 Open Access
Open Access
ARTICLE
Advancing Wound Filling Extraction on 3D Faces: An Auto-Segmentation and Wound Face Regeneration Approach
1 Department of Mathematics and Statistics, Quy Nhon University, Quy Nhon City, 55100, Viet Nam
2 Applied Research Institute for Science and Technology, Quy Nhon University, Quy Nhon City, 55100, Viet Nam
3 CIRTECH Institute, HUTECH University, Ho Chi Minh City, 72308, Viet Nam
* Corresponding Author: H. Nguyen-Xuan. Email:
Computer Modeling in Engineering & Sciences 2024, 139(2), 2197-2214. https://doi.org/10.32604/cmes.2023.043992
Received 18 July 2023; Accepted 23 October 2023; Issue published 29 January 2024
Abstract
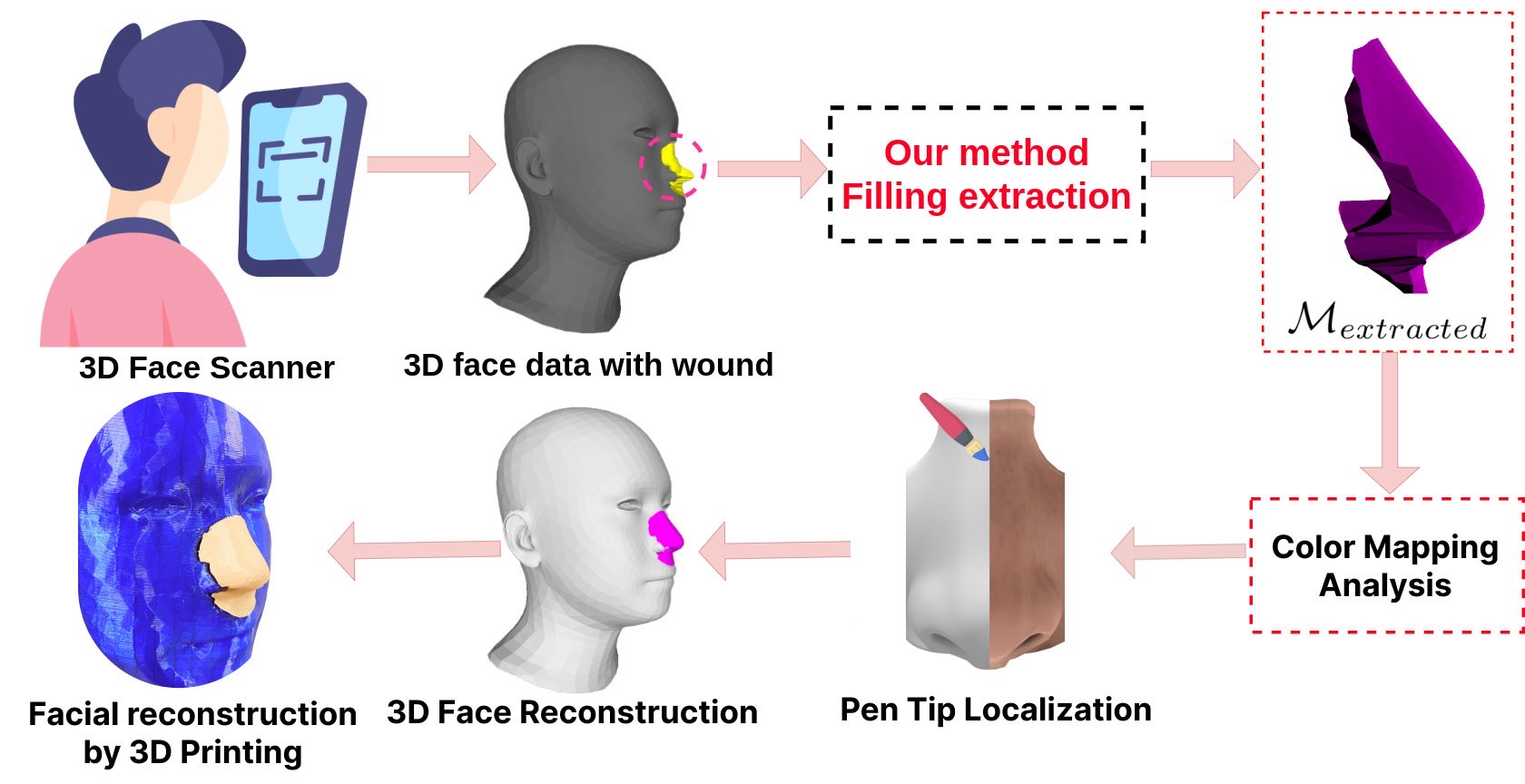
Facial wound segmentation plays a crucial role in preoperative planning and optimizing patient outcomes in various medical applications. In this paper, we propose an efficient approach for automating 3D facial wound segmentation using a two-stream graph convolutional network. Our method leverages the Cir3D-FaIR dataset and addresses the challenge of data imbalance through extensive experimentation with different loss functions. To achieve accurate segmentation, we conducted thorough experiments and selected a high-performing model from the trained models. The selected model demonstrates exceptional segmentation performance for complex 3D facial wounds. Furthermore, based on the segmentation model, we propose an improved approach for extracting 3D facial wound fillers and compare it to the results of the previous study. Our method achieved a remarkable accuracy of 0.9999993% on the test suite, surpassing the performance of the previous method. From this result, we use 3D printing technology to illustrate the shape of the wound filling. The outcomes of this study have significant implications for physicians involved in preoperative planning and intervention design. By automating facial wound segmentation and improving the accuracy of wound-filling extraction, our approach can assist in carefully assessing and optimizing interventions, leading to enhanced patient outcomes. Additionally, it contributes to advancing facial reconstruction techniques by utilizing machine learning and 3D bioprinting for printing skin tissue implants. Our source code is available at https://github.com/SIMOGroup/WoundFilling3D.Graphic Abstract
Keywords
Nowadays, people are injured by traffic accidents, occupational accidents, birth defects, diseases that have made them lose a part of their body. In which, defects when injured in the head and face areas account for a relatively high rate [1]. Wound regeneration is an important aspect of medical care, aimed at restoring damaged tissues and promoting wound healing in patients with complex wounds [2]. However, the treatment of craniofacial and facial defects can be challenging due to the many specific requirements of the tissue and the complexity of the anatomical structure of that region [3]. Traditional methods used for wound reconstruction often involve grafting techniques using automated grafts (from the patient’s own body) or allogeneic grafts (from a donor) [4]. However, these methods have limitations such as availability, donor morbidity, and potential for rejection. In recent years, the development of additive manufacturing technology has promoted the creation of advanced techniques in several healthcare industries [5–7]. The implementation of 3D printing technology in the preoperative phase enables clinicians to establish a meticulous surgical strategy by generating an anatomical model that accurately reflects the patient’s unique anatomy. This approach facilitates the development of customized drilling and cutting instructions, precisely tailored to the patient’s specific anatomical features, thereby accommodating the potential incorporation of a pre-formed implant [8]. Moreover, the integration of 3D printing technology and biomaterials plays a crucial role in advancing remedies within the field of regenerative medicine, addressing the pressing demand for novel therapeutic modalities [9–12]. The significance of wound reconstruction using 3D bioprinting in the domain of regenerative medicine is underscored by several key highlights, as outlined below:
- Customization and Precision: 3D bioprinting allows for the creation of patient-specific constructs, tailored to match the individual’s wound geometry and requirements. This level of customization ensures a better fit and promotes improved healing outcomes.
- Tissue Regeneration: The ability to fabricate living tissues using 3D bioprinting holds great promise for wound reconstruction. The technique enables the deposition of cells and growth factors in a controlled manner, facilitating tissue regeneration and functional restoration [13,14].
- Reduced Donor Dependency: The scarcity of donor tissues and the associated risks of graft rejection are significant challenges in traditional wound reconstruction methods. 3D bioprinting can alleviate these limitations by providing an alternative approach that relies on the patient’s own cells or bioinks derived from natural or synthetic sources [15].
- Complex Wound Healing: Certain wounds, such as large burns, chronic ulcers, or extensive tissue loss, pose significant challenges to conventional wound reconstruction methods. 3D bioprinting offers the potential to address these complex wound scenarios by creating intricate tissue architectures that closely resemble native tissues.
- Accelerated Healing: By precisely designing the structural and cellular components of the printed constructs, 3D bioprinting can potentially enhance the healing process. This technology can incorporate growth factors, bioactive molecules, and other therapeutic agents, creating an environment that stimulates tissue regeneration and accelerates wound healing [16].
Consequently, 3D bioprinting technology presents a promising avenue for enhancing craniofacial reconstruction modalities in individuals afflicted by head trauma.
Wound dimensions, including length, width, and depth, are crucial parameters for assessing wound healing progress and guiding appropriate treatment interventions [17]. For effective facial reconstruction, measuring the dimensions of a wound accurately can pose significant challenges in clinical and scientific settings [18]. Firstly, wound irregularity presents a common obstacle. Wounds rarely exhibit regular shapes, often characterized by uneven edges, irregular contours, or irregular surfaces. Such irregularity complicates defining clear boundaries and determining consistent reference points for measurement. Secondly, wound depth measurement proves challenging due to undermined tissue or tunnels. These features, commonly found in chronic or complex wounds, can extend beneath the surface, making it difficult to assess the wound’s true depth accurately. Furthermore, the presence of necrotic tissue or excessive exudate can obscure the wound bed, further hindering depth measurement. Additionally, wound moisture and fluid dynamics pose significant difficulties. Wound exudate, which may vary in viscosity and volume, can accumulate and distort measurements. Excessive moisture or the presence of dressing materials can alter the wound’s appearance, potentially leading to inaccurate measurements. Moreover, the lack of standardization in wound measurement techniques and tools adds to the complexity.
Currently, deep learning has indeed emerged as a predominant technique for wound image segmentation as well as various other applications in medical imaging and computer vision [19–21]. Based on the characteristics of the input data [22,23], three deep learning methods are used for segmentation and wound measurement, as shown in Fig. 1. The study of Anisuzzaman et al. [23] presented case studies of these three methods. The methods used to segment the wound based on the characteristics of the input data are as follows:
- 2D image segmentation: Deep learning methods in 2D for wound segmentation offer several advantages. Firstly, they are a well-established and widely used technique in the field. Additionally, large annotated 2D wound segmentation datasets are available, facilitating model training and evaluation. These methods exhibit efficient computational processing compared to their 3D counterparts, enabling faster inference times and improved scalability. Furthermore, deep learning architectures, such as convolutional neural networks, can be leveraged for effective feature extraction, enhancing the accuracy of segmentation results. However, certain disadvantages are associated with deep learning methods in 2D for wound segmentation. One limitation is the lack of depth information, which can restrict segmentation accuracy, particularly for complex wounds with intricate shapes and depth variations. Additionally, capturing the wound’s full spatial context and shape information can be challenging in 2D, as depth cues are not explicitly available. Furthermore, these methods are susceptible to variations in lighting conditions, image quality, and perspectives, which can introduce noise and affect the segmentation performance.
- 2D to 3D reconstruction: By incorporating depth information, the conversion to 3D enables a better capture of wounds’ shape and spatial characteristics, facilitating a more comprehensive analysis. Moreover, there is a potential for improved segmentation accuracy compared to 2D methods, as the additional dimension can provide richer information for delineating complex wound boundaries. Nevertheless, certain disadvantages are associated with converting from 2D to 3D for wound segmentation. The conversion process itself may introduce artifacts and distortions in the resulting 3D representation, which can impact the accuracy of the segmentation. Additionally, this approach necessitates additional computational resources and time due to the complexity of converting 2D data into a 3D representation [24]. Furthermore, the converted 3D method may not completely overcome the limitations of the 2D method.
- 3D mesh or point cloud segmentation: Directly extracting wound segmentation from 3D data (mesh/point cloud) offers several advantages. One notable advantage is the retention of complete 3D information on the wound, enabling accurate and precise segmentation. By working directly with the 3D data, this method effectively captures the wound’s intricate shape, volume, and depth details, surpassing the capabilities of both 2D approaches and converted 3D methods. Furthermore, the direct utilization of 3D data allows for a comprehensive analysis of the wound’s spatial characteristics, facilitating a deeper understanding of its structure and morphology.

Figure 1: Methods of using deep learning in wound measurement by segmentation
Hence, employing a 3D (mesh or point cloud) segmentation method on specialized 3D data, such as those obtained from 3D scanners or depth sensors, can significantly improve accuracy compared to the other two methods. The use of specialized 3D imaging technologies enables the capture of shape, volume, and depth details with higher fidelity and accuracy [25]. Consequently, the segmentation results obtained from this method are expected to provide a more precise delineation of wound boundaries and a more accurate assessment of wound characteristics. Therefore, this method can enhance wound segmentation accuracy and advance wound assessment techniques.
Besides, facial wounds and defects present unique challenges in reconstructive surgery, requiring accurate localization of the wound and precise estimation of the defect area [26]. The advent of 3D imaging technologies has revolutionized the field, enabling detailed capture of facial structures. However, reconstructing a complete face from a 3D model with a wound remains a complex task that demands advanced computational methods. Accurately reconstructing facial defects is crucial for surgical planning, as it provides essential information for appropriate interventions and enhances patient outcomes [27]. Some prominent studies, such as Sutradhar et al. [28] utilized a unique approach based on topology optimization to create patient-specific craniofacial implants using 3D printing technology; Nuseir et al. [29] proposed the utilization of direct 3D printing for the fabrication of a pliable nasal prosthesis, accompanied by the introduction of an optimized digital workflow spanning from the scanning process to the achievement of an appropriate fit; and some other prominent studies presented in survey studies such as [30,31]. However, these methods often require a lot of manual intervention and are prone to subjectivity and variability. To solve this problem, the method proposed in [32,33] leverages the power of modeling [34] to automate the process of 3D facial reconstruction with wounds, minimizing human error and improving efficiency. To extract the filling for the wound, the study [32] proposed the method of using the reconstructed 3D face and the 3D face of the patient without the wound. This method is called outlier extraction by the authors. These advancements can be leveraged to expedite surgical procedures, enhance precision, and augment patient outcomes, thereby propelling the progression of technology-driven studies on facial tissue reconstruction, particularly in bio 3D printing. However, this method still has some limitations as follows:
- The method of extracting filler for the wound after 3D facial reconstruction has not yet reached high accuracy.
- In order to extract the wound filling, the method proposed by [32] necessitated the availability of the patient’s pre-injury 3D facial ground truth. This requirement represents a significant limitation of the proposed wound filling extraction approach, as obtaining the patient’s pre-injury 3D facial data is challenging in real-world clinical settings.
To overcome these limitations, the present study aims to address the following objective:
- Train the 3D facial wound container segmentation automatic model using a variety of appropriate loss functions to solve the data imbalance problem.
- Propose an efficient approach to extract the 3D facial wound filling by leveraging the face regeneration model in the study [32] combined with the wound segmentation model.
- Evaluate the experimental results of our proposed method and the method described in the study by Nguyen et al. [32]. One case study will be selected to be illustrated through 3D printing.
Research reported by Nguyen et al. [32] has proposed a method to extract the filling for the wound for 3D face reconstruction. However, as we analyzed in Section 1, study [32] still has certain limitations. To address those limitations, we propose a unique approach to 3D face reconstruction with the combination of segmentation on injured 3D face data. This section introduces the structure of the 3D segmentation model and presents our proposed method.
2.1 Architecture of Two-Stream Graph Convolutional Network
Recent years have witnessed remarkable advancements in deep learning research within the domain of 3D shape analysis, as highlighted by Ioannidou et al. [35]. This progress has catalyzed the investigation into translation-invariant geometric attributes extracted from mesh data, facilitating the precise labeling of vertices or cells on 3D surfaces. Along with the development of 3D shape analysis, the field of 3D segmentation has advanced tremendously and brought about many applications across various fields, including computer vision and medical imaging [36]. Geometrically grounded approaches typically leverage pre-defined geometric attributes, such as 3D coordinates, normal vectors, and curvatures, to differentiate between distinct mesh cells. Several noteworthy models have emerged, including PointNet [37], PointNet++ [38], PointCNN [39], MeshSegNet [40], and DGCNN [41]. While these methods have demonstrated efficiency, they often employ a straightforward strategy of concatenating diverse raw attributes into an input vector for training a single segmentation network. Consequently, this strategy can generate isolated erroneous predictions. The root cause lies in the inherent dissimilarity between various raw attributes, such as cell spatial positions (coordinates) and cell morphological structures (normal vectors), which leads to confusion when merged as input. Therefore, the seamless fusion of their complementary insights for acquiring comprehensive high-level multi-view representations faces hindrance. Furthermore, the use of low-level predetermined attributes in these geometry-centric techniques is susceptible to significant variations. To address this challenge, the two-stream graph convolutional network (TSGCNet) [42] for 3D segmentation emerges as an exceptional technique, showcasing outstanding performance and potential in the field. This network harnesses the powerful geometric features available in the mesh to execute segmentation tasks. Consequently, in this study, we have selected this model as the focal point to investigate its applicability and effectiveness in the context of our research objectives. In [42], the proposed methodology employs two parallel streams, namely the C stream and the N stream. TSGCNet incorporates input-specific graph-learning layers to extract high-level geometric representations from the coordinates and normal vectors. Subsequently, the features obtained from these two complementary streams are fused in the feature-fusion branch to facilitate the acquisition of discriminative multi-view representations, specifically for segmentation purposes. An overview of the architecture of the two-stream graph convolutional network is shown in Fig. 2.

Figure 2: Architectural overview of the TSGCNet model for segmentation on injured 3D face data
The C-stream is designed to capture the essential topological characteristics derived from the coordinates of all vertices of a mesh. The C-stream receives an input denoted as
The TSGCNet model employs three layers in each stream to extract features. Subsequently, the features from these layers are combined as follows:
In order for the model to comprehensively understand the 3D mesh structure, Zhang et al. [42] combined
where
We utilize the TSGCNet model, as presented in study [42], to perform segmentation of the wound area on the patient’s 3D face. This model demonstrates a remarkable capacity for accurately discriminating boundaries between regions harboring distinct classes. Our dataset comprises two distinct classes, namely facial abnormalities and normal regions. Due to the significantly smaller proportion of facial wounds compared to the normal area, an appropriate training strategy is necessary to address the data imbalance phenomenon effectively. To address this challenge, we utilize specific functions that effectively handle data imbalance within the semantic segmentation task. These functions include focal loss [43], dice loss [44], cross-entropy loss and weighted cross-entropy loss [45].
1) Focal loss is defined as:
where
2) Dice loss also known as the Sorensen-Dice coefficient is defined as:
where
3) Cross-entropy segmentation loss is defined as:
where
4) Weighted Cross-Entropy loss is as follows:
where
We implement a training strategy utilizing the TSGCNet model combined with various loss functions to achieve the best model performance for the wound segmentation task. This training strategy is described in detail in Algorithm 1.

After identifying the optimal wound segmentation model, we proceed to extract the mesh containing the area that needs filling on the 3D face. Let

Figure 3: Several filling extraction results

Figure 4: An illustration of the wound-filling extraction algorithm

We utilize a dataset of 3D faces with craniofacial injuries called Cir3D-FaIR [32]. The dataset used in this study is generated through simulations within a virtual environment, replicating realistic facial wound locations. A set of 3,678 3D mesh representations of uninjured human faces is employed to simulate facial wounds. Specifically, each face in the dataset is simulated with ten distinct wound locations. Consequently, the dataset comprises 40,458 human head meshes, encompassing uninjured faces and wounds in various positions. In practice, the acquired data undergoes mesh data processing by reducing the sample to 15,000 cells of the mesh, eliminating redundant information while preserving the original topology. Each 3D face mesh consists of 15,000 cells of the mesh and is labeled according to the location of wounds on the face, specifically indicating the presence of the wounds. This simulation dataset has been evaluated by expert physicians to assess the complexity associated with the injuries. Fig. 5 showcases several illustrative examples of typical cases from the dataset. The dataset is randomly partitioned into distinct subsets, with 80% of the data assigned to training and 20% designated for validation. The objective is to perform automated segmentation of the 3D facial wound region and integrate it with the findings of Nguyen et al. [32] regarding defect face reconstruction to extract the wound-filling part specific to the analyzed face.

Figure 5: Illustrations of the face dataset with wounds
The wound area segmentation model on the patient’s 3D face is trained through experiments with different loss functions to select the most effective model, as outlined in Algorithm 1. The training process was conducted using a single NVIDIA Quadro RTX 6000 GPU over the course of 50 epochs. The Adam optimizer is employed in conjunction with a mini-batch size of 4. The initial learning rate was set at
In this study, the quantitative evaluation of segmentation performance on a 3D grid is accomplished through two metrics: (1) Overall Accuracy (OA), which is obtained by dividing the number of correctly segmented cells by the total number of cells; and (2) the calculation of Intersection-over-Union (IoU) for each class, followed by the calculation of the mean Intersection-over-Union (mIoU). The IoU is a vital metric used in 3D segmentation to assess the accuracy and quality of segmentation results. It quantifies the degree of overlap between the segmented region and the ground truth, providing insights into the model’s ability to accurately delineate objects or regions of interest within a 3D space. Training 3D models is always associated with challenges related to hardware requirements, processing speed, and cost. Processing and analyzing 3D data is more computationally intensive compared to 2D data. The hardware requirements for 3D segmentation are typically higher, including more powerful CPUs or GPUs, more RAM, and potentially specialized hardware for accelerated processing. Especially, performing segmentation on 3D data takes more time due to the increased complexity. In essence, the right loss function can lead to faster convergence, better model performance, and improved interpretability. Therefore, experimentation and thorough evaluation are crucial to determining which loss function works best for data. The model was trained on the dataset using four iterations of experiments, wherein different loss functions were employed. The outcomes of these experiments are presented in Table 1. The utilized loss functions demonstrate excellent performance in the training phase, yielding highly satisfactory outcomes on large-scale unbalanced datasets. Specifically, we observe that the model integrated with cross-entropy segmentation loss exhibits rapid convergence, requiring only 16 epochs to achieve highly favorable outcomes. As outlined in Section 2.2, the model exhibiting the most favorable outcomes, as determined by the cross-entropy segmentation loss function, was selected for the segmentation task. This particular model achieved an impressive

Furthermore, in the context of limited 3D data for training segmentation models in dentistry, Zhang et al. [42] showcased the remarkable efficacy of the TSGCNet model. Their training approach involved the utilization of 80 dental data meshes, culminating in an impressive performance outcome of 95.25%. To investigate the effectiveness of the TSGCNet model with a small amount of face data with injuries, we train the TSGCNet model including 100 meshes for training and 20 meshes for testing. The TSGCNet model was trained for 50 epochs, employing a cross-entropy segmentation loss function. This approach achieved an overall accuracy of 97.69%. This result underlines the effectiveness of the two-stream graph convolutional network in accurately segmenting complex and minor wounds, demonstrating its ability to capture geometric feature information from the 3D data. However, training the model with a substantial dataset is crucial to ensure a comprehensive understanding of facial features and achieve a high level of accuracy. Consequently, we selected a model that achieved an
From the above segmentation result, our primary objective is to conduct a comparative analysis between our proposed wound fill extraction method and a method with similar objectives as discussed in the studies by Nguyen et al. [32,33]. A notable characteristic of the Cir3D-FaIR dataset is that all meshes possess a consistent vertex order. This enables us to streamline the extraction process of the wound filler. Utilizing the test dataset, we employ the model trained in the study by Nguyen et al. [32] for the reconstruction of the 3D face. Subsequently, we apply our proposed method to extract the wound fill from the reconstructed 3D face. As previously stated, we introduce a methodology for the extraction of wound filling. The details of this methodology are explained in Algorithm 2 and Fig. 4.

For the purpose of notational convenience, we designate the filling extraction method presented in the study by Nguyen et al. [32] as the ”old proposal”. We conduct a performance evaluation of both our proposed method and the old proposal method on a dataset consisting of 8090 meshes, which corresponds to 20% of the total dataset. A comprehensive description of the process for comparing the two methods is provided in Algorithm 3. The results show that our proposal has an average accuracy of 0.9999993%, while the method in the old proposal is 0.9715684%. The accuracy of the fill extraction method has been improved, which is very practical in the medical reconstruction problem. After that, the study randomly extracted the method outputs from the test set, depicted in Fig. 3. We have used 3D printing technology to illustrate the results of the actual model, which is significantly improved compared to the old method, as shown in Fig. 6 and illustrate a 3D printed model to extract the wound filling as shown in Fig. 7.

Figure 6: Filling extraction results with 3D printing

Figure 7: A 3D-printed pattern to fill a wound
The results of this study emphasize the potential of utilizing appropriate 3D printing technology for facial reconstruction in patients. This can involve prosthetic soft tissue reconstruction or 3D printing of facial biological tissue [46]. 3D bioprinting for skin tissue implants requires specialized materials and methods to create customized skin constructs for a range of applications, including wound healing and reconstructive surgery. The choice of materials and fusion methods may vary based on the specific site (e.g., face or body) and the desired characteristics of the skin tissue implant. In the realm of 3D printing for biological soft tissue engineering, a diverse array of materials is strategically employed to emulate the intricate structures and properties inherent to native soft tissues. Hydrogels, such as alginate, gelatin, and fibrin, stand out as popular choices, primarily owing to their high water content and excellent biocompatibility. Alginate, derived from seaweed, exhibits favorable characteristics such as good printability and high cell viability, making it an attractive option. Gelatin, a denatured form of collagen, closely replicates the extracellular matrix, providing a biomimetic environment conducive to cellular growth. Fibrin, a key protein in blood clotting, offers a natural scaffold for cell attachment and proliferation. Additionally, synthetic polymers like polycaprolactone (PCL) and poly(lactic-co-glycolic acid) (PLGA) provide the benefit of customizable mechanical properties and degradation rates. Studies [47–49] have presented detailed surveys of practical applications of many types of materials for 3D printing of biological tissue. Our research is limited to proposing an efficient wound-filling extraction method with high accuracy. In the future, we will consider implementing the application of this research in conjunction with physician experts at hospitals in Vietnam. By harnessing 3D printing technology, as illustrated in Fig. 8, healthcare professionals can craft highly tailored and precise facial prosthetics, considering each patient’s unique anatomy and needs. This high level of customization contributes to achieving a more natural appearance and better functional outcomes, addressing both aesthetic and functional aspects of facial reconstruction [14]. This approach holds significant promise for enhancing facial reconstruction procedures and improving the overall quality of life for patients who have undergone facial trauma or have congenital facial abnormalities. Moreover, high-quality 3D facial scanning applications on phones are becoming popular. We are able to implement our proposal integration into smartphones to support sketching the reconstruction process on the injured face. This matter is further considered in our forthcoming research endeavors.

Figure 8: 3D printing process of 3D models for geometric visualization
Although our 3D facial wound reconstruction method achieves high performance, it still has certain limitations. Real-world facial data remains limited due to ethics in medical research. Therefore, we amalgamate scarce MRI data from patients who consented to share their personal data with the data generated from the MICA model to create a dataset. Our proposal primarily focuses on automatically extracting the region to be filled in a 3D face, addressing a domain similar to practical scenarios. We intend to address these limitations in future studies when we have access to a more realistic volume of 3D facial data from patients.
Furthermore, challenges related to unwanted artifacts, obstructions, and limited contrast in biomedical 3D scanning need to be considered. To tackle these challenges, we utilize cutting-edge 3D scanning technology equipped with enhanced hardware and software capabilities. This enables us to effectively mitigate artifacts and obstructions during data collection. We implement rigorous quality assurance protocols throughout the 3D scanning process, ensuring the highest standards of image quality. Additionally, we pay careful attention to patient positioning and provide guidance to minimize motion artifacts. Moreover, we employ advanced 3D scanning techniques, such as multi-modal imaging that combines various imaging modalities like CT and MRI. This approach significantly enhances image quality and improves contrast, which is essential for accurate medical image interpretation.
This study explored the benefits of using a TSGCNet to segment 3D facial trauma defects automatically. Furthermore, we have proposed an improved method to extract the wound filling for the face. The results show the most prominent features as follows:
- An auto-segmentation model was trained to ascertain the precise location and shape of 3D facial wounds. We have experimented with different loss functions to give the most effective model in case of data imbalance. The results show that the model works well for complex wounds on the Cir3D-FaIR face dataset with an accuracy of 0.9999993%.
- Concurrently, we have proposed a methodology to enhance wound-filling extraction performance by leveraging both a segmentation model and a 3D face reconstruction model. By employing this approach, we achieve higher accuracy than previous studies on the same problem. Additionally, this method obviates the necessity of possessing a pre-injury 3D model of the patient’s face. Instead, it enables the precise determination of the wound’s position, shape, and complexity, facilitating the rapid extraction of the filling material.
- This research proposal aims to contribute to advancing facial reconstruction techniques using AI and 3D bioprinting technology to print skin tissue implants. Printing skin tissue for transplants has the potential to revolutionize facial reconstruction procedures by providing personalized, functional, and readily available solutions. By harnessing the power of 3D bioprinting technology, facial defects can be effectively addressed, enhancing both cosmetic and functional patient outcomes.
- From this research direction, our proposed approach offers a promising avenue for advancing surgical support systems and enhancing patient outcomes by addressing the challenges associated with facial defect reconstruction. Combining machine learning, 3D imaging, and segmentation techniques provides a comprehensive solution that empowers surgeons with precise information and facilitates personalized interventions in treating facial wounds.
Acknowledgement: We would like to thank Vietnam Institute for Advanced Study in Mathematics (VIASM) for hospitality during our visit in 2023, when we started to work on this paper.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Duong Q. Nguyen, H. Nguyen-Xuan, Nga T.K. Le; data collection: Thinh D. Le; analysis and interpretation of results: Duong Q. Nguyen, Thinh D. Le, Phuong D. Nguyen; draft manuscript preparation: Duong Q. Nguyen, Thinh D. Le, H. Nguyen-Xuan. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Our source code and data can be accessed at https://github.com/SIMOGroup/WoundFilling3D.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Asscheman, S., Versteeg, M., Panneman, M., Kemler, E. (2023). Reconsidering injury severity: Looking beyond the maximum abbreviated injury score. Accident Analysis & Prevention, 186, 107045. [Google Scholar]
2. Han, S. K. (2023). Innovations and advances in wound healing. Springer Berlin, Heidelberg: Springer Nature. [Google Scholar]
3. Nyberg, E. L., Farris, A. L., Hung, B. P., Dias, M., Garcia, J. R. et al. (2016). 3D-printing technologies for craniofacial rehabilitation, reconstruction, and regeneration. Annals of Biomedical Engineering, 45(1), 45–57. [Google Scholar] [PubMed]
4. Larsson, A. P., Briheim, K., Hanna, V., Gustafsson, K., Starkenberg, A. et al. (2021). Transplantation of autologous cells and porous gelatin microcarriers to promote wound healing. Burns, 47(3), 601–610. [Google Scholar] [PubMed]
5. Tabriz, A. G., Douroumis, D. (2022). Recent advances in 3D printing for wound healing: A systematic review. Journal of Drug Delivery Science and Technology, 74, 103564. [Google Scholar]
6. Tan, G., Ioannou, N., Mathew, E., Tagalakis, A. D., Lamprou, D. A. et al. (2022). 3D printing in ophthalmology: From medical implants to personalised medicine. International Journal of Pharmaceutics, 625, 122094. [Google Scholar] [PubMed]
7. Relano, R. J., Concepcion, R., Francisco, K., Enriquez, M. L., Co, H. et al. (2021). A bibliometric and trend analysis of applied technologies in bioengineering for additive manufacturing of human organs. 2021 IEEE 13th International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment, and Management (HNICEM), Manila, Philippines. [Google Scholar]
8. Lal, H., Patralekh, M. K. (2018). 3D printing and its applications in orthopaedic trauma: Atechnological marvel. Journal of Clinical Orthopaedics and Trauma, 9(3), 260–268. [Google Scholar] [PubMed]
9. Tack, P., Victor, J., Gemmel, P., Annemans, L. (2016). 3D-printing techniques in a medical setting: A systematic literature review. BioMedical Engineering Online, 15(1), 115. [Google Scholar] [PubMed]
10. Hoang, D., Perrault, D., Stevanovic, M., Ghiassi, A. (2016). Surgical applications of three-dimensional printing: A review of the current literature and how to get started. Annals of Translational Medicine, 4(23). [Google Scholar]
11. Liu, N., Ye, X., Yao, B., Zhao, M., Wu, P. et al. (2021). Advances in 3D bioprinting technology for cardiac tissue engineering and regeneration. Bioactive Materials, 6(5), 1388–1401. [Google Scholar] [PubMed]
12. Wallace, E. R., Yue, Z., Dottori, M., Wood, F. M., Fear, M. et al. (2023). Point of care approaches to 3D bioprinting for wound healing applications. Progress in Biomedical Engineering, 5(2), 23002. [Google Scholar]
13. Cubo, N., Garcia, M., del Canizo, J. F., Velasco, D., Jorcano, J. L. (2016). 3D bioprinting of functional human skin: Production and in vivo analysis. Biofabrication, 9(1), 15006. [Google Scholar]
14. Gao, C., Lu, C., Jian, Z., Zhang, T., Chen, Z. et al. (2021). 3D bioprinting for fabricating artificial skin tissue. Colloids and Surfaces B: Biointerfaces, 208, 112041. [Google Scholar] [PubMed]
15. Jain, P., Kathuria, H., Dubey, N. (2022). Advances in 3D bioprinting of tissues/organs for regenerative medicine and in-vitro models. Biomaterials, 287, 121639. [Google Scholar] [PubMed]
16. Li, C., Cui, W. (2021). 3D bioprinting of cell-laden constructs for regenerative medicine. Engineered Regeneration, 2, 195–205. [Google Scholar]
17. Grey, J. E., Enoch, S., Harding, K. G. (2006). Wound assessment. BMJ, 332(7536), 285–288. [Google Scholar] [PubMed]
18. Musa, D., Guido-Sanz, F., Anderson, M., Daher, S. (2022). Reliability of wound measurement methods. IEEE Open Journal of Instrumentation and Measurement, 1, 1–9. [Google Scholar]
19. Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F. et al. (2017). A survey on deep learning in medical image analysis. Medical Image Analysis, 42, 60–88. [Google Scholar] [PubMed]
20. Scebba, G., Zhang, J., Catanzaro, S., Mihai, C., Distler, O. et al. (2022). Detect-and-segment: A deep learning approach to automate wound image segmentation. Informatics in Medicine Unlocked, 29, 100884. [Google Scholar]
21. Juhong, T., Hui Peng, J. Z. (2021). Mri brain tumor segmentation using 3D U-Net with dense encoder blocks and residual decoder blocks. Computer Modeling in Engineering & Sciences, 128(2), 427–445. https://doi.org/10.32604/cmes.2021.014107 [Google Scholar] [CrossRef]
22. Zhang, R., Tian, D., Xu, D., Qian, W., Yao, Y. (2022). A survey of wound image analysis using deep learning: Classification, detection, and segmentation. IEEE Access, 10, 79502–79515. [Google Scholar]
23. Anisuzzaman, D., Wang, C., Rostami, B., Gopalakrishnan, S., Niezgoda, J. et al. (2022). Image-based artificial intelligence in wound assessment: A systematic review. Advances in Wound Care, 11(12), 687–709. [Google Scholar] [PubMed]
24. Zhang, X., Xu, F., Sun, W., Jiang, Y., Cao, Y. (2023). Fast mesh reconstruction from single view based on gcn and topology modification. Computer Systems Science and Engineering, 45(2), 1695–1709. [Google Scholar]
25. Shah, A., Wollak, C., Shah, J. (2015). Wound measurement techniques: Comparing the use of ruler method, 2D imaging and 3D scanner. Journal of the American College of Clinical Wound Specialists, 5, 52–57. [Google Scholar] [PubMed]
26. Das, A., Awasthi, P., Jain, V., Banerjee, S. S. (2023). 3D printing of maxillofacial prosthesis materials: Challenges and opportunities. Bioprinting, 32, e00282. [Google Scholar]
27. Maroulakos, M., Kamperos, G., Tayebi, L., Halazonetis, D., Ren, Y. (2019). Applications of 3D printing on craniofacial bone repair: A systematic review. Journal of Dentistry, 80, 1–14. [Google Scholar] [PubMed]
28. Sutradhar, A., Park, J., Carrau, D., Nguyen, T. H., Miller, M. J. et al. (2016). Designing patient-specific 3D printed craniofacial implants using a novel topology optimization method. Medical & Biological Engineering & Computing, 54(7), 1123–1135. [Google Scholar]
29. Nuseir, A., Hatamleh, M. M., Alnazzawi, A., Al-Rabab’ah, M., Kamel, B. et al. (2018). Direct 3D printing of flexible nasal prosthesis: Optimized digital workflow from scan to fit. Journal of Prosthodontics, 28(1), 10–14. [Google Scholar] [PubMed]
30. Ghai, S., Sharma, Y., Jain, N., Satpathy, M., Pillai, A. K. (2018). Use of 3-D printing technologies in craniomaxillofacial surgery: A review. Oral and Maxillofacial Surgery, 22(3), 249–259. [Google Scholar] [PubMed]
31. Salah, M., Tayebi, L., Moharamzadeh, K., Naini, F. B. (2020). Three-dimensional bio-printing and bone tissue engineering: Technical innovations and potential applications in maxillofacial reconstructive surgery. Maxillofacial Plastic and Reconstructive Surgery, 42(1), 18. [Google Scholar] [PubMed]
32. Nguyen, P. D., Le, T. D., Nguyen, D. Q., Nguyen, T. Q., Chou, L. W. et al. (2023). 3D facial imperfection regeneration: Deep learning approach and 3D printing prototypes. arXiv preprint arXiv:2303.14381. [Google Scholar]
33. Nguyen, P. D., Le, T. D., Nguyen, D. Q., Nguyen, B., Nguyen-Xuan, H. (2023). Application of self-supervised learning to MICA model for reconstructing imperfect 3D facial structures. arXiv preprint arXiv: 2304.04060. [Google Scholar]
34. Zhou, Y., Wu, C., Li, Z., Cao, C., Ye, Y. et al. (2020). Fully convolutional mesh autoencoder using efficient spatially varying kernels. Proceedings of the 34th International Conference on Neural Information Processing Systems, Red Hook, NY, USA, Curran Associates Inc. [Google Scholar]
35. Ioannidou, A., Chatzilari, E., Nikolopoulos, S., Kompatsiaris, I. (2017). Deep learning advances in computer vision with 3D data: A survey. ACM Computing Surveys, 50(2), 1–38. [Google Scholar]
36. Gezawa, A. S., Wang, Q., Chiroma, H., Lei, Y. (2023). A deep learning approach to mesh segmentation. Computer Modeling in Engineering & Sciences, 135(2), 1745–1763. https://doi.org/10.32604/cmes.2022.021351 [Google Scholar] [CrossRef]
37. Charles, R., Su, H., Kaichun, M., Guibas, L. J. (2017). Pointnet: Deep learning on point sets for 3D classification and segmentation. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Los Alamitos, CA, USA, IEEE Computer Society. [Google Scholar]
38. Qi, C. R., Yi, L., Su, H., Guibas, L. J. (2017). PointNet++: Deep hierarchical feature learning on point sets in a metric space. Proceedings of the 31st International Conference on Neural Information Processing Systems, Red Hook, NY, USA, Curran Associates Inc. [Google Scholar]
39. Li, Y., Bu, R., Sun, M., Wu, W., Di, X. et al. (2018). PointCNN: Convolution on x-transformed points. In: Bengio, S., Wallach, H., Larochelle, H., Grauman, K., Cesa-Bianchi, N. et al. (Eds.Advances in neural information processing systems, vol. 31. Montréal, Canada: Curran Associates, Inc. [Google Scholar]
40. Lian, C., Wang, L., Wu, T. H., Liu, M., Durán, F. et al. (2019). Meshsnet: Deep multi-scale mesh feature learning for end-to-end tooth labeling on 3D dental surfaces. International Conference on Medical Image Computing and Computer-Assisted Intervention, Cham. [Google Scholar]
41. Wang, Y., Sun, Y., Liu, Z., Sarma, S. E., Bronstein, M. M. et al. (2019). Dynamic graph cnn for learning on point clouds. ACM Transactions on Graphics, 38(5), 1–12. [Google Scholar]
42. Zhao, Y., Zhang, L., Liu, Y., Meng, D., Cui, Z. et al. (2022). Two-stream graph convolutional network for intra-oral scanner image segmentation. IEEE Transactions on Medical Imaging, 41(4), 826–835. [Google Scholar] [PubMed]
43. Lin, T. Y., Goyal, P., Girshick, R., He, K., Dollár, P. (2017). Focal loss for dense object detection. 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy. [Google Scholar]
44. Sudre, C. H., Li, W., Vercauteren, T., Ourselin, S., Jorge Cardoso, M. et al. (2017). Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Cardoso, M. J., Arbel, T., Carneiro, G., Syeda-Mahmood, T., Tavares, J. M. R. et al. (Eds.Deep learning in medical image analysis and multimodal learning for clinical decision support. Cham: Springer International Publishing. [Google Scholar]
45. Jadon, S. (2020). A survey of loss functions for semantic segmentation. 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Vina del Mar, Chile. [Google Scholar]
46. Yan, Q., Dong, H., Su, J., Han, J., Song, B. et al. (2018). A review of 3D printing technology for medical applications. Engineering, 4(5), 729–742. [Google Scholar]
47. Derakhshanfar, S., Mbeleck, R., Xu, K., Zhang, X., Zhong, W. et al. (2018). 3D bioprinting for biomedical devices and tissue engineering: A review of recent trends and advances. Bioactive Materials, 3(2), 144–156. [Google Scholar] [PubMed]
48. Mao, H., Yang, L., Zhu, H., Wu, L., Ji, P. et al. (2020). Recent advances and challenges in materials for 3D bioprinting. Progress in Natural Science: Materials International, 30(5), 618–634. [Google Scholar]
49. Mallakpour, S., Azadi, E., Hussain, C. M. (2021). State-of-the-art of 3D printing technology of alginate-based hydrogels-an emerging technique for industrial applications. Advances in Colloid and Interface Science, 293, 102436. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools