 Open Access
Open Access
ARTICLE
Enhanced Temporal Correlation for Universal Lesion Detection
1 School of Computer Science and Technology, Shandong University of Finance and Economics, Jinan, China
2 School of Information Science and Technology, Linyi University, Linyi, China
3 School of Creative Technologies, University of Portsmouth, Portsmouth, UK
* Corresponding Author: Muwei Jian. Email:
Computer Modeling in Engineering & Sciences 2024, 138(3), 3051-3063. https://doi.org/10.32604/cmes.2023.030236
Received 28 March 2023; Accepted 01 August 2023; Issue published 15 December 2023
Abstract
Universal lesion detection (ULD) methods for computed tomography (CT) images play a vital role in the modern clinical medicine and intelligent automation. It is well known that single 2D CT slices lack spatial-temporal characteristics and contextual information compared to 3D CT blocks. However, 3D CT blocks necessitate significantly higher hardware resources during the learning phase. Therefore, efficiently exploiting temporal correlation and spatial-temporal features of 2D CT slices is crucial for ULD tasks. In this paper, we propose a ULD network with the enhanced temporal correlation for this purpose, named TCE-Net. The designed TCE module is applied to enrich the discriminate feature representation of multiple sequential CT slices. Besides, we employ multi-scale feature maps to facilitate the localization and detection of lesions in various sizes. Extensive experiments are conducted on the DeepLesion benchmark demonstrate that this method achieves 66.84% and 78.18% for FS@0.5 and FS@1.0, respectively, outperforming compared state-of-the-art methods.Keywords
Computed tomography (CT), a widely used medical aid in diagnosis, scans and creates images of body parts from multiple directions and cross-sections. As the number of CT images in the medical system explosively grows, the limited number of professional radiologists makes it an urgent task to research medical diagnostic applications. With the continuous development of deep learning, which integrating with medical treatments to improve medical efficiency and solve the problem of uneven distribution of medical resources. At the same time, excellent lesion detection models have been proposed, including methods for single organs, such as lung nodule detection [1–10], lymph nodes [11–13], and liver tumors [14–39]. However, these methods cannot meet the clinical demand for detecting multiple lesioned organs at the same time. Therefore, universal lesion detection has gradually gained the attention of researchers, while many promising achievements have been obtained, such as [1–3,5,15,16], etc.
In recent years, Yan et al. [4] proposed a large dataset DeepLesion [4], which had greatly contributed to the development of universal lesion detection models. Based on the DeepLesion, some researchers [15–23] have proposed valid detection methods and have shown better metrics, respectively. Among the existing ULD methods, the accuracy is generally improved by the following aspects: anchor-free mechanism [24,25] or enhancing feature representation [1,16,20,26,27], optimizing anchor mechanism [15,28,29], and domain knowledge augment [30]. We focus on the idea of enhancing feature representation, which mainly include: (1) adding attention mechanism [1,26,27,31–34], the context information inter slices and the spatial information intra slices can be concentrated in the most prominent regions to enrich the feature representation; (2) introducing deformable convolution in the network [20], which can increase the receptive field of feature extraction; (3) using pseudo-3D [34] to process 3D CT input data reduces the number of network parameters to some extent. Although these schemes are effective, there is still room for improvement in terms of detection properties and efficiency.
We show the effectiveness of several methods for detecting single-organ lesions on a multi-organ lesion detection task in Fig. 1. We can see that a universal lesion detection network has more potential and values. Inspired by camouflaged object detection in video [17], enhancing the temporal correlation of multiple sequential slices contributes to the capture of lesions between slices by the network. Accordingly, by enhancing the temporal correlation, the feature representation of slices can be improved for improving the detection accuracy. Through the above observation, we find that the existing universal lesion detection methods neglect to focus on the correlation between CT slices, which can provide richer features for complex lesion detection tasks. Therefore, we propose an enhanced temporal correlation detection network (i.e., TCE-Net), in which the temporal correlation enhancement (TCE) module is introduced to extract temporal correlations of multiple sequential CT slices. In addition, multi-scale feature maps of sequential slices are extracted using a pre-trained Feature Pyramid Network (FPN) [31] for detecting lesions of different sizes.
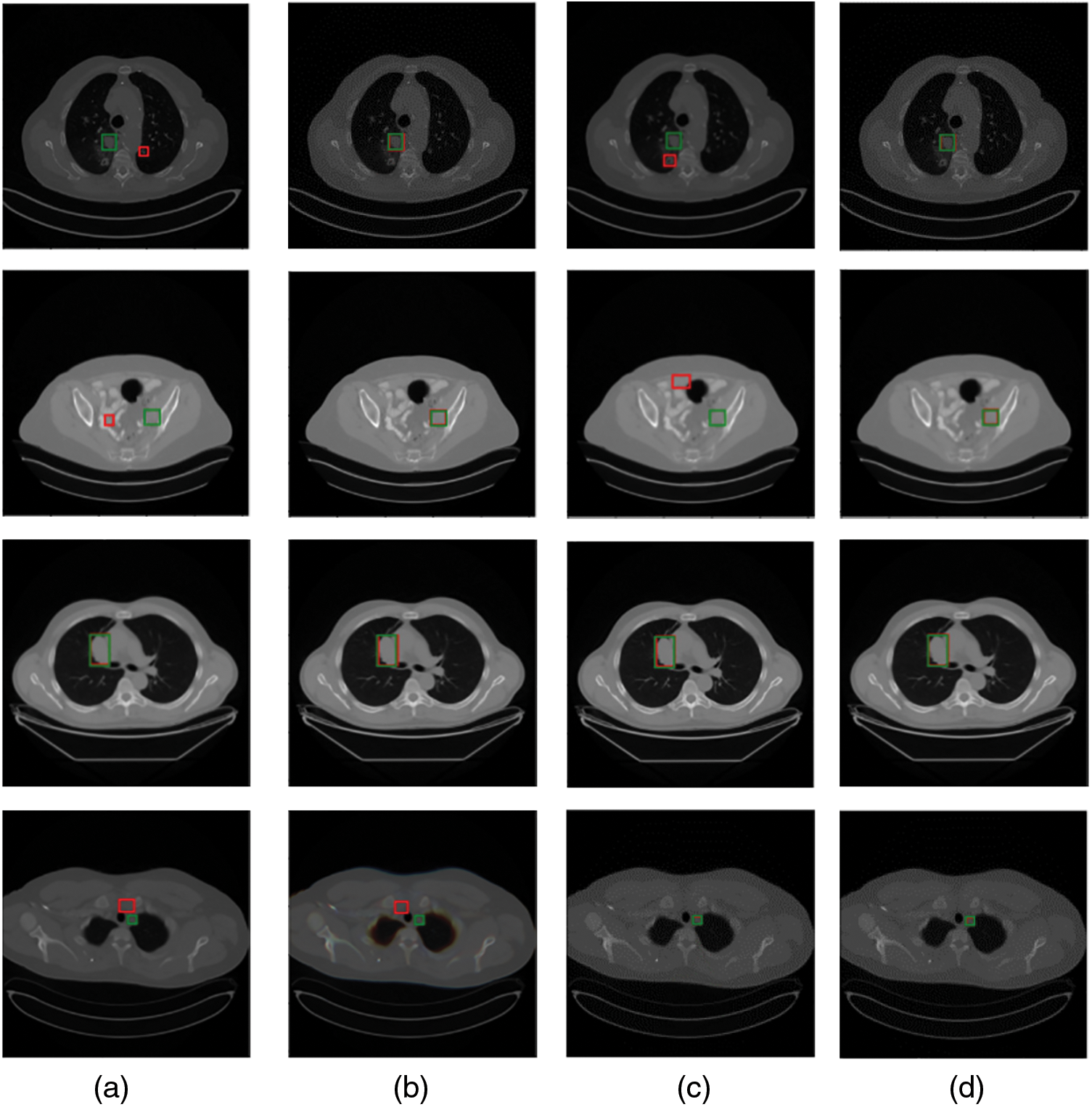
Figure 1: Comparison of visualization results of several single organ detection networks with ours. Green and red boxes correspond to ground-truths in the test set and predicted true positives. (a) The method proposed in [37]; (b) a modified SSD model by [38]; (c) MSCR [39]; (d) ours
The contributions of this work are threefold:
(i) An end-to-end network called TCE-Net is proposed for universal lesion detection on CT images.
(ii) A temporal correlation enhancement module was designed for lesion detection by extracting the temporal correlation inter multiple sequential CT slices.
(iii) The experimental outcomes demonstrate that the developed framework performs favourably against state-of-the-art approaches on the DeepLesion benchmarks.
The remainder of this article are organized as follows. Section 2 introduces related work. The exploited TCE-Net for universal lesion detection is described in details in Section 3. Then, experimental results are provided in Section 4. Finally, we draw a conclusion of this paper in Section 5.
A Universal Lesion Detection (ULD) is a task that has great potential for development in object detection and is in a high clinical demand in clinical [40–51]. In recent years, the ULD task has received great attention, and related research works have been proposed one after another. Since 3D convolutional neural networks are very high memory consuming and expensive to train, and it is also a complex and difficult task to label 3D boxes for 3D CT dataset, Yan et al. [5] proposed a 3D context enhanced region-based CNN, called 3DCE, incorporating 3D context into a 2D CNN network to extract features and using a 2D detection network to make the final prediction. Shao et al. [1] proposed a Multi-Scale Booster, called MSB, which is used different dilation rates to capture the lesion responses from each pyramid feature map, respectively. In [27], with the aim of enriching feature representations and improving feature discrimination, they introduced a cross-slice context attention that selectively aggregated information from different slices and an intra-slice spatial attention to focus feature learning on the most salient regions. Meanwhile, pseudo-3D has also been applied to lesion detection, such as MP3D FPN proposed by Zhang et al. [16], which can efficiently extract 3D context for universal lesion detection in CT slices. As in [1], the MVP-Net introduced by Li et al. [3] also extracts multi-scale features, with the difference that the MVP-Net used three FPNs corresponding to three window widths simultaneously to suit different detection parts. In addition to the above methods, there are also approaches based on domain knowledge enhancement, such as Sheoran et al. [30]. Meanwhile, refining the anchor mechanism is also a common mindset [15,24,25,28,29].
Currently, enhancing feature representation is still the majority route. Yet existing universal lesion detection models ignore the hidden temporal correlations between sequential slices which facilitate the catching of lesions when enhancing feature representation. With this motivation as a beginning point, we explore TCE-Net, where the TCE module focuses on extracting temporal feature descriptions, and has achieved impressive outcomes. More details are illustrated in Sections 3 and 4.
This section introduces a framework for universal lesion detection based on CT slices, named the TCE-Net method. Fig. 2 shows the framework of our proposed method, which will be detailed in the following subsections. First of all, an overall description of our approach is given in Section 3.1. In this framework, we develop the temporal correlation enhancement (TCE) module (see Section 3.2), whose structure is shown in Fig. 3. Then, Faster R-CNN [21] is used as a detector to demonstrate the effectiveness of our module. As for the total loss of the training phase, a detailed description is given in Section 3.3.
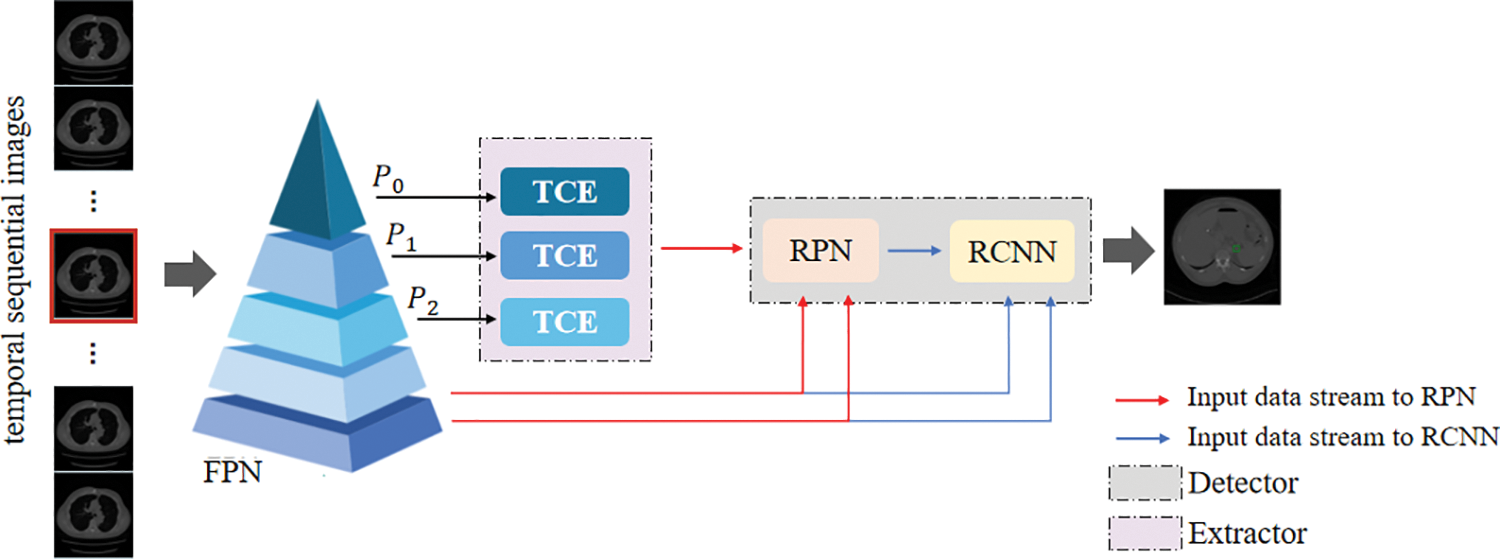
Figure 2: The framework of the proposed TCE-Net. TCE module learns the temporal correlation representation from features of sequential slices
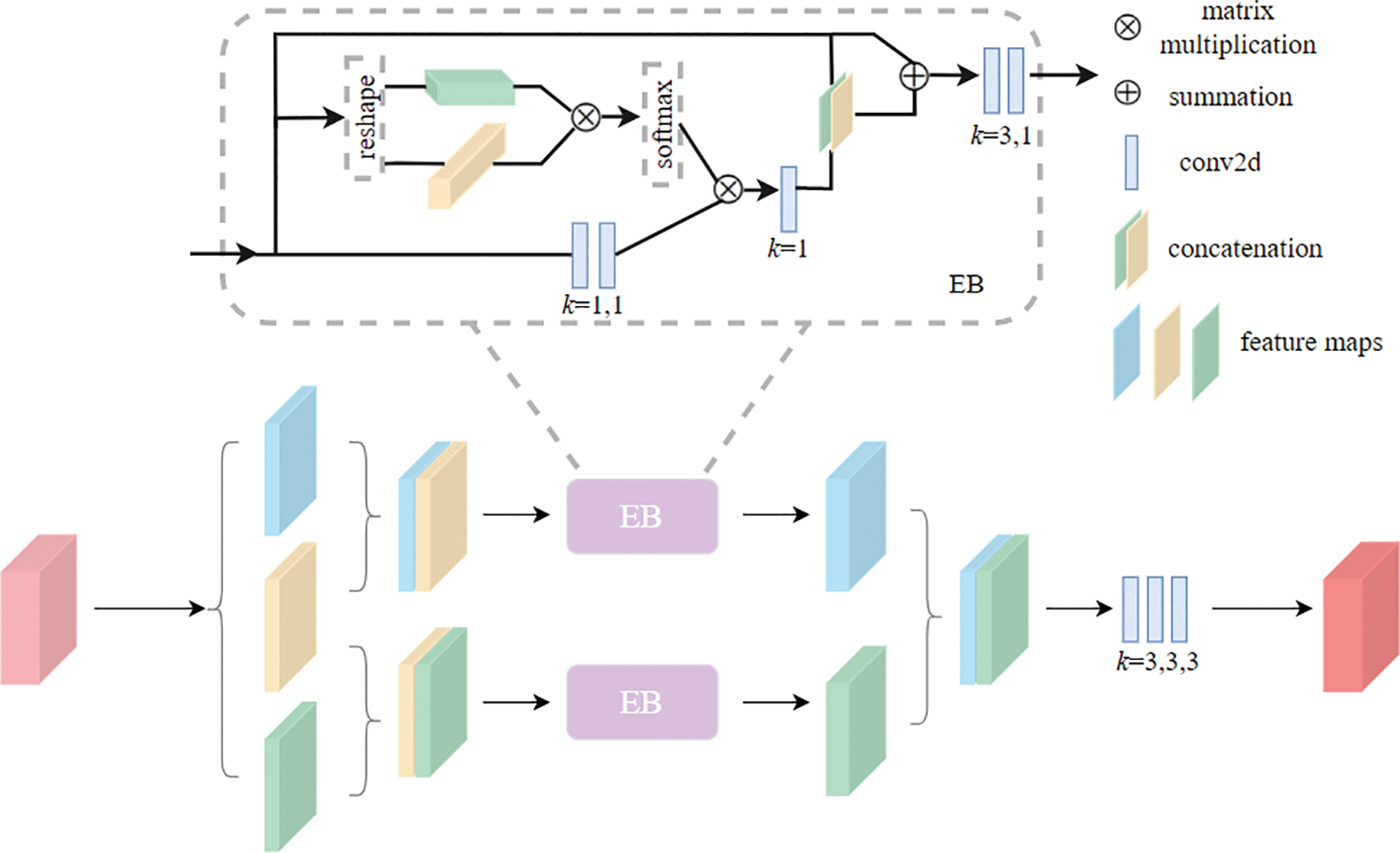
Figure 3: The pipeline of the TCE module, where the Enhanced Block (EB) helps to obtain correlation relationship between neighbouring slices group. The
Given a set of sequential CT slices from a single subject, we learn temporal correlation and spatial-temporal features from multiple sequential slices to improve the characterization of the input data for accurate lesion detection. During the training stage, we match the predicted bounding box with the ground truth using the total loss. At the inference time, the overall network generates the final prediction directly.
Here, assume that the location of the target slice is
Firstly, the pre-trained FPN is cited in [31], including bottom-up, top-down and lateral connection components and is employed to extract multi-scale feature maps from the input data to prepare for the detection of lesions of different sizes. We define the feature maps of the output of each layer of the pyramid from top to bottom as
Subsequently, the
where
Finally, the multi-scale enhanced temporal correlation features obtained by the TCE module cascade shallow features
where
In this section, we give detailed descriptions about the proposed TCE module for temporal correlation enhancement.
As shown in Fig. 3, in the TCE module, to aggregate correlations between multiple neighboring slices and reinforce the temporal correlation representation between the whole sequential slices, the study first divides the obtained feature maps of multiple slices G into N groups (N is set to 3 in this paper). Arithmetically, we define the features of N groups in G:
In Eq. (3),
where,
Enhanced Block For learning temporal correlation representations from adjacent slices, we adopt an architecture, called Enhanced Block, i.e., EB.
The study begins by extracting the context information in the x × y-plane and the correlation information in the z-axis of the input feature map x entered into the Enhanced Block, to obtain temporal correlation and spatial-temporal feature between adjacent slices, defined as
where
where the cat
To train the TCE-Net, we use lesion classification loss and bounding-box regression loss in Faster R-CNN.
where
4.1 Datasets and Evaluation Metrics
DeepLesion [4], is the largest multi-category, lesion-level annotated dataset of clinical medical CT images ever published by the NIH Clinical Center, which collects a total of 10594 CT scans before and after 4427 individual anonymous patients, including 32735 instances of labeled lesions. DeepLesion divides all data into a 70% training set, a 15% validation set, and a 15% test set. The study uses DeepLesion’s training set to train the model and tests the model in the test set to obtain the average sensitivity under different false positives, serving as an evaluation metric to compare performance with other models.
We train the model on an NVIDIA GeForce RTX 3090 GPU, using the FPN with ResNet-50 that pre-trained on ImageNet [23] in our framework. We set five anchor scales (16, 32, 64, 128, 256) and three anchor ratios (0.5, 1.0, 2.0) in RPN while the threshold value of IOU is set to 0.7, in order to balance learning speed and learning effectiveness of model. The CT slice is resized to
We adopt the stochastic gradient descent (SGD) optimizer with momentum 0.9 and weight decay 0.0001 to train the model for 14 epochs. The base learning rate is set at 0.002, reduced by a factor of 10 after the 11th and 13th epochs.
4.2 Comparison with Other Methods
We compare the proposed TCE-Net with some state-of-the-art methods including Faster R-CNN [21], 3DCE [5], ULDOR [19], GPN [18], MVP-Net [3], LENS [22] and Deformable Dilated Faster R-CNN [20]. To be fair comparison, we have re-implemented 3DCE using the FPN backbone. We perform the evaluation by computing the sensitivity values at different false positives rates and mean average precision (mAP) as illustrated in Table 1.
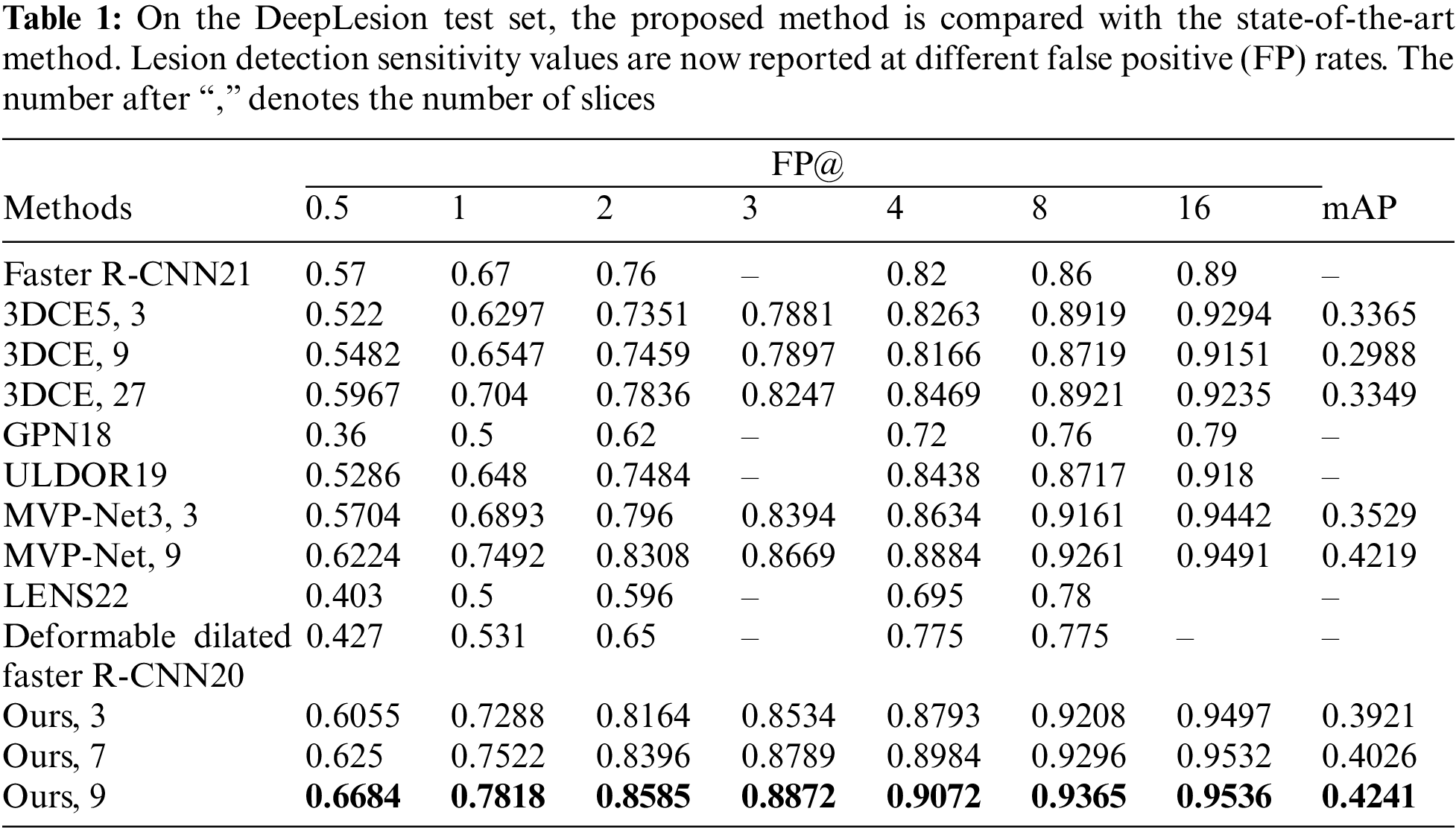
From Table 1 and Fig. 4, we can see that GPN uses lesion detection under a single-scale feature map, and the performance is significantly poorer than that of multi-scale feature-based lesion detection models like MVP-Net and ours, which illustrates the essential need for multi-scale feature extraction. Our model outperforms 3DCE 3 slices by 9.91% and MVP-Net 3 slices by 3.95% for FPs@4.0 for the same 3 slices case. More importantly, our proposed TCE-Net 9 slices achieves a performance boost of over 7.78% for FPs@1.0 and 4.6% for FPs@0.5 compared to 3DCE 27 slices and MVP-Net 9 slices, respectively. Meanwhile, the indicator mAP reaches its greatest at TCE-Net 9 slices.
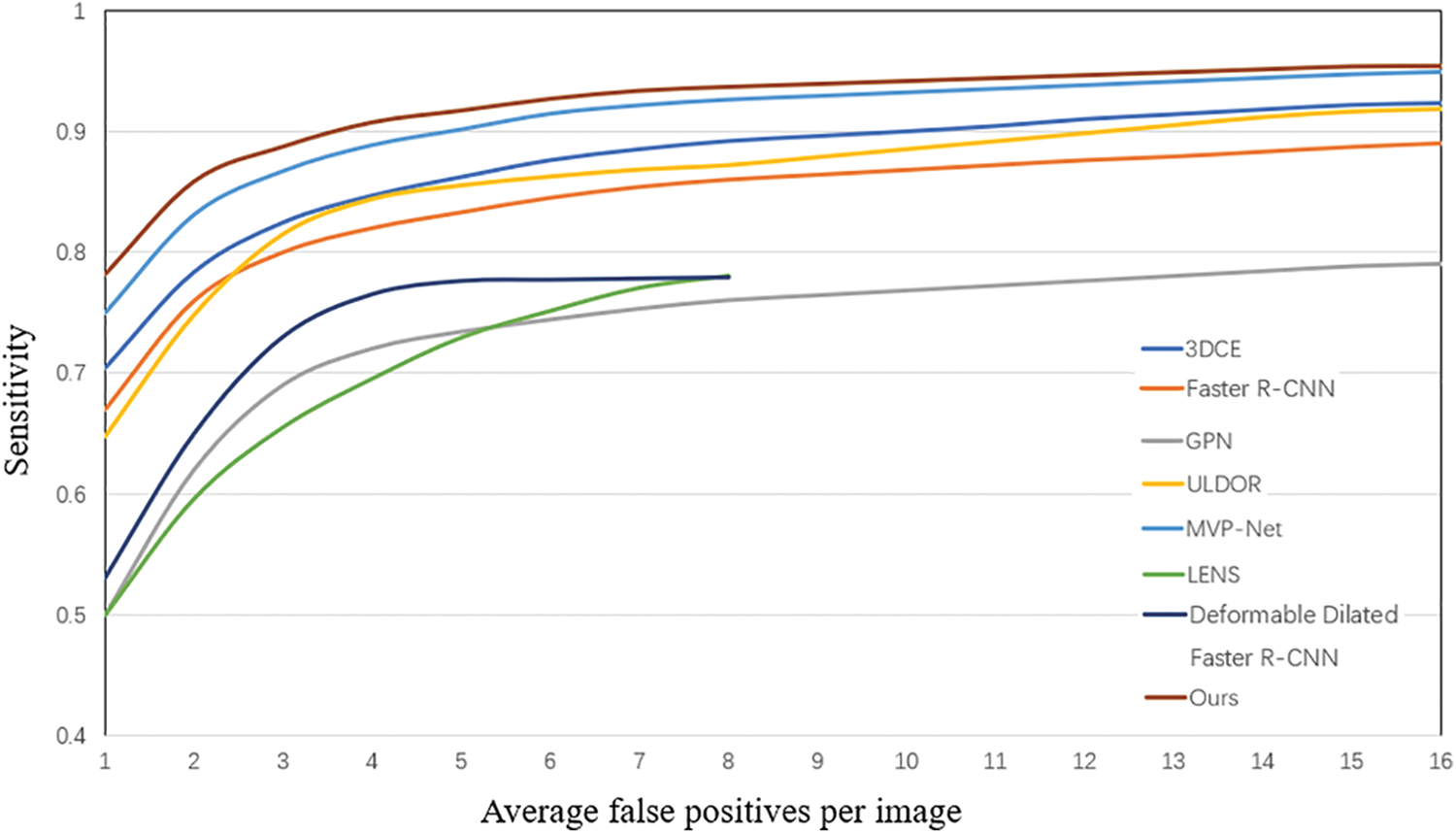
Figure 4: FROC curves of various methods on the test set
Predicted bounding boxes generated from several compared models are illustrated in Fig. 5. Our model shows superior capability to predict both the lesion location and size when compared with other methods. It is evident that the proposed method enhances the performance in detecting very small-sized or larger lesions. However, the insufficient training data for certain organ lesions resulted in inadequate generalization for the detection of these lesions as can be seen in Fig. 6.
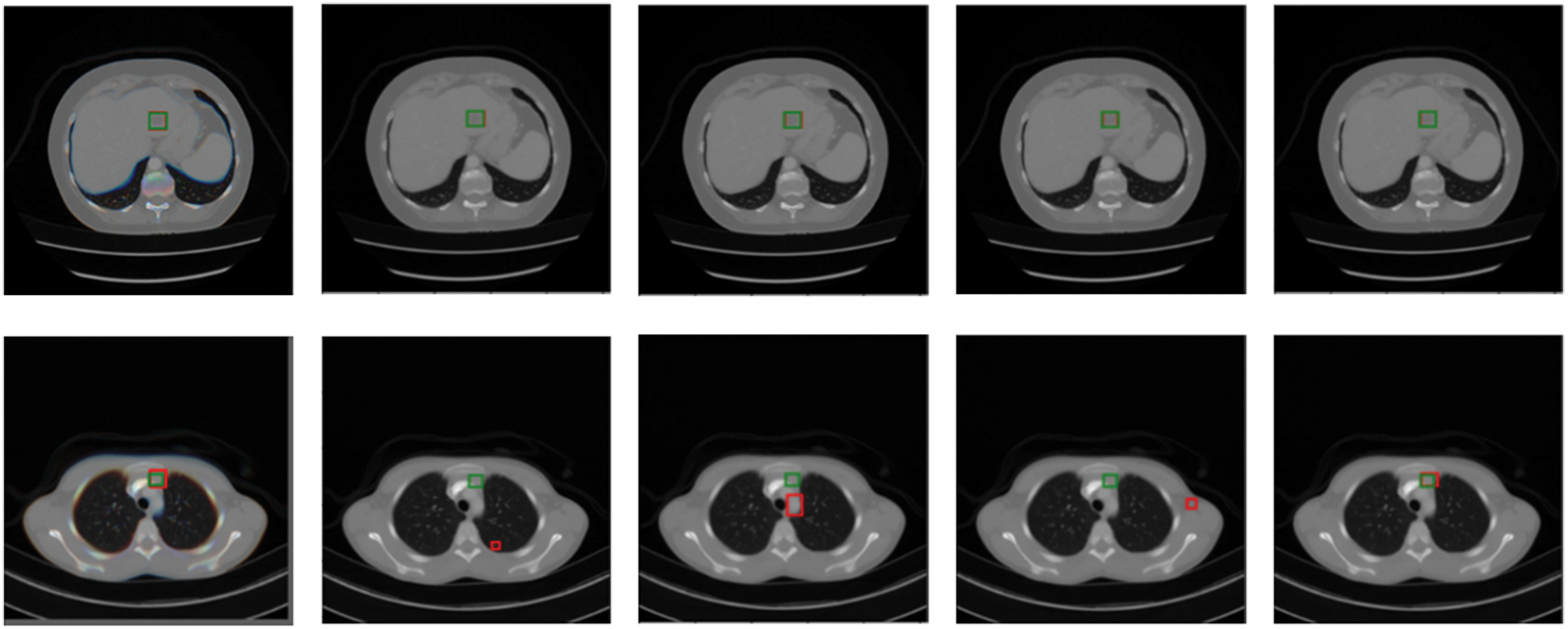
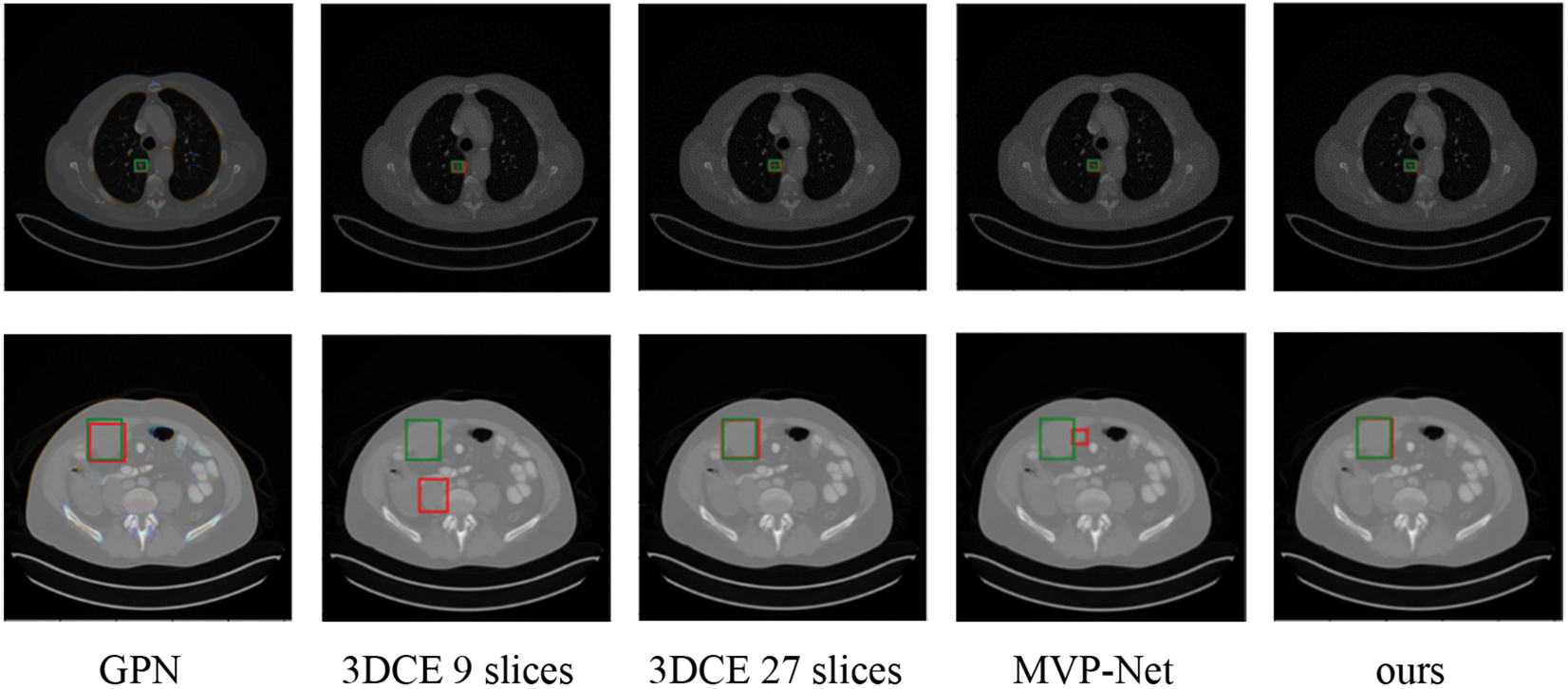
Figure 5: Qualitative comparison of several existing universal lesion detection models and ours on the testing set of DeepLesion. Green and red boxes correspond to ground-truths in the test set and predicted true positives
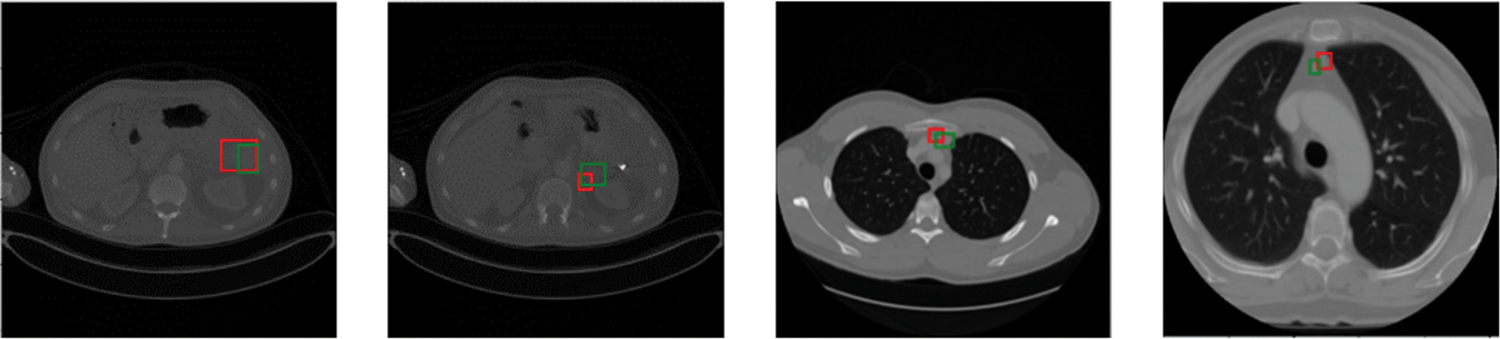
Figure 6: Visualization of inaccurate lesion detection results. Green and red boxes correspond to ground-truths in the test set and predicted true positives
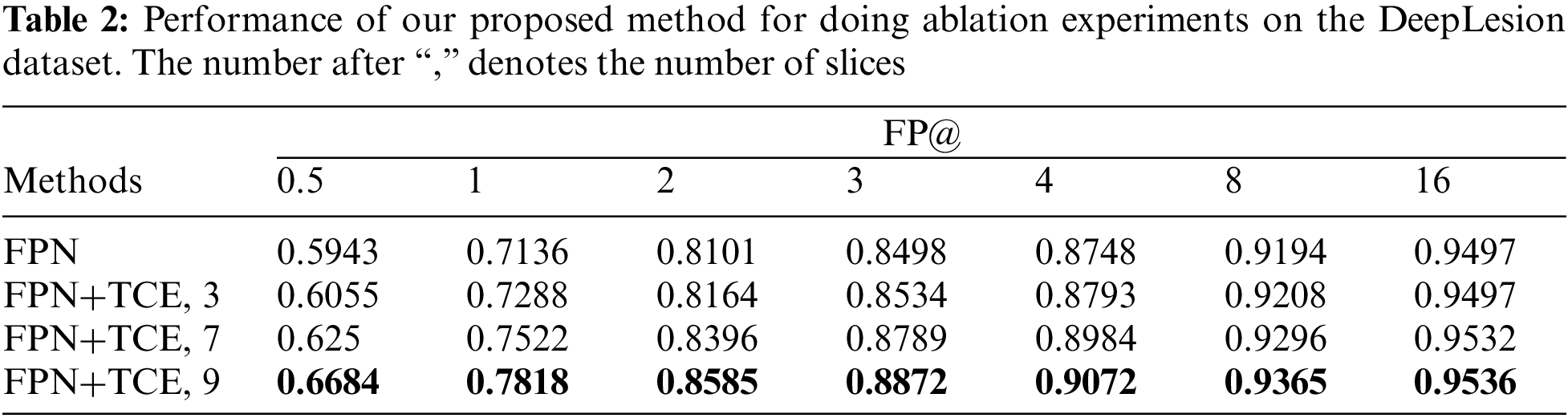
We perform a number of ablations to probe into our TCE-Net. As shown in Table 2, it shows that the use of the temporal correlation enhancement module has improved the metric by 1.52% compared to the baseline FPN, on the basis of which the feature information is increased by adding the number of input slices to 7 and 9 slices, respectively. And it can be seen that the accuracy of the model was improved by 3.86% and 7.41%, respectively, and the best results are achieved when using the combination of TCE and input 9 slices.
This work investigates the effectiveness of the enhanced temporal correlation detection network (TCE-Net) for universal lesion detection (ULD) in CT scans. The temporal correlation enhancement module has been proposed to enhance the effective fusion of multi-slice features through temporal correlation between multiple adjacent slices. We validate the effectiveness of our method with various experimental and analytic results. It shows that the proposed method brings a performance boost in the lesion detection tasks in CT scans. Universal lesion detection is a challenging task, partly because lesions in some organs have minimal differences from adjacent tissues. Therefore, false positives are high for detection in these organs. Meanwhile, the imbalance of data samples from various types of organs also limits the generalization ability of TCE-Net. In the future enriched organ CT data will be needed to train TCE-Net to improve the performance.
Acknowledgement: We would like to thank the editor and anonymous reviewers for their helpful remarks.
Funding Statement: This research was funded by Taishan Young Scholars Program of Shandong Province; and Key Development Program for Basic Research of Shandong Province (ZR2020ZD44).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Muwei Jian, Yue Jin and Hui Yu; data collection: Muwei Jian, Yue Jin; analysis and interpretation of results: Muwei Jian, Yue Jin and Hui Yu; draft manuscript preparation: Muwei Jian, Yue Jin and Hui Yu; supervision and funding acquisition: Muwei Jian. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Preprocessed data are available on request from the first author.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Shao, Q., Gong, L., Ma, K., Liu, H., Zheng, Y. et al. (2019). Attentive CT lesion detection using deep pyramid inference with multi-scale booster. In: Medical image computing and computer assisted Intervention, pp. 301–309. Shenzhen, China. [Google Scholar]
2. Yan, K., Tang, Y. B., Peng, Y. F., Sandfort, V., Bagheri, M. et al. (2019). MULAN: Multitask universal lesion analysis network for joint lesion detection, tagging, and segmentation. In: Medical image computing and computer assisted Intervention, pp. 194–202. Shenzhen, China. [Google Scholar]
3. Li, Z. H., Zhang, S., Zhang, J. G., Huang, K. Q., Wang, Y. Z. et al. (2019). MVP-Net: Multi-view FPN with position-aware attention for deep universal lesion detection. In: Medical image computing and computer assisted Intervention, pp. 13–21. Shenzhen, China. [Google Scholar]
4. Yan, K., Wang, X. S., Lu, L., Zhang, L., Harrison, A. P. et al. (2018). Deep lesion graphs in the wild: Relationship learning and organization of significant radiology image findings in a diverse large-scale lesion database. In: Computer vision and pattern recognition, vol. 17, pp. 9261–9270. Salt Lake City, USA. [Google Scholar]
5. Yan, K., Bagheri, M., Summers, R. M. (2018). 3D context enhanced region-based convolutional neural network for end-to-end lesion detection. In: Medical image computing and computer assisted Intervention, pp. 511–519, Granada Spain. [Google Scholar]
6. Ding, J., Li, A., Hu, Z. Q., Wang, L. W. (2017). Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks. In: Medical image computing and computer assisted Intervention, pp. 559–567. Quebec, Canada. [Google Scholar]
7. Zhu, W. T., Liu, C., Fan, W., Xie, X. H. (2018). DeepLung: Deep 3D dual path nets for automated pulmonary nodule detection and classification. IEEE Winter Conference on Applications of Computer Vision, pp. 673–681. California, USA. [Google Scholar]
8. Xie, H. T., Yang, D. B., Sun, N. N., Chen, Z. N., Zhang, Y. D. (2019). Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recognition, 85, 109–119. [Google Scholar]
9. Zhu, W. T., Vang, Y. S., Huang, Y. F., Xie, X. H. (2018). DeepEM: Deep 3D ConvNets with EM for weakly supervised pulmonary nodule detection. In: Medical image computing and computer assisted Intervention, pp. 812–820. Granada Spain. [Google Scholar]
10. Khosravan, N., Bagci, U. (2018). S4 ND: Single-shot single-scale lung nodule detection. 21st International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 794–802. Granada Spain. [Google Scholar]
11. Zhu, Z. T., Yan, K., Jin, D., Cai, J. Z., Ho, T. Y. et al. (2020). Detecting scatteredly-distributed, small, and critically important objects in 3D oncology imaging via decision stratification. Computer Vision and Pattern Recognition, Washington Seattle, USA. [Google Scholar]
12. Shin, H. C., Roth, H. R., Gao, M. C., Lu, L., Xu, Z. Y. et al. (2016). Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Transactions on Medical Imaging, 35, 1285–1298. [Google Scholar] [PubMed]
13. Roth, H. R., Lu, L., Liu, J., Yao, J., Seff, A. et al. (2016). Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Transactions on Medical Imaging, 35, 1170–1181. [Google Scholar] [PubMed]
14. Wang, X. D., Han, S. Z., Chen, Y. Q., Gao, D. S., Vasconcelos, N. (2019). Volumetric attention for 3D medical image segmentation and detection. In: Medical image computing and computer assisted Intervention, pp. 175–184. Shenzhen, China. [Google Scholar]
15. Zlocha, M., Dou, Q., Glocker, B. (2019). Improving RetinaNet for CT lesion detection with dense masks from weak RECIST labels. In: Medical image computing and computer assisted Intervention, pp. 402–410. Shenzhen, China. [Google Scholar]
16. Zhang, S., Xu, J. C., Chen, Y. C., Ma, J. C., Yu, Y. Z. (2020). Revisiting 3D context modeling with supervised pre-training for universal lesion detection in CT slices. In: Medical image computing and computer assisted Intervention, pp. 542–551. Lima, Peru. [Google Scholar]
17. Cheng, X. L., Xiong, H., Fan, D. P., Zhong, Y. R., Harandi, M. et al. (2022). Implicit motion handling for video camouflaged object detection. Conference on Computer Vision and Pattern Recognition, pp. 13854–13863. New Orleans, Louisiana, USA. [Google Scholar]
18. Li, Y. (2019). Detecting lesion bounding ellipses with Gaussian proposal networks. Machine Learning in Medical Imaging, 11861, 337–344. [Google Scholar]
19. Tang, Y. B., Yan, K., Tang, Y. X., Liu, J. M., Xiao, J. et al. (2019). ULDor: A universal lesion detector For Ct scans with Pseudo masks and hard negative example mining. IEEE 16th International Symposium on Biomedical Imaging, pp. 833–836. Venezia, Italy. [Google Scholar]
20. Hellmann, F., Ren, Z., André, E., Schuller, B. W. (2021). Deformable dilated faster R-CNN for universal lesion detection in CT images. 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society, pp. 2896–2902. Guadalajara, Mexico. [Google Scholar]
21. Ren, S., He, K., Girshick, R., Sun, J. (2016). Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Transactions on Pattern Analysis and Machine Intelligence, 39(6), 1137–1149. [Google Scholar] [PubMed]
22. Yan, K., Cai, J. Z., Zheng, Y. J., Harrison, A. P., Jin, D. K. et al. (2021). Learning from multiple datasets with heterogeneous and partial labels for universal lesion detection in CT. IEEE Transactions on Medical Imaging, 40, 2759–2770. [Google Scholar] [PubMed]
23. Deng, J., Dong, W., Socher, R., Li, L. J., Li, K. et al. (2009). ImageNet: A large-scale hierarchical image database. IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255. Miami, FL, USA. [Google Scholar]
24. Sheoran, M., Dani, M., Sharma, M., Vig, L. (2022). An efficient anchor-free universal lesion detection in CT-scans. IEEE 19th International Symposium on Biomedical Imaging, pp. 1–4. Kolkata, India. [Google Scholar]
25. Cai, J. Z., Yan, K., Cheng, C. T., Xiao, J., Liao, C. H. et al. (2020). Deep volumetric universal lesion detection using light-weight Pseudo 3D convolution and surface point regression. Medical Image Computing and Computer Assisted Intervention, 12264, 3–13.–> [Google Scholar]
26. Liu, Z., Han, K., Xue, K. F., Song, Y. Q., Liu, L. et al. (2022). Improving CT-image universal lesion detection with comprehensive data and feature enhancements. Multimedia Systems, 28, 1741–1752. [Google Scholar]
27. Tao, Q. Y., Ge, Z. Y., Cai, J. F., Yin, J. X., See, S. (2019). Improving deep lesion detection using 3D contextual and spatial attention. In: Medical image computing and computer assisted Intervention, pp. 185–193. Shenzhen, China. [Google Scholar]
28. Li, H., Han, H., Zhou, S. K. (2020). Bounding maps for universal lesion detection. Medical Image Computing and Computer Assisted Intervention, vol. 12264, pp. 417–428. [Google Scholar]
29. Li, H., Chen, L., Han, H., Chi, Y., Zhou, S. K. (2021). Conditional training with bounding map for universal lesion detection. Medical Image Computing and Computer Assisted Intervention, vol. 12905, pp. 141–152. [Google Scholar]
30. Sheoran, M., Dani, M., Sharma, M., Vig, L. (2022). DKMA-ULD: Domain knowledge augmented multi-head attention based robust universal lesion detection. arXiv:2203.06886. [Google Scholar]
31. Lin, T. Y., Dollar, P., Girshick, R., He, K., Hariharan, B. et al. (2017). Feature pyramid networks for object detection. IEEE Conference on Computer Vision and Pattern Recognition, pp. 936–944. Hawaii, USA. [Google Scholar]
32. He, K., Zhang, X., Ren, S., Sun, J. (2016). Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778. Las Vegas, USA. [Google Scholar]
33. Yu, F., Koltun, V. (2016). Multi-scale context aggregation by dilated convolutions. International Conference on Learning Representations, San Juan, Puerto Rico. [Google Scholar]
34. Qiu, Z., Yao, T., Mei, T. (2017). Learning spatio-temporal representation with Pseudo-3D residual networks. Proceedings of the IEEE International Conference on Computer Vision, pp. 5533–5541. Hawaii, USA. [Google Scholar]
35. Storn, R., Price, K. (1997). Differential evolution—A simple and efficient heuristic for global optimization over continuous spaces. Journal of Global Optimization, 11, 341–359. [Google Scholar]
36. Lin, T. Y., Goyal, P., Girshick, R., He, K. M., Dollár, P. (2020). Focal loss for dense object detection. IEEE Transactions on Pattern Analysis and Machine Intelligence, 42, 318–327. [Google Scholar] [PubMed]
37. Zhang, H., Chen, Y. R., Song, Y. N., Xiong, Z. L., Yang, Y. M. et al. (2019). Automatic kidney lesion detection for CT images using morphological cascade convolutional neural networks. IEEE Access, 7, 83001–83011. [Google Scholar]
38. Lee, S. G., Bae, J. S., Kim, H., Kim, J. H., Yoon, S. (2018). Liver lesion detection from weakly-labeled multi-phase CT volumes with a grouped single shot MultiBox detector. In: Medical image computing and computer assisted Intervention, pp. 693–701. [Google Scholar]
39. Liang, D., Lin, L. F., Chen, X., Hu, H. J., Zhang, Q. et al. (2019). Multi-stream scale-insensitive convolutional and recurrent neural networks for liver tumor detection in dynamic CT images. IEEE International Conference on Image Processing, pp. 794–798. Taipei, Taiwan. [Google Scholar]
40. Sheng, M., Li, J., Bhatti, U. A., Liu, J., Huang, M. et al. (2023). Zero watermarking algorithm for medical image based on Resnet50-DCT. Computers, Materials & Continua, 75(1), 293–309. https://doi.org/10.32604/cmc.2023.036438 [Google Scholar] [CrossRef]
41. Karakis, R. (2023). MI-STEG: A medical image steganalysis framework based on ensemble deep learning. Computers, Materials & Continua, 74(3), 4649–4666. https://doi.org/10.32604/cmc.2023.035881 [Google Scholar] [CrossRef]
42. Geman, O., Postolache, O. A., Chiuchisan, I., Prelipceanu, M., Ritambhara (2019). An intelligent assistive tool using exergaming and response surface methodology for patients with brain disorders. IEEE Access, 7, 21502–21513. [Google Scholar]
43. Shao, J., Chen, S., Zhou, J., Zhu, H., Wang, Z. et al. (2023). Application of U-Net and optimized clustering in medical image segmentation: A review. Computer Modeling in Engineering & Sciences, 136(3), 2173–2219. https://doi.org/10.32604/cmes.2023.025499 [Google Scholar] [CrossRef]
44. Postolache, O., Hemanth, D. J., Alexandre, R., Gupta, D., Geman, O. et al. (2021). Remote monitoring of physical rehabilitation of stroke patients using IoT and virtual reality. IEEE Journal on Selected Areas in Communications, 39(2), 562–573. [Google Scholar]
45. Hemanth, D. J., Anitha, J., Naaji, A., Geman, O., Popescu, D. E. et al. (2019). A modified deep convolutional neural network for abnormal brain image classification. IEEE Access, 7, 4275–4283. [Google Scholar]
46. Jian, M., Wu, R., Chen, H., Fu, L., Yang, C. (2023). Dual-Branch-UNet: A dual-branch convolutional neural network for medical image segmentation. Computer Modeling in Engineering & Sciences, 137(1), 705–716. [Google Scholar]
47. Jian, M., Wang, J., Yu, H., Wang, G. (2021). Integrating object proposal with attention networks for video saliency detection. Information Sciences, 576, 819–830. [Google Scholar]
48. Yin, Y., Han, Z., Jian, M., Wang, G., Chen, L. et al. (2023). AMSUnet: A neural network using atrous multi-scale convolution for medical image segmentation. Computers in Biology and Medicine, 162, 107120. [Google Scholar] [PubMed]
49. Rahi, B., Li, M. Z., Qi, M. (2023). A review of techniques on gait-based person re-identification. International Journal of Network Dynamics and Intelligence, 2, 66–92. [Google Scholar]
50. Shakiba, F. M., Shojaee, M., Azizi, S. M., Zhou, M. (2022). Real-time sensing and fault diagnosis for transmission lines. International Journal of Network Dynamics and Intelligence, 1, 36–47. [Google Scholar]
51. Li, X., Li, M. L., Yan, P. F., Li, G. Y., Jiang, Y. C. et al. (2023). Deep learning attention mechanism in medical image analysis: Basics and beyonds. International Journal of Network Dynamics and Intelligence, 2, 93–116. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools