 Open Access
Open Access
ARTICLE
An Efficient Numerical Scheme for Biological Models in the Frame of Bernoulli Wavelets
1
Department of Scientific Research, Yunnan Normal University, Kunming, 650500, China
2
Department of Mathematics and Science Education, Harran University, Sanliurfa, 63050, Türkiye
3
Department of Mathematics, Bangalore University, Bengaluru, 560056, India
4
School of Information Science and Technology, Yunnan Normal University, Kunming, 650500, China
5
Faculty of Engineering and Architecture, Kirsehir Ahi Evran University, Kırsehir, 40500, Türkiye
* Corresponding Author: Haci Mehmet Baskonus. Email:
(This article belongs to the Special Issue: Recent Developments on Computational Biology-I)
Computer Modeling in Engineering & Sciences 2023, 137(3), 2381-2408. https://doi.org/10.32604/cmes.2023.028069
Received 28 November 2022; Accepted 17 March 2023; Issue published 03 August 2023
Abstract
This article considers three types of biological systems: the dengue fever disease model, the COVID-19 virus model, and the transmission of Tuberculosis model. The new technique of creating the integration matrix for the Bernoulli wavelets is applied. Also, the novel method proposed in this paper is called the Bernoulli wavelet collocation scheme (BWCM). All three models are in the form system of coupled ordinary differential equations without an exact solution. These systems are converted into a system of algebraic equations using the Bernoulli wavelet collocation scheme. The numerical wave distributions of these governing models are obtained by solving the algebraic equations via the Newton-Raphson method. The results obtained from the developed strategy are compared to several schemes such as the Runge Kutta method, and ND solver in mathematical software. The convergence analyses are discussed through theorems. The newly implemented Bernoulli wavelet method improves the accuracy and converges when it is compared with the existing methods in the literature.Keywords
The evolution and development of the contemporary world can’t be separated from mathematics. Mathematics is related to nearly all human movement. A mathematical model is a helpful tool for solving real-life problems. Application to determine the blowout of the transmittable disease model in a particular region is one of the finest examples of this. Researchers started studying biological systems over many years. These biological systems are usually in the form of a system of coupled ordinary differential equations. Only a few of these biological models have exact solutions and are typically intricate. Therefore, there is a necessity for a decent approximated solution. Numerical methods are often the best choice for these systems as they will produce nonnegative solutions and describe these systems’ dynamic behavior. In these systems, the specified variables commonly represent nonnegative quantities, such as the size of the population, number of chemical components, concentration, and physical properties.
Due to technological advances in medical research, like vaccination and antibodies to viral diseases, it was predictable that the spread of transmissible diseases would be overcome. But it didn’t. The reason is that most of the developed countries have concentrated their research on cancer and other incurable diseases. Consequently, infectious diseases still cause intense suffering and a high mortality rate. Ebola, Yellow fever, Aids, Malaria, and other names have left a deep scar on the history of humanity forever.
Among all the above viruses, Dengue fever is mainly extensive in Southern Asia. Dengue is a complicated disease transmitted to humans through mosquitoes belonging to the Genus Aedes. They exist in two forms: Dengue Haemorrhagic Fever (DHF) and classical dengue. The furthermost problematic characteristic of dengue is that it can be triggered by four different serotypes known as DEN1, DEN2, DEN3, and DEN4. If the person is infected with any one of the types will never be infected by the same type again, a phenomenon called homologous immunity. Though, once the human is infected by any of the above types, in about 12 weeks, he loses immunity to the other types, known as heterologous immunity.
The mathematical modelling of Dengue fever is [1]:
where
Coronavirus disease (COVID-19) is a new disease generated by a recently recognized virus, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) was first reported in Wuhan, China, in December 2019 and was declared a pandemic by the World Health Organisation (WHO) on 11th March 2020. The transmission of viruses in human beings from the compartment of S(t) to the I(t) and from the compartment of I(t) to the R(t) can be described by the set of three systems of non-linear ordinary differential equations with only two constraint approximations.
The simple SIR model for COVID-19 virus transmission is [6]:
where S(t) represents the fraction of Susceptible persons, I(t) represents the fraction of Infected persons, R(t) represents the fraction of recovered persons,
TB (Tuberculosis) is an infectious epidemic usually produced by bacteria named Mycobacteria Tuberculosis which outbreaks human lungs. In some situations, it can also affect the other parts of the body, like the kidney, spine, skin, brain, etc. TB is spread from an infected person to an average person through air droplets that are contaminated with germs. A person will become easily contaminated when they breathe a few microorganism germs. TB-infected individuals can be diagnosed through blood and skin tests. TB creates a high risk for HIV and diabetes patients. The signs of the live TB disease include a cough that tests for more than three weeks, loss in weight, coughing up blood, felt tiredness, exhaustion, chest pain, and night sweats. The World Health Organisation states that Tuberculosis has infected one-third of the world population. In 2018, an estimated 10 million new cases of TB disease were described, and a total of 1.5 million people died from TB worldwide. Apart from this, 1.2 million estimated TB patients died HIV-negative, and 0.25 million died HIV-positive in 2018. A large number of TB patients died in the region of low- and middle-income countries (LMICs), including India (27%), Nigeria (4%), Indonesia (8%), Pakistan (6%), Philippine (6%), Bangladesh (4%) and South Africa (24%). Southeast Asia (44%) has reported most of the occurred cases of TB [11], even though TB is a treatable transferrable disease using drug therapy [12].
The SIR Model of Tuberculosis is as follows [13]:
where N = S + I + R, S(t) is the number of vulnerable individuals in the population at the time t, I(t) is the number of infected individuals in the population at time t, R(t) is the number of individuals who recovered in the population at time t, r is the rate of transmission of disease from susceptible become infectious (
The subject of wavelet appeared in the mid-1980s, influenced by ideas from both pure and applied mathematics. In 1807, Joseph Fourier developed a method for representing signals with a series of coefficients based on an analysis function. He laid the mathematical basis from which the wavelet theory was developed. The first to mention wavelets was Alfred Haar in 1909 in his Ph.D. thesis. The wavelet theory is a relatively renewed and emerging theory in mathematical research. wavelet analysis is a great tool that significantly impacts study and engineering. The primary criteria that draw researchers to the wavelet are orthogonality and effective localization. With the help of wavelets that are mathematical operations, data may be divided into several frequency components, each of which can then be analyzed at a different resolution. It is possible to employ wavelets as a mathematical tool to extract information from a variety of data formats. Wavelets are based on the fundamental theory of expressing a complicated function by a set of self-similar functions by super positioning, a principle introduced by Joseph Fourier in the 1800s. They have advantages over traditional Fourier methods in analyzing physical situations where the signal contains discontinuities and sharp spikes. Bernoulli wavelet is one of the continuous basis wavelets with orthogonality, compact support, etc. Due to these properties, wavelet methods yield better accuracy in the solution. Some of the problems tackled by the wavelets collocation method on mathematical models arising in science and technology are Laguerre, Hermite, and Legendre wavelets methods to solve some of the mathematical models [23–31], the Bernoulli wavelets scheme for fractional delay differential equations [32] and Fractional pantograph differential equations [33], Non-linear second-order Lane–Emden pantograph delay differential mode solved by Bernoulli wavelets [34], Fractional Riccati differential equation [35], Fractional order model of novel coronavirus outbreak [36], non-linear singular Lane–Emden type equations arising in astrophysics are successfully tackled by Bernoulli wavelet [37], Designing of Morlet wavelet as a neural network for a novel prevention category in the HIV system [38], Bernstein wavelets for SIR epidemic infectious model [39], Bernstein wavelets for predator-prey dynamical system [40], Bernoulli wavelets collocation method newly applied for different mathematical problems [41–44], Bernoulli wavelets for Glucose-Insulin regulatory dynamical system [45], and Fibonacci wavelets [46]. The results of BWCS are compared with other methods, like the Runge Kutta method and ND Solver. The results obtained in this paper are new and do not exist in the literature. This paper is organized as follows: Section 2 gives the preliminaries of the Bernoulli wavelet, and its properties, Section 3 deals with the functional matrix of the Bernoulli wavelet, and Section 4 reflects the method of solution and application of the proposed method deals with Section 5. Lastly, Section 6 presents conclusions on the new BWCS.
2 Preliminaries of Bernoulli Wavelet and Its Properties
The Bernoulli wavelets are defined as [47]:
with
where
The coefficient
Some initial Bernoulli numbers are:
with
Some initial Bernoulli polynomials are:
Theorem 1 [48]: Let H be a Hilbert space and W be a closed subspace of H such that
Theorem 2 [48,49]: Let
3 Functional Matrix of Bernoulli Wavelets
Some Bernoulli wavelet basis at
On integrating the above basis concerning x limit from
Hence,
where,
In general, the Bernoulli wavelet’s first integration can be depicted as;
Here, we would like to bring the solution of the Dengue Fever Model (1.1) to the Bernoulli wavelet space. Assume that,
Integrate Eqs. (5)–(7) concerning ‘
From the Eq. (4) along with initial conditions expressed in terms of
where D, E, and F are the known vectors. Substitute Eqs. (5)–(8) in (1) we get
Now collocate each Eq. (9) with the following grid points:
We get a non-linear system of algebraic equations from the above procedure:
Eq. (10) can be solved by Newton’s Raphson method. This yields the values of Bernoulli’s unknown coefficients and substituting these values on (8) yields Bernoulli’s wavelet-based numerical solutions for the defined model.
Next, we would like to bring the solution of the Coronavirus Model (2) to the Bernoulli wavelet space. Substitute Eqs. (5)–(8) in (2) we get
Now collocate each equation in (11) with the following grid points:
We get a non-linear system of algebraic equations from the above procedure:
Eq. (12) can be solved by Newton’s Raphson method. This yields the values of Bernoulli’s unknown coefficients, after substituting these values on (8) delivers Bernoulli’s wavelet-based numerical solutions for the defined model.
Next, we would like to bring the solution of the Tuber coulis model (3) to the Bernoulli wavelet space. Substitute Eqs. (5)–(8) in (3) we get
Now collocate each equation in (13) with the following grid points:
We get a non-linear system of algebraic equations from the above procedure:
Eq. (14) can be solved by Newton’s Raphson method. This yields the values of unknown coefficients, after substituting these values on (8) produces Bernoulli’s wavelet-based numerical solutions for the defined model.
5 Numerical Implementation of the Biological Models
5.1 SIR Model of Dengue Fever [1]
Here we present our computational results and provide a discussion of the results. Considering the mathematical model (1) with the initial conditions:






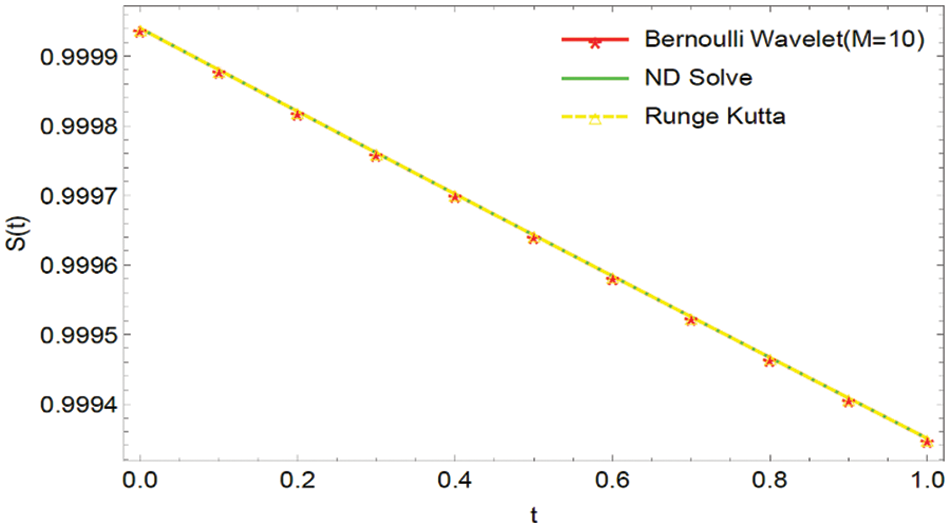
Figure 1: Graphical comparison of the BWCS solution, Runge-Kutta method solution with ND Solver solution for
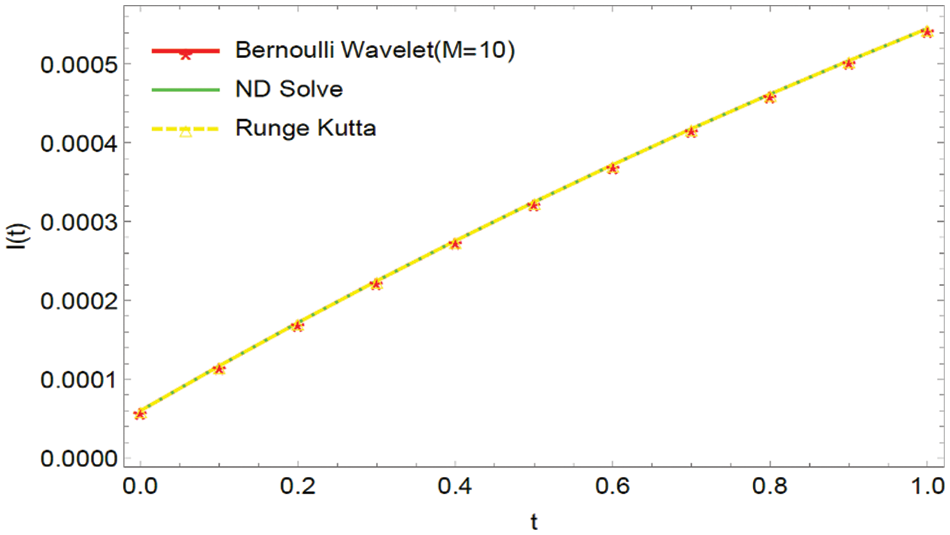
Figure 2: Graphical comparison of the BWCS solution, Runge-Kutta method solution with ND Solver solution for
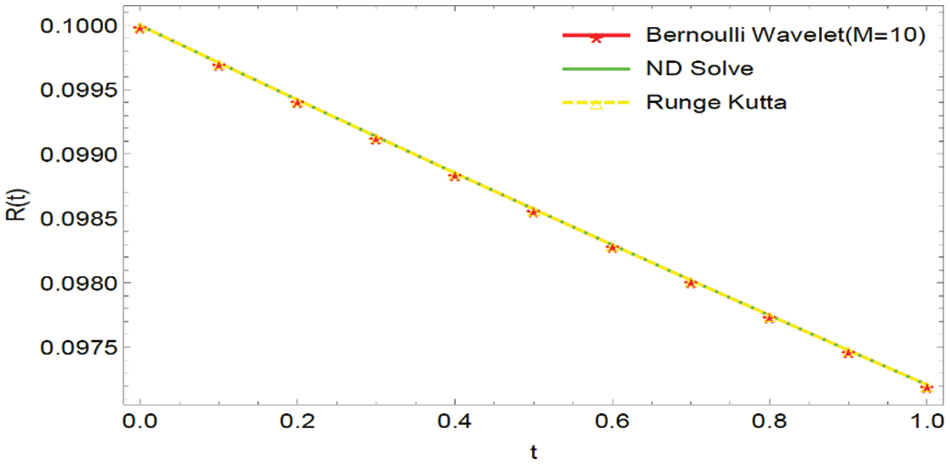
Figure 3: Graphical comparison of the BWCS, Runge-Kutta method solution with ND Solver solution for

Figure 4: Graphical representation of Absolute Errors (AE) for

Figure 5: Graphical representation of Absolute Errors (AE) for

Figure 6: Graphical representation of Absolute Errors (AE) for
5.2 Sir Model of COVID-19 Virus
Here we are considering the mathematical model of the COVID-19 virus (2) with the initial conditions:





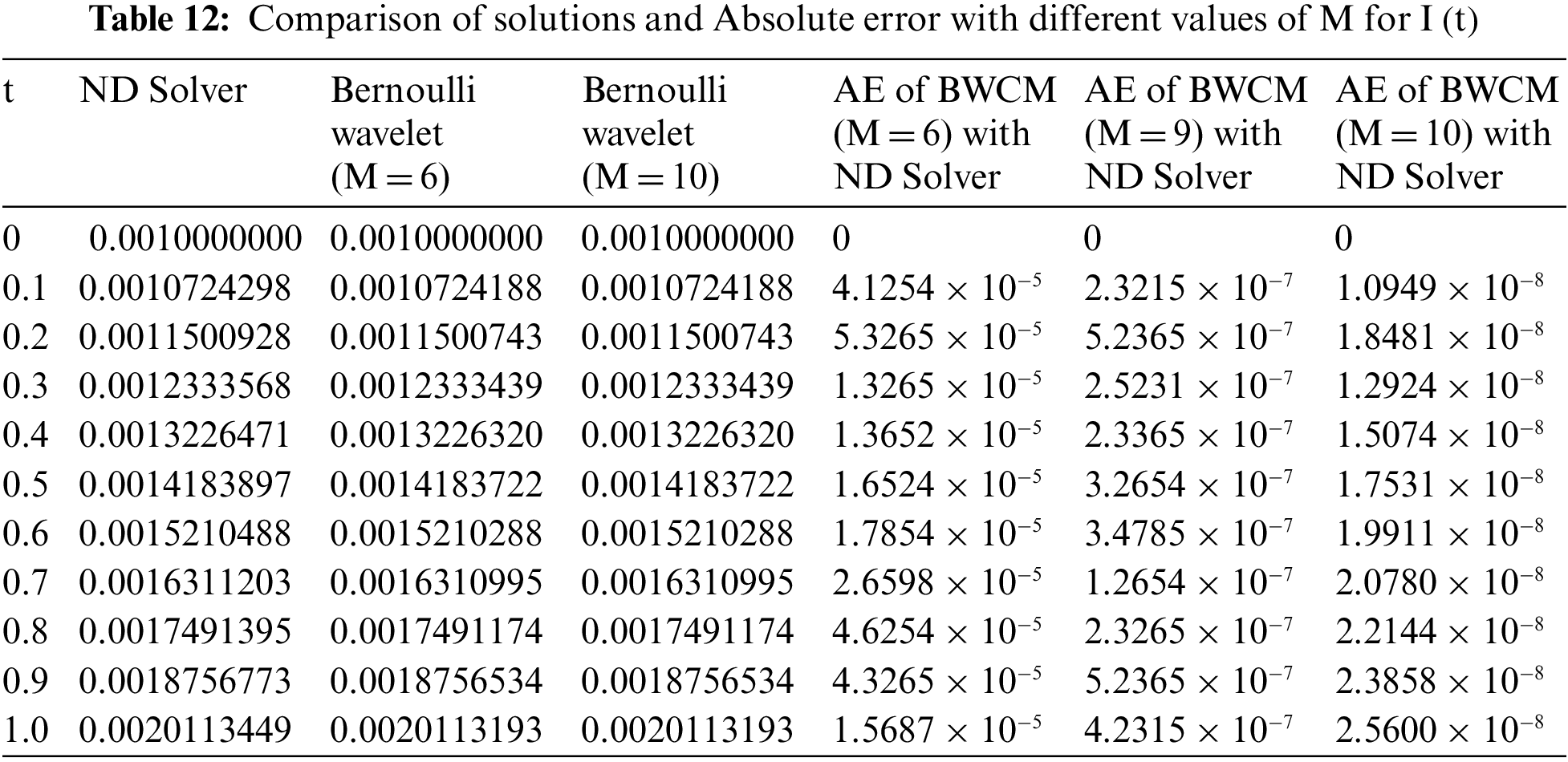
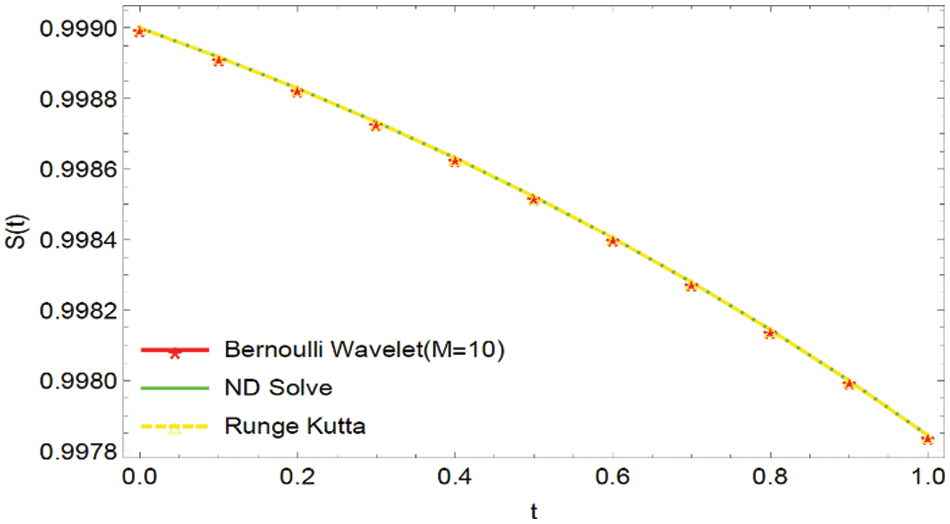
Figure 7: Graphical comparison of the BWCS solution, Runge-Kutta method solution with ND Solver solution for

Figure 8: Graphical comparison of the BWCS solution, Runge-Kutta method solution with ND Solver solution for

Figure 9: Graphical comparison of the BWCS solution, Runge-Kutta method solution with ND Solver solution for

Figure 10: Graphical representation of Absolute Errors (AE) for

Figure 11: Graphical representation of Absolute Errors (AE) for
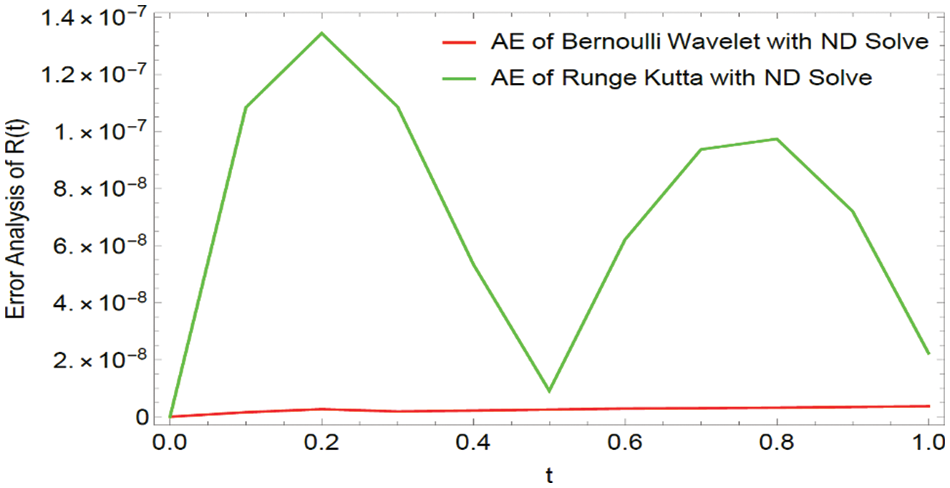
Figure 12: Graphical representation of Absolute Errors (AE) for
5.3 SIR Model for Transmission of Tuberculosis
In this section, we are considering the mathematical model for transmission of Tuberculosis (3) with the initial conditions:







Figure 13: Graphical comparison of the BWCS, Runge-Kutta method solution with ND Solver solution for

Figure 14: Graphical comparison of the BWCS solution, Runge-Kutta method solution with ND Solver solution for

Figure 15: Graphical comparison of the BWCS solution, Runge-Kutta method solution with ND Solver solution for

Figure 16: Graphical representation of Absolute Errors (AE) for

Figure 17: Graphical representation of Absolute Errors (AE) for

Figure 18: Graphical representation of Absolute Errors (AE) for
In this article, we primarily concentrated on the numerical computations of three biological models: the Dengue fever model, the COVID-19 model, and the transmission of Tuberculosis model. We developed the functional integration matrix for the Bernoulli wavelet and generated a novel technique called BWCS. We used this functional matrix to estimate the mathematical model’s numerical solution, which is in the form of a non-linear system of ordinary differential equations. Our numerical results are compared with the Runge-Kutta method and ND solver solution through tables and Figures. Tables and Figures exposed the efficiency of our projected approach and compared it with different techniques. The received computational result indicates that our projected procedure is valuable and specific in contrast with other techniques. Also, we have introduced some theorems on convergence analysis. Tables 1–9 reveal that the BWCM solution is better than the Runge-Kutta method solution as compared with the ND solver solution. Figs. 1–3, 7–9, and 13–15 show that the BWCS solution is very close to the ND solver solution. Figs. 4–6, 10–12, and 16–18 reveal that the absolute error of the proposed method is better than the absolute error of the Runge-Kutta method when compared with the ND solver solution. Tables and Figures reveal that the BWCS converges rapidly compared to the Runge-Kutta method. This concludes that the developed strategy is a well-suited technique for the numerical approximations of biological models.
Acknowledgement: Authors thank editors and reviewers for helping the enrichment of this article. Also, Kumbinarasaiah S thanks Bangalore University for its support.
Funding Statement: The authors received no specific support for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
REFERENCES
1. Khalid, M., Sultana, M., Khan, F. S. (2015). Numerical solution of SIR model of dengue fever. International Journal of Computer Applications, 118(21). [Google Scholar]
2. Rangkuti, Y. M., Side, S., Noorani, M. S. M. (2014). The numerical analytic solution of SIR model of dengue fever disease in South Sulawesi uses homotopy perturbation and variational iteration methods. Journal of Mathematical and Fundamental Sciences, 46(1), 91–105. [Google Scholar]
3. Mungkasi, S. (2020). Improved variational iteration solutions to the SIR model of dengue fever disease for the case of South Sulawesi. Journal of Mathematical & Fundamental Sciences, 52(3), 297–311. [Google Scholar]
4. Umar, M., Sabir, Z., Raja, M. A. Z., Sánchez, Y. G. (2020). A stochastic numerical computing heuristic of SIR nonlinear model based on dengue fever. Results in Physics, 19, 103585. [Google Scholar]
5. Lede, Y. K., Mungkasi, S. (2019). Performance of the Runge-Kutta methods in solving a mathematical model for the spread of dengue fever disease. AIP Conference Proceedings, 2202, 020044. [Google Scholar]
6. Suba, M., Shanmugapriya, R., Balamuralitharan, S., Joseph, G. A. (2020). Current mathematical models and numerical simulation of sir model for coronavirus disease-2019 (COVID-19). European Journal of Molecular & Clinical Medicine, 7(5), 41–54. [Google Scholar]
7. Zeb, A., Alzahrani, E., Erturk, V. S., Zaman, G. (2020). Mathematical model for coronavirus disease 2019 (COVID-19) containing isolation class. BioMed Research International, 2020, 3452402. [Google Scholar] [PubMed]
8. Annas, S., Pratama, M. I., Rifandi, M., Sanusi, W., Side, S. (2020). Stability analysis and numerical simulation of SEIR model for pandemic COVID-19 spread in Indonesia. Chaos, Solitons & Fractals, 139, 110072. [Google Scholar]
9. ud Din, R., Algehyne, E. A. (2021). Mathematical analysis of COVID-19 by using SIR model with convex incidence rate. Results in Physics, 23, 103970. [Google Scholar]
10. Shahrear, P., Rahman, S. S., Nahid, M. M. H. (2021). Prediction and mathematical analysis of the outbreak of coronavirus (COVID-19) in Bangladesh. Results in Applied Mathematics, 10, 100145. [Google Scholar]
11. Bai, C. Z., Fang, J. X. (2004). The existence of a positive solution for a singular coupled system of nonlinear fractional differential equations. Applied Mathematics and Computation, 150(3), 611–621. [Google Scholar]
12. Goodrich, C. S. (2011). Existence and uniqueness of solutions to a fractional difference equation with nonlocal conditions. Computers & Mathematics with Applications, 61(2), 191–202. [Google Scholar]
13. Side, S., Utami, A. M., Pratama, M. I. (2018). Numerical solution of SIR model for transmission of tuberculosis by runge-kutta method. Journal of Physics: Conference Series, 1040, 012021. [Google Scholar]
14. Kanwal, S., Siddiqui, M. K., Bonyah, E., Sarwar, K., Shaikh, T. S. et al. (2022). Analysis of the epidemic biological model of tuberculosis (TB) via numerical schemes. Complexity, 2022, 5147951. [Google Scholar]
15. Ibrahim, M. O., Egbetade, S. A. (2013). On the homotopy analysis method for an seir tuberculosis model. American Journal of Applied Mathematics and Statistics, 1(4), 71–75. [Google Scholar]
16. Das, K., Murthy, B. S. N., Samad, S. A., Biswas, M. H. A. (2021). Mathematical transmission analysis of SEIR tuberculosis disease model. Sensors International, 2, 100120. [Google Scholar]
17. Ningsi, G., Mungkasi, S. (2020). Variational iteration method used to solve a SIR epidemic model of tuberculosis. Proceedings of the 2nd International Conference of Science and Technology for the Internet of Things, Yogyakarta, Indonesia. [Google Scholar]
18. Veeresha, P., Malagi, N. S., Prakasha, D. G., Baskonus, H. M. (2022). An efficient technique to analyze the fractional model of vector-borne diseases. Physica Scripta, 97(5), 054004. [Google Scholar]
19. Veeresha, P., Ilhan, E., Prakasha, D. G., Baskonus, H. M., Gao, W. (2022). A new numerical investigation of fractional order susceptible-infected-recovered epidemic model of childhood disease. Alexandria Engineering Journal, 61(2), 1747–1756. [Google Scholar]
20. Veeresha, P., Ilhan, E., Prakasha, D. G., Baskonus, H. M., Gao, W. (2021). Regarding on the fractional mathematical model of tumour invasion and metastasis. Computer Modeling in Engineering & Sciences, 127(3), 1013–1036. https://doi.org/10.32604/cmes.2021.014988 [Google Scholar] [CrossRef]
21. Wang, Y., Veeresha, P., Prakasha, D. G., Baskonus, H. M., Gao, W. (2022). Regarding deeper properties of the fractional order kundu-eckhaus equation and massive thirring model. Computer Modeling in Engineering & Sciences, 133(3), 697–717. https://doi.org/10.32604/cmes.2022.021865 [Google Scholar] [CrossRef]
22. Yan, L. (2023). Design of a computational heuristic to solve the non-linear liénard differential model non-linear liénard differential model. Computer Modeling in Engineering & Sciences, 136(1), 201–221. https://doi.org/10.32604/cmes.2023.025094 [Google Scholar] [CrossRef]
23. Shiralashetti, S. C., Kumbinarasaiah, S. (2019). Laguerre wavelets collocation method for the numerical solution of the Benjamina–Bona–Mohany equations. Journal of Taibah University for Science, 13(1), 9–15. [Google Scholar]
24. Shiralashetti, S. C., Kumbinarasaiah, S. (2020). Laguerre wavelets exact parseval frame-based numerical method for the solution of system of differential equations. International Journal of Applied and Computational Mathematics, 6(4), 1–16. [Google Scholar]
25. Shiralashetti, S. C., Hoogar, B. S., Kumbinarasaiah, S. (2019). Laguerre wavelet based numerical method for the solution of third order non-linear delay differential equations with damping. International Journal of Management, Technology and Engineering, 9(1), 3640–3647. [Google Scholar]
26. Shiralashetti, S. C., Kumbinarasaiah, S. (2017). Theoretical study on continuous polynomial wavelet bases through wavelet series collocation method for nonlinear Lane–Emden type equations. Applied Mathematics and Computation, 315, 591–602. [Google Scholar]
27. Kumbinarasaiah, S., Adel, W. (2021). Hermite wavelet method for solving nonlinear Rosenau–Hyman equation. Partial Differential Equations in Applied Mathematics, 4, 100062. [Google Scholar]
28. Saeed, U., ur Rehman, M. (2014). Hermite wavelet method for fractional delay differential equations. Journal of Difference Equations, 2014, 1–8. [Google Scholar]
29. Mundewadi, R. A., Kumbinarasaiah, S. (2019). Numerical solution of Abel’s integral equations using Hermite Wavelet. Applied Mathematics and Nonlinear Sciences, 4(1), 169–180. [Google Scholar]
30. ur Rehman, M., Khan, R. A. (2011). The legendre wavelet method for solving fractional differential equations. Communications in Nonlinear Science and Numerical Simulation, 16(11), 4163–4173. [Google Scholar]
31. Yuttanan, B., Razzaghi, M. (2019). Legendre wavelets approach for numerical solutions of distributed order fractional differential equations. Applied Mathematical Modelling, 70, 350–364. [Google Scholar]
32. Rahimkhani, P., Ordokhani, Y., Babolian, E. (2017). A new operational matrix based on Bernoulli wavelets for solving fractional delay differential equations. Numerical Algorithms, 74(1), 223–245. [Google Scholar]
33. Rahimkhani, P., Ordokhani, Y., Babolian, E. (2017). Numerical solution of fractional pantograph differential equations by using generalized fractional-order Bernoulli wavelet. Journal of Computational and Applied Mathematics, 309, 493–510. [Google Scholar]
34. Adel, W., Sabir, Z. (2020). Solving a new design of nonlinear second-order Lane–Emden pantograph delay differential model via Bernoulli collocation method. The European Physical Journal Plus, 135(5), 1–12. [Google Scholar]
35. Ordokhani, Y., Rahimkhani, P., Babolian, E. (2017). Application of fractional-order Bernoulli functions for solving fractional Riccati differential equation. International Journal of Nonlinear Analysis and Applications, 8(2), 277–292. [Google Scholar]
36. Hedayati, M., Ezzati, R., Noeiaghdam, S. (2021). New procedures of a fractional order model of novel coronavirus (COVID-19) outbreak via wavelets method. Axioms, 10(2), 122. [Google Scholar]
37. Balaji, S. (2016). A new Bernoulli wavelet operational matrix of derivative method for the solution of nonlinear singular Lane–Emden type equations arising in astrophysics. Journal of Computational and Nonlinear Dynamics, 11(5). [Google Scholar]
38. Sabir, Z., Umar, M., Raja, M. A. Z., Baskonus, H. M., Gao, W. (2022). Designing of morlet wavelet as a neural network for a novel prevention category in the HIV system. International Journal of Biomathematics, 15(4), 2250012. [Google Scholar]
39. Kumar, S., Ahmadian, A., Kumar, R., Kumar, D., Singh, J. et al. (2020). An efficient numerical method for fractional SIR epidemic model of infectious disease by using bernstein wavelets. Mathematics, 8(4), 1–22. [Google Scholar]
40. Kumar, S., Kumar, R., Cattani, C., Samet, B. (2020). Chaotic behaviour of fractional predator-prey dynamical system. Chaos, Solitons & Fractals, 135, 109811. [Google Scholar]
41. Kumbinarasaiah, S., Manohara, G. (2022). Modified Bernoulli wavelets functional matrix approach for the HIV infection of CD4+ T cells model. Results in Control and Optimization, 100197. [Google Scholar]
42. Kumbinarasaiah, S., Preetham, S. (2022). Applications of the Bernoulli wavelet collocation method in the analysis of MHD boundary layer flow of a viscous fluid. Journal of Umm Al-Qura University for Applied Sciences,9(1), 1–14. [Google Scholar]
43. Kumbinarasaiah, S., Raghunatha, K. R., Preetham, M. P. (2022). Applications of Bernoulli wavelet collocation method in the analysis of Jeffery–Hamel flow and heat transfer in Eyring–Powell fluid. Journal of Thermal Analysis and Calorimetry,148(3), 1–17. [Google Scholar]
44. Soltanpour Moghadam, A., Arabameri, M., Baleanu, D., Barfeie, M. (2020). Numerical solution of variable fractional order advection-dispersion equation using Bernoulli wavelet method and new operational matrix of fractional order derivative. Mathematical Methods in the Applied Sciences, 43(7), 3936–3953. [Google Scholar]
45. Agrawal, K., Kumar, R., Kumar, S., Hadid, S., Momani, S. (2022). Bernoulli wavelet method for non-linear fractional glucose–Insulin regulatory dynamical system. Chaos, Solitons & Fractals, 164, 112632. [Google Scholar]
46. Kumbinarasaiah, S., Mulimani, M. (2022). A novel scheme for the hyperbolic partial differential equation through fibonacci wavelets. Journal of Taibah University for Science, 16(1), 1112–1132. [Google Scholar]
47. Kumbinarasaiah, S., Manohara, G. (2023). Modified Bernoulli wavelets functional matrix approach for the HIV infection of CD4+ T cells model. Results in Control and Optimization, 10, 100197. [Google Scholar]
48. Kumbinarasaiah, S., Manohara, G., Hariharan, G. (2023). Bernoulli wavelets functional matrix technique for a system of non-linear singular lane emden equations. Mathematics and Computers in Simulation, 204, 133–165. [Google Scholar]
49. Srinivasa, K., Baskonus, H. M., Guerrero Sánchez, Y. (2021). Numerical solutions of the mathematical models on the digestive system and COVID-19 pandemic by hermite wavelet technique. Symmetry, 13(12), 2428. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools