that closely align with the annotations of clinical experts. Throughout the research, the effectiveness of our approach was systematically evaluated against multiple metrics, including precision, recall, F1-score, and accuracy. The model’s performance was meticulously recorded over numerous training epochs, revealing a consistent improvement in learning and an increase in the capacity to generalize from the training data to validation data.
The comparison of regions highlighted by the LIME algorithm against those marked by clinicians showed a significant overlap, substantiating the model’s diagnostic accuracy. This alignment is particularly noteworthy, as it suggests that the model is not only recognizing the correct features for COVID-19 identification but is also aligning with the clinical understanding of the disease’s radiographic manifestations. Our approach advanced the implementation of XAI in medical imaging, addressing the pressing need for models that are both accurate and transparent in their decision-making processes. The transparency achieved by applying LIME techniques facilitated a deeper understanding of the model’s reasoning, enabling clinicians to trust and interpret AI-driven diagnoses.
Validation of the model on a publicly available dataset confirmed its efficacy, showcasing the potential to integrate such AI systems into healthcare environments to support and enhance diagnostic processes. The results obtained suggest that our approach can serve as a benchmark for future studies aiming to refine the application of AI in detecting and understanding COVID-19 and other pathologies through medical imaging. Our research underscored the importance of XAI in the medical field, where explainability is not a luxury but a necessity. Our study contributes to the body of knowledge by providing a robust model that not only excels in diagnostic accuracy but also the clarity of its interpretative output, thus paving the way for future developments in AI-assisted healthcare diagnostics. We suggest future studies to compare the efficacy of LIME with other XAI techniques, which could provide a broader understanding of the model’s interpretability. Expanding the application of our model to other diseases presents another avenue for research, potentially broadening the scope of AI-assisted diagnostics in the medical field.
Acknowledgement: We would like to extend our sincere thanks to Dr. Sylveea Mannan, a certified radiologist from Canada, for her expert annotation of regions of interest in COVID-19 CT images, a crucial aspect for our research in XAI.
Funding Statement: The authors extend their appreciation to the Deanship for Research Innovation, Ministry of Education in Saudi Arabia, for funding this research work through project number IFKSUDR-H122.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Mohammad Mehedi Hassan and Salman A. AlQahtani; data collection: Mabrook S. AlRakhami and Ahmed Zohier; analysis and interpretation of results: Mohammad Mehedi Hassan and Salman A. AlQahtani; draft manuscript preparation: Mohammad Mehedi Hassan, Salman A. AlQahtani, Mabrook S. AlRakhami and Ahmed Zohier. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data used to support the findings of the study are available in the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Organization, W. H. (2024). WHO COVID-19 dashboard. https://data.who.int/dashboards/COVID19/cases?n=c (accessed on 07/01/2024). [Google Scholar]
2. Wang, W., Xu, Y., Gao, R., Lu, R., Han, K. et al. (2020). Detection of SARS-CoV-2 in different types of clinical specimens. Jama, 323(18), 1843–1844. [Google Scholar] [PubMed]
3. Farjana, A., Liza, F. T., Al Mamun, M., Das, M. C., Hasan, M. M. (2023). SARS COVIDAID: Automatic detection of SARS CoV-19 cases from CT scan images with pretrained transfer learning model (VGG19, RESNet50 and DenseNet169) architecture. 2023 International Conference on Smart Applications, Communications and Networking (SmartNets), pp. 1–6. Istanbul, Turkey, IEEE. [Google Scholar]
4. Pathak, Y., Shukla, P. K., Tiwari, A., Stalin, S., Singh, S. (2022). Deep transfer learning based classification model for COVID-19 disease. Irbm, 43(2), 87–92. [Google Scholar] [PubMed]
5. Zu, Z. Y., Jiang, M. D., Xu, P. P., Chen, W., Ni, Q. Q. et al. (2020). Coronavirus disease 2019 (COVID-19A perspective from China. Radiology, 296(2), E15–E25. [Google Scholar] [PubMed]
6. Xie, X., Zhong, Z., Zhao, W., Zheng, C., Wang, F. et al. (2020). Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: Relationship to negative RT-PCR testing. Radiology, 296(2), E41–E45. [Google Scholar] [PubMed]
7. Singh, D., Kumar, V., Vaishali, Kaur, M. (2020). Classification of COVID-19 patients from chest CT images using multi-objective differential evolution-based convolutional neural networks. European Journal of Clinical Microbiology & Infectious Diseases, 39, 1379–1389. [Google Scholar]
8. Yu, T. T., Wang, J. Q., Wu, L. T., Xu, Y. (2019). Three-stage network for age estimation. CAAI Transactions on Intelligence Technology, 4(2), 122–126. [Google Scholar]
9. Kaur, J., Kaur, P. (2023). A CNN transfer learning-based automated diagnosis of COVID-19 from lung computerized tomography scan slices. New Generation Computing, 41(4), 795–838. [Google Scholar]
10. Joshi, K. K., Gupta, K., Agrawal, J. (2023). An efficient transfer learning approach for prediction and classification of SARS–COVID-19. Multimedia Tools and Applications. https://doi.org/10.1007/s11042-023-17086-y [Google Scholar] [CrossRef]
11. Mishra, A. K., Das, S. K., Roy, P., Bandyopadhyay, S. (2020). Identifying COVID19 from chest CT images: A deep convolutional neural networks based approach. Journal of Healthcare Engineering, 2020, 1–7. [Google Scholar]
12. Jaiswal, A., Gianchandani, N., Singh, D., Kumar, V., Kaur, M. (2021). Classification of the COVID-19 infected patients using densenet201 based deep transfer learning. Journal of Biomolecular Structure and Dynamics, 39(15), 5682–5689. [Google Scholar] [PubMed]
13. Li, C., Yang, Y., Liang, H., Wu, B. (2021). Transfer learning for establishment of recognition of COVID-19 on ct imaging using small-sized training datasets. Knowledge-Based Systems, 218, 106849. [Google Scholar] [PubMed]
14. Wang, S., Kang, B., Ma, J., Zeng, X., Xiao, M. et al. (2021). A deep learning algorithm using CT images to screen for corona virus disease (COVID-19). European Radiology, 31, 6096–6104. [Google Scholar] [PubMed]
15. Islam, M. M., Hannan, T., Sarker, L., Ahmed, Z. (2023). COVID-denseNet: A deep learning architecture to detect COVID-19 from chest radiology images. Proceedings of International Conference on Data Science and Applications: ICDSA 2022, vol. 2, pp. 397–415. Singapore, Springer. [Google Scholar]
16. Kaur, T., Gandhi, T. K. (2022). Classifier fusion for detection of COVID-19 from CT scans. Circuits, Systems, and Signal Processing, 41(6), 3397–3414. [Google Scholar] [PubMed]
17. Mercaldo, F., Belfiore, M. P., Reginelli, A., Brunese, L., Santone, A. (2023). Coronavirus COVID-19 detection by means of explainable deep learning. Scientific Reports, 13(1), 462. [Google Scholar] [PubMed]
18. Volkov, E. N., Averkin, A. N. (2023). Explainable artificial intelligence in medical image analysis: State of the art and prospects. 2023 XXVI International Conference on Soft Computing and Measurements (SCM), pp. 134–137. Saint Petersburg, Russia, IEEE. [Google Scholar]
19. Chadaga, K., Prabhu, S., Bhat, V., Sampathila, N., Umakanth, S. et al. (2023). A decision support system for diagnosis of COVID-19 from non-COVID-19 influenza-like illness using explainable artificial intelligence. Bioengineering, 10(4), 439. [Google Scholar] [PubMed]
20. Prasad Koyyada, S., Singh, T. P. (2023). An explainable artificial intelligence model for identifying local indicators and detecting lung disease from chest X-ray images. Healthcare Analytics, 4, 100206. [Google Scholar]
21. Sarp, S., Catak, F. O., Kuzlu, M., Cali, U., Kusetogullari, H. et al. (2023). An XAI approach for COVID-19 detection using transfer learning with X-ray images. Heliyon, 9(4), e15137. [Google Scholar] [PubMed]
22. Ye, Q., Xia, J., Yang, G. (2021). Explainable AI for COVID-19 CT classifiers: An initial comparison study. 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), pp. 521–526. Aveiro, Portugal, IEEE. [Google Scholar]
23. Mahmoudi, S. A., Stassin, S., Daho, M. E. H., Lessage, X., Mahmoudi, S. (2022). Explainable deep learning for COVID-19 detection using chest X-ray and CT-scan images. In: Healthcare informatics for fighting COVID-19 and future epidemics, pp. 311–336. https://doi.org/10.1007/978-3-030-72752-9_16 [Google Scholar] [CrossRef]
24. Boutorh, A., Rahim, H., Bendoumia, Y. (2021). Explainable AI models for COVID-19 diagnosis using CT-scan images and clinical data. International Meeting on Computational Intelligence Methods for Bioinformatics and Biostatistics, pp. 185–199. Springer. [Google Scholar]
25. Budhiraja, I., Garg, D., Kumar, N. (2023). Choquet integral based deep learning model for COVID-19 diagnosis using eXplainable AI for NG-IOT models. Computer Communications, 212, 227–238. [Google Scholar]
26. Sarkar, O., Islam, M. R., Syfullah, M. K., Islam, M. T., Ahamed, M. F. et al. (2023). Multi-scale CNN: An explainable AI-integrated unique deep learning framework for lung-affected disease classification. Technologies, 11(5), 134. [Google Scholar]
27. Wani, N. A., Kumar, R., Bedi, J. (2024). Deepxplainer: An interpretable deep learning based approach for lung cancer detection using explainable artificial intelligence. Computer Methods and Programs in Biomedicine, 243, 107879. [Google Scholar] [PubMed]
28. Soares, E., Angelov, P., Biaso, S., Froes, M. H., Abe, D. K. (2020). SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification. medRxiv. https://doi.org/10.1101/2020.04.24.20078584 [Google Scholar] [CrossRef]
29. Gupta, K., Bajaj, V. (2023). Deep learning models-based ct-scan image classification for automated screening of COVID-19. Biomedical Signal Processing and Control, 80, 104268. [Google Scholar] [PubMed]
30. Perumal, M., Srinivas, M. (2023). DenSplitnet: Classifier-invariant neural network method to detect COVID-19 in chest CT data. Journal of Visual Communication and Image Representation, 97, 103949. [Google Scholar]
31. Ibrahim, M. R., Youssef, S. M., Fathalla, K. M. (2023). Abnormality detection and intelligent severity assessment of human chest computed tomography scans using deep learning: A case study on SARS-CoV-2 assessment. Journal of Ambient Intelligence and Humanized Computing, 14(5), 5665–5688. [Google Scholar] [PubMed]
32. Gaur, P., Malaviya, V., Gupta, A., Bhatia, G., Pachori, R. B. et al. (2022). COVID-19 disease identification from chest CT images using empirical wavelet transformation and transfer learning. Biomedical Signal Processing and Control, 71, 103076. [Google Scholar] [PubMed]
33. Lu, S. Y., Zhang, Z., Zhang, Y. D., Wang, S. H. (2021). CGENet: A deep graph model for COVID-19 detection based on chest CT. Biology, 11(1), 33. [Google Scholar] [PubMed]
34. Basu, A., Sheikh, K. H., Cuevas, E., Sarkar, R. (2022). COVID-19 detection from ct scans using a two-stage framework. Expert Systems with Applications, 193, 116377. [Google Scholar] [PubMed]
35. Rostami, M., Oussalah, M. (2022). A novel explainable COVID-19 diagnosis method by integration of feature selection with random forest. Informatics in Medicine Unlocked, 30, 100941. [Google Scholar] [PubMed]
36. Uddin, M. Z., Khaksar, W., Torresen, J. (2017). Facial expression recognition using salient features and convolutional neural network. IEEE Access, 5, 26146–26161. [Google Scholar]
37. Neerincx, M. A., van der Waa, J., Kaptein, F., van Diggelen, J. (2018). Using perceptual and cognitive explanations for enhanced human-agent team performance. Engineering Psychology and Cognitive Ergonomics, pp. 204–214. as Vegas, NV, USA, Springer. [Google Scholar]
38. Huang, G., Liu, Z., van der Maaten, L., Weinberger, K. Q. (2017). Densely connected convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708. Honolulu, USA. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools