 Open Access
Open Access
ARTICLE
TC-Fuse: A Transformers Fusing CNNs Network for Medical Image Segmentation
1
School of Information Sciences and Technology, Shijiazhuang Tiedao University, Shijiazhuang, 050043, China
2
College of Resources and Environment, Xingtai University, Xingtai, 054001, China
3
School of Electrical and Electronic Engineering, Shijiazhuang Tiedao University, Shijiazhuang, 050043, China
* Corresponding Author: Ying Zhang. Email:
(This article belongs to the Special Issue: Computer Modeling of Artificial Intelligence and Medical Imaging)
Computer Modeling in Engineering & Sciences 2023, 137(2), 2001-2023. https://doi.org/10.32604/cmes.2023.027127
Received 17 October 2022; Accepted 02 February 2023; Issue published 26 June 2023
Abstract
In medical image segmentation task, convolutional neural networks (CNNs) are difficult to capture long-range dependencies, but transformers can model the long-range dependencies effectively. However, transformers have a flexible structure and seldom assume the structural bias of input data, so it is difficult for transformers to learn positional encoding of the medical images when using fewer images for training. To solve these problems, a dual branch structure is proposed. In one branch, Mix-Feed-Forward Network (Mix-FFN) and axial attention are adopted to capture long-range dependencies and keep the translation invariance of the model. Mix-FFN whose depth-wise convolutions can provide position information is better than ordinary positional encoding. In the other branch, traditional convolutional neural networks (CNNs) are used to extract different features of fewer medical images. In addition, the attention fusion module BiFusion is used to effectively integrate the information from the CNN branch and Transformer branch, and the fused features can effectively capture the global and local context of the current spatial resolution. On the public standard datasets Gland Segmentation (GlaS), Colorectal adenocarcinoma gland (CRAG) and COVID-19 CT Images Segmentation, the F1-score, Intersection over Union (IoU) and parameters of the proposed TC-Fuse are superior to those by Axial Attention U-Net, U-Net, Medical Transformer and other methods. And F1-score increased respectively by 2.99%, 3.42% and 3.95% compared with Medical Transformer.Keywords
Medical image segmentation is an important field of medical image analysis, and it is also a very important part of computer-aided diagnosis, monitoring, intervention and treatment. The key to medical image segmentation is to segment the objects of interest (such as organs or lesions) in medical images. The analysis and measurement methods based on image segmentation can meet various medical needs and help doctors make a more accurate diagnosis [1,2]. Nowadays, medical image segmentation methods have been widely used in the fields of heart segmentation, gland segmentation, brain tumor detection, and so on. With the researchers’ exploration of deep learning, convolutional neural networks (CNNs) have achieved remarkable performance in many medical image segmentation tasks. CNNs are also used in most of the latest medical image segmentation models. However, CNNs also have obvious disadvantages. For example, convolutional layers used in CNNs are difficult to capture long-range dependencies because they aggregate local information in the filter region across the current layer to the next layer. To capture the long-range dependencies, a deeper network or a very large filter is used to make the parameters of the model increase sharply and make the training more difficult. Moreover, the increase in the depth of the CNN model may lead to the disappearance of the gradient of the low-level network, and make the convergence speed of the deep neural network become slower and slower [3]. Some works, such as atrous convolutions [4], image pyramids [5], and attention mechanisms [6], have been proposed to capture the long-range dependencies of convolutional networks. However, the atrous convolutions could cause a gridding effect, which will weaken the segmentation performance of the model. And the image pyramids increase the number of parameters of the model. In addition, the global receptive field obtained by the model that uses an attention mechanism is generally through a global pooling operation, which is difficult to provide pixel-level attention. So there is still room for improvement in the aspect of capturing long-range dependencies.
When the background of the image is scattered and accounts for a large proportion of the image, if the network is not strong enough to capture the long-range dependencies, it is easy to mistakenly classify the pixels in the background as masks [7]. Learning the long-range dependencies between the pixels corresponding to the background can effectively reduce false positives. Similarly, when the segmentation mask is large, learning the long-range dependencies between the pixels corresponding to the mask is also helpful in making prediction more effective. Transformers [8] in the applications of natural language processing (NLP) can find the dependencies between the given sequence inputs so as to effectively model the long-range dependencies. It is a pioneering self-attention deep learning technology that enables self-attention mechanisms to be realized on the global scale. However, the transformer has a flexible structure and seldom assumes the structural bias of input data. So it is difficult to learn image position encoding by using few images for training. It is difficult to train on small-scale data [9]. The number of images that can be used for training and the corresponding labels in the medical datasets is relatively few. Moreover, labeling mask areas in medical datasets needs professional medical knowledge. Medical datasets are very difficult to be expanded to large-scale data. Therefore, pre-training technology is often used when transformers are used in medical image segmentation tasks. However, pre-training of these transformers has high demands for computer hardware.
In order to solve the existing problems in the current medical image segmentation tasks, inspired by Medical Transformer [7], a medical image segmentation model TC-Fuse is proposed. Different from Medical Transformer [7] with two transformer branches, TC-Fuse is a dual branch structure composed of one transformer branch and one CNN branch. TC-Fuse not only has the ability of excellent long-range dependencies learning like a transformer, but also can improve the generalization ability of the model, so that it can achieve excellent performances on small-scale datasets such as medical image datasets, and accurately segment the target objects. Moreover, TC-Fuse solves the problems faced by the convolution neural network in modeling long-range dependencies to a certain extent. On the other hand, axial attention used by TC-Fuse provides pixel-level attention. The main innovations of our model are:
(1) A network structure composed of paralleled transformer branch and CNN branch is proposed to capture high-level semantic context and low-level spatial features, respectively. The attention fusion module is used to fuse the final output features of these two branches. It can give full play to the advantages of transformers and CNNs. And there is no need to build a very deep network, which avoids the overstaffing of the model and alleviates the problems of gradient disappearance and diminishing feature reuse.
(2) A transformer branch composed of Mix-FFN and axial attention is proposed to provide location information and keep the translation invariance of the model. Compared with the original transformer, the proposed transformer has a stronger ability of perceiving location information.
(3) On the public standard datasets GlaS, CRAG, and COVID-19 CT Segmentation datasets, the proposed model achieves good performance for medical image segmentation, which proves the effectiveness of the proposed method. The F1-score (F1) in GlaS, CRAG, and COVID-19 CT Segmentation datasets are 84.01%, 83.13%, and 72.10%, respectively. And the IoU values in GlaS, CRAG, and COVID-19 CT Segmentation datasets are 73.80%, 71.13%, and 56.37%, respectively.
2.1 Medical Image Segmentation Based on CNNs
In recent years, medical image segmentation methods based on CNNs have made some progress, and excellent image segmentation models such as FCN [10], U-Net [11], and DeepLabV3+ [12] have emerged. Because the encoder-decoder architecture proposed in U-Net is popular due to its excellent performance, many improvements and extensions of U-Net have been proposed. For example, some models replace the vanilla convolutional layer of U-Net with other backbone networks such as Residual U-Net [13] with ResNet [14] as the backbone network and Dense U-Net [15] with DenseNet [16] as the backbone network. Some models adopted more skip connections between the encoder and decoder of U-Net to construct U-Net++ [17] and U-Net 3+ [18]. These CNN-based medical image segmentation models have stronger generalization ability, higher segmentation accuracy and efficiency for medical images segmentation task. UNeXt [19] replaced the deepest two-layer convolution blocks of U-Net with a multi-layer perceptron (MLP) block, which improved the performance of medical image segmentation, and reduced the number of parameters and computational complexity. Xie et al. [20] proposed a semi-supervised model based on pairwise relation for gland segmentation and used unlabeled data for training to alleviate the lack of gland datasets. Graham et al. [21] used convolutional neural network and spatial pyramid pooling to segment the gland images and achieved state-of-the-art performance. Yu et al. [22] redesigned the skip connection of U-Net and the internal connection between decoder sub-networks to enhance the extraction ability of semantic features at different levels and the fusion of multi-scale features in U-Net. Combining the advantages of DenseNet and ResNet, Tie et al. [23] proposed an improved 3D U-Net, which used dense blocks in the encoder part and residual blocks in the decoder part.
Inspired by the strong encoding ability of the transformer [8] for long-range dependencies in the applications of natural language processing, the transformer has also been widely used in computer vision tasks recently. ViT [24] proved for the first time that the pure transformer can have the most advanced image classification performance when there are enough training data. In order to achieve better performance in medical image segmentation tasks, Valanarasu et al. [7] proposed gated axial attention on the basis of Axial Deeplab [25]. SETR [26] introduced a transformer into the encoder part of the network, which achieved a better segmentation effect. Swin Transformer [27] proposed the shifted window based on self-attention with linear computational complexity, which not only reduced the computational overhead but also had the flexibility as a general backbone network. Swin Transformer achieved SOTA performance in image classification, object detection and semantic segmentation tasks. SegFormer [28] inserted the depth-wise convolution between the fully-connected layers of the feed-forward network in the transformer block to replace the absolute position encoding so as to resist the damage to the translation invariance of the model due to the absolute position encoding. Mahajan et al. [29] proposed a new hybrid method using Aquila optimizer (AO) and arithmetic optimization algorithm (AOA), and this method could be applied in vision transformer to make the network converge faster and achieve high-quality results.
2.3 Medical Image Segmentation Based on Transformers and CNNs
In order to give full play to the advantages of transformers and CNNs at the same time, some researchers have proposed some hybrid models with transformers and CNNs. TransUnet [30] has the advantages of transformer and U-Net. First, it used CNNs to extract low-level features, then used transformer blocks to extract the global context information, and finally used skip connections and decoder to enhance the detail information. TransUnet achieved excellent performance in multi-organ segmentation and heart segmentation. TransClaw U-Net [31] upsampled the bottom of TransUnet, and combined the encoding part, upsampling part and decoding part of the corresponding layers to achieve more accurate organ segmentation. TransFuse [32] used transformers and CNNs in parallel to obtain multi-level feature representation, and then fused them to improve the efficiency of global context modeling. TransFuse achieved SOTA in the polyp segmentation task. In FAT-Net [33] as a classic encoder-decoder architecture, and a transformer branch was added to its encoder in parallel to capture long-range dependencies and global context information. In addition, a memory-efficient decoder and a adaptive feature module were used In FAT-Net to enhance the ability of feature fusion. Nevertheless, all of these methods need pre-trained the model to achieve better effect in medical image segmentation.
As shown in Fig. 1, the proposed TC-Fuse model consists of one transformer branch and one CNN branch. The transformer branch is used to extract global features and give full play to the advantages of the transformer in learning long-range dependencies. The CNN branch does not conduct down-sampled operations in order to better preserve the details, so as to achieve better performance on small-scale datasets. Finally, the features extracted from the transformer branch and CNN branch are fused through the BiFusion module, and then
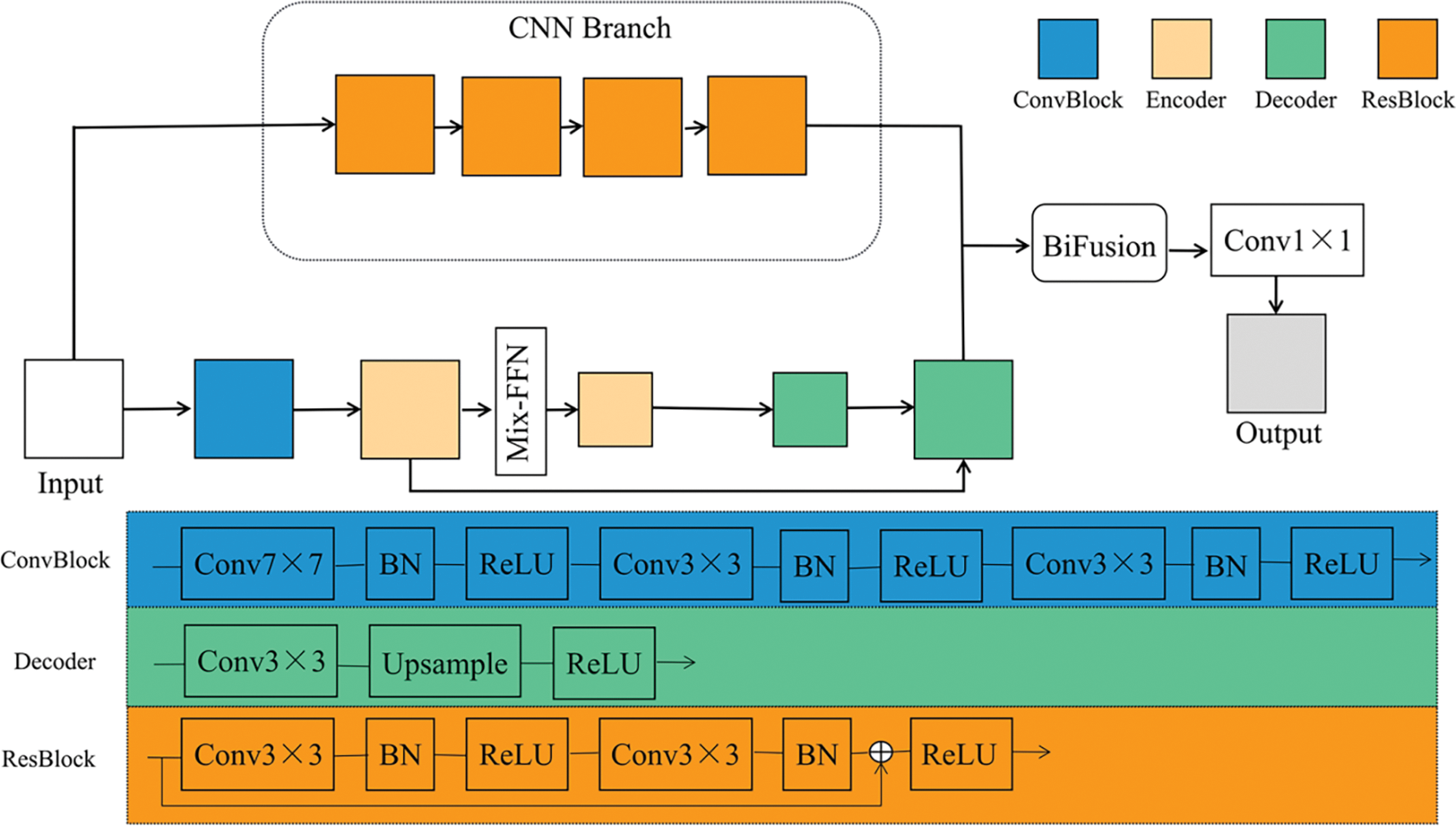
Figure 1: Architecture of TC-Fuse
As shown in Fig. 1, the transformer branch consists of convolutional block, encoders, Mix-FFN, and decoders. The convolution block contains three convolutional layers. After every convolutional layer, there are batch normalization and ReLU activation function. The encoder is an axial transformer layer, as shown in Fig. 2. So as to overcome the complexity of the original self-attention, the axial attention used in Medical Transformer [7] is adopted to decompose the self-attention into two self-attention modules [25]. And Mix-FFN is used to replace the relative position encoding of axial attention. The decoder block consists of a convolutional layer, followed by an upsampling layer and a ReLU activation function.

Figure 2: The architecture of the encoder
The self-attention mechanism can effectively use all the feature information of the images, learn the input context information and capture their relationship, so as to deal with the long-range dependencies in the images and better generate the details of each position in the images. Self-attention mechanism can be expressed as:
where
However, the original self-attention needs to calculate the relationship between each token with all other tokens, which makes the cost of calculation very high. In order to reduce the complexity of the original self-attention, the self-attention is decomposed into axial attention that performs self-attention along the height axis and width axis of the feature maps. There are two self-attention modules in the encoder. The first module performs self-attention on the height axis of the feature maps, which is represented by Eq. (2). The second module operates on the width axis and is represented by Eq. (3).
where i and j are the pixel position along the width and height axes. The complexity is reduced from
Translation invariance is very important for the semantic segmentation tasks because the pixels in the original images should correspond with the pixels in the labeled images in the semantic segmentation tasks. Nevertheless, absolute position encoding may destroy the translation invariance of the model. Although the relative position encoding has the advantage of translation invariance, the relative position encoding carries out additional calculation, and the standard transformer implementation needs to be modified [35]. Inspired by Xie et al. [28], Mix-FFN which transmits position information through
where

Figure 3: The structure of Mix-FFN
Usually, deep CNNs use hundreds of convolutional layers to extract information, which may lead to the overstaffing of the model, gradient disappearance and feature reuse. To address these problems, only four ResBlocks [14] were introduced into the CNN branch instead of the usual five ResBlocks found in the ResNet-based model. This modification aims to reduce computation and memory consumption. There are two convolution operations in each ResBlock. After each convolutional layer, there are batch normalization and ReLU activation function, and there are skip connections in each ResBlock to solve the problem of the degradation of network. In addition, the CNN branch is not down-sampled to better extract the details. The purpose of this design is to maintain the balance between the consumption of computation and memory with the segmentation performance of the network. In addition, ResBlock can be replaced by other blocks such as the vanilla convolutional layer [11] and DenseBlock [16]. Hence, CNN branch is flexible and convenient for ablation experiments to verify the performance of ResBlock.
Although it is simple to directly add the outputs of CNN branch with the outputs of the transformer branch, directly adding the pixels value representing the masks with the pixels value representing the background may cause mis-segmentation. Hence, the output features of the two branches cannot be fused effectively. To solve the problem, inspired by Zhang et al. [32], the BiFusion fusion module is introduced to fuse the output features of the two branches. The BiFusion fusion module combines complementary information, and improves data quality [36], and can effectively fuse the encoding features of CNN and transformer. The detailed configurations of BiFusion are shown in Table 1. Its structure is shown in Fig. 4 and can be expressed as follows:
where
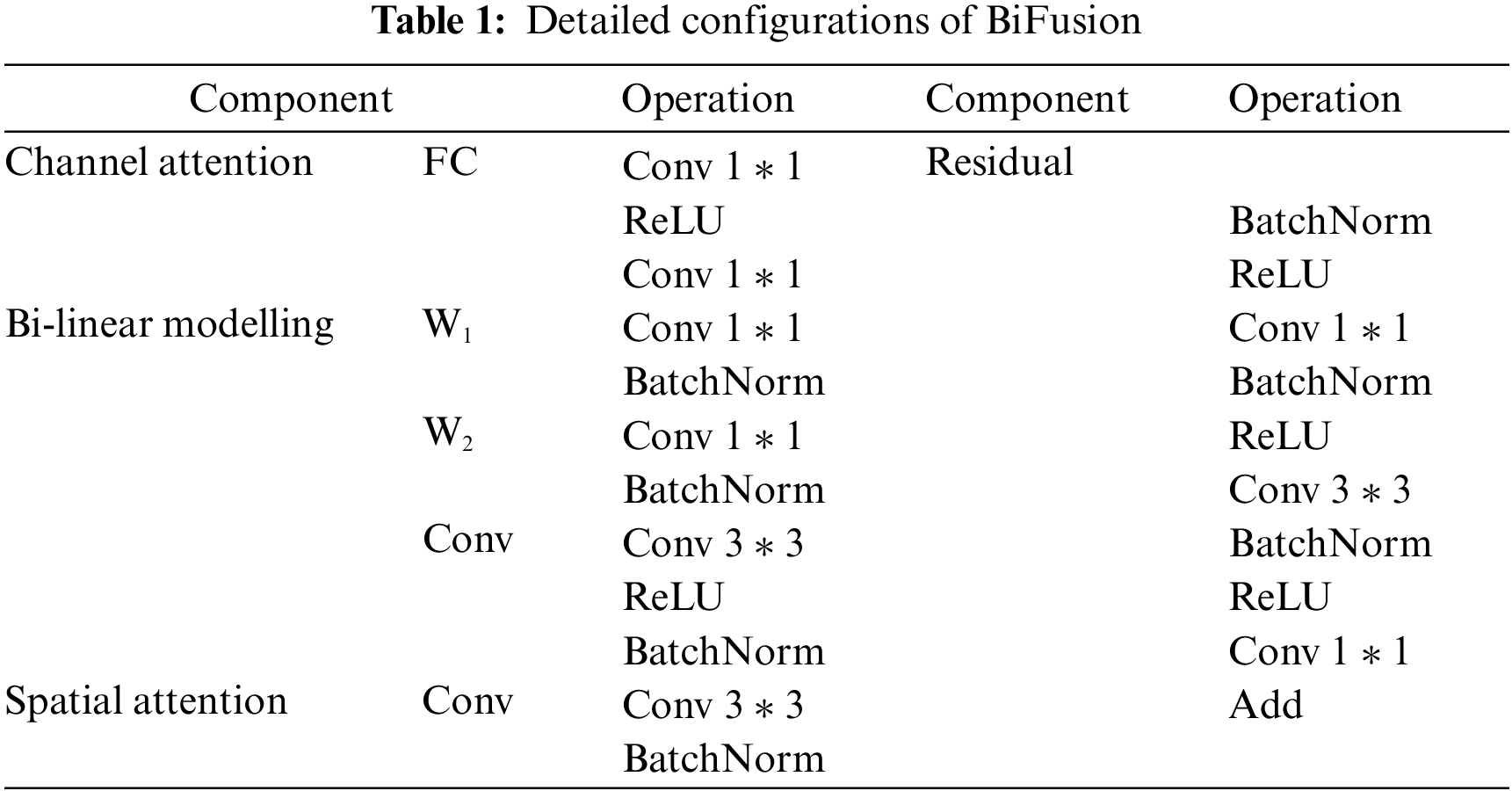

Figure 4: BiFusion module
So as to evaluate the performance of TC-Fuse, this study adopts three datasets: Gland Segmentation (GlaS) dataset [39], Colorectal adenocarcinoma gland (CRAG) dataset [21] and COVID-19 CT Images Segmentation dataset [40]. The Gland Segmentation (GlaS) dataset consists of 165 H&E stained histopathological images, 65 images of which are used for training, 20 images for validation and 80 images for testing. The resolution of the images is adjusted to 128

The proposed TC-Fuse model is implemented based on the deep learning framework PyTorch, and trained and tested on NVIDIA GeForce RTX 3080Ti GPU and Intel Xeon E5-2686 v4 CPUs. Adam optimizer with a learning rate of 0.0001 is adopted in training. The batchsize is set to 4 and the epoch is set to 400. The data augment method of random horizontal flipping is applied to the training stage. In addition, the whole network is trained end-to-end by the binary cross entropy loss function, which can be written as:
where w and h are the width and height of the images,
To further illustrate the performance of the proposed TC-Fuse, F1-score (F1), Intersection over Union (IoU), Hausdorff distance 95% (HD) and Pixel Accuracy (PA) are used as the metrics in the comparison with different methods. In addition to the above metrics, Sensitivity is introduced into ablation experiments. F1, IoU and Sensitivity are expressed by Eqs. (7)–(9), respectively:
where TP, FP and FN devote true-positive, false-positive, and false-negative, respectively.
The calculation equation of PA is:
where
The calculation equation of HD is:
where H is Hausdorff distance 95%, P is prediction map,
4.4.1 Ablation Experiments on the Structure of TC-Fuse
In order to verify the effects of transformer branch, CNN branch and BiFusion module in TC-Fuse on improving the performance of gland segmentation, ablation experiments are conducted on GlaS and CRAG datasets, and the results are shown in Tables 3 and 4. At first, Mix-FFN in the transformer branch is removed. The experimental results show that Mix-FFN greatly improves the performance of the model with 6.78%, 7.38%, 15.17% and 2.62% increasement in F1, IoU, Sensitivity and PA on GlaS dataset, respectively and 0.92%, 1.34% and 4.95% increasement in F1, IoU and Sensitivity on CRAG dataset, respectively, because Mix-FFN can effectively improve the transformer’s perception of location information and make up for the lack of location information in the self-attention mechanism. Then, without Mix-FFN, we replace the encoder with Gated Axial-Attention [7] to verify the effectiveness of the traditional relative position encoding. The results show that relative position encoding causes 1.34%, 0.20% and 3.16% decrease in F1, IoU and Sensitivity on GlaS dataset, and increases 0.87%, 1.26% and 0.25% in F1, IoU and Sensitivity on CRAG dataset, but not as good as TC-Fuse. This indicates that Mix-FFN is more effective than relative position encoding in gland segmentation tasks. Secondly, the effectiveness of the transformer branch and CNN branch are respectively verified. F1, IoU, Sensitivity and PA by the proposed model without CNN branch on GlaS dataset is 3.27%, 7.08%, 3.26% and 4.66% lower than those without the transformer branch. The F1, IoU and PA scores of the one without CNN branch on CRAG dataset are 10.44%, 13.50% and 15.49% lower than those of without the transformer branch, while the Sensitivity scores improve just 1.51%. This indicates that the transformer may perform worse than CNN on small-scale datasets, which highlights the importance of combining transformer with CNN. Finally, BiFusion module is removed. This comparison demonstrates that the BiFusion module improves 2.00%, 2.41%, 2.19% and 2.20% in F1, IoU, Sensitivity and PA on GlaS dataset and 3.24%, 4.62%, 8.08% and 0.55% in F1, IoU, Sensitivity and PA on CRAG dataset. It shows that BiFusion module can effectively combine the encoding features of CNN and transformer.
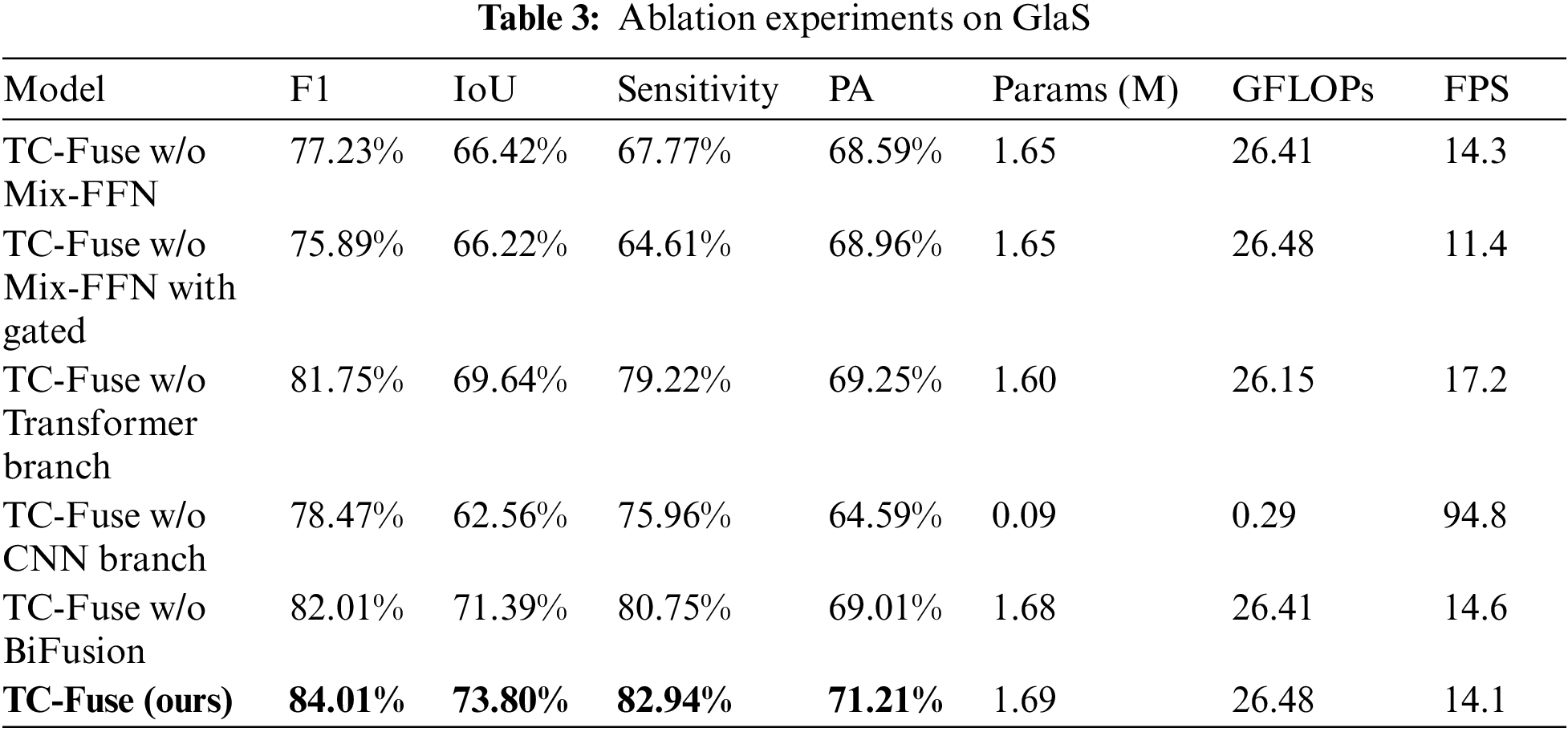

From Table 3, it can be seen that the number of parameters of TC-Fuse is 0.04 M slightly higher than that of TC-Fuse without Mix-FFN. The GFLOPs and FPS of them are very close. These indicate that the structure of Mix-FFN is simple but the performance of the model can be well improved by it. The FPS of TC-Fuse w/o Mix-FFN with Gated is 2.9 lower than 14.3 of TC-Fuse w/o Mix-FFN, which indicates that the computational complexity of relative position encoding is higher than that of Mix-FFN. Although GFLOPs and FPS of TC-Fuse w/o Transformer Branch have increased by 0.33 and decreased by 3.1, respectively after adding transformer branch, the segmentation effect has been improved significantly. This shows that the long-range dependencies extracted by transformer branch can significantly improve the segmentation performance, and there is little impact on the computational complexity. After adding BiFusion to the TC-Fuse w/o BiFusion, the Params, GFLOPs have increased by 0.01 and 0.07, respectively, and FPS decreased by 0.5. But the segmentation performance improved greatly by BiFusion indicating the effectiveness of this fusion module.
4.4.2 Ablation Experiments on the Location of Mix-FFN
In order to verify the influence of Mix-FFN at different locations of transformer branch, the following ablation experiments are conducted on GlaS and CRAG datasets. The experiments results are shown in Tables 5 and 6, respectively. The first encoder is called as TransBlock1 and the second encoder is called as TransBlock2 in the transformer branch of TC-Fuse. The Mix-FFN is inserted after TransBlock1, after TransBlock2, after TransBlock1 and after TransBlock2, respectively. The results on GlaS and CRAG datasets show that the best results are obtained by Mix-FFN after TransBlock1.On GlaS dataset, F1, IoU, Sensitivity and PA respectively increased by 2.85%, 3.90%, 7.14% and 1.58% compared with placing Mix-FFN after TransBlock2, as well as 2.55%, 5.01%, 1.33% and 3.86% compared with placing Mix-FFN after TransBlock1 and after TransBlock2. On CRAG dataset, F1, IoU and Sensitivity respectively increased by 4.54%, 6.40% and 13.24% compared with placing Mix-FFN after TransBlock1 and after TransBlock2. Hence, the proposed TC-Fuse is placed in this way. The reason is that if Mix-FFN is placed after TransBlock2, the resolution of the input is not large enough to provide sufficient position information. If Mix-FFN is placed after TransBlock1 and TransBlock2, the model has not better performance due to the location information provided by twice Mix-FFN. The location information provided by Mix-FFN placed after TransBlock1 is just enough.


4.4.3 Ablation Experiments on the Type of CNN Backbones
CNN can help the network to have better performance on small-scale datasets. Therefore, CNN branch is introduced into the network to extract features together with the transformer branch. So as to verify the impact of different kinds of CNN backbones, three different CNN backbones are verified on GlaS and CRAG datasets. The results of the ablation experiments are shown in Tables 7 and 8. The structures of the residual convolution block [14], vanilla convolution block [11] and dense convolution block [16] are shown in Fig. 5. Vanilla convolution block is simple in structure and powerful in performance. However, as its depth increases, it will lead to the degradation of network. Residual convolution block by adding a skip connection on the basis of vanilla convolution block can solve the degradation of network while retaining the advantages of vanilla convolution block [14]. Dense convolution block uses a large number of skip connections, but the number of its parameter is less than that of residual convolution block, and its generalization performance is stronger. The results in Tables 7 and 8 show that the effect of residual convolution block is better than those of vanilla convolution block and dense connection block. On the GlaS dataset, F1, IoU, Sensitivity and PA are 2.42%, 4.90%, 6.05% and 1.54% higher than those using dense connection block, and are 5.20%, 8.78%, 13.22% and 1.05% higher than those using vanilla convolution block. On CRAG dataset, F1, IoU and Sensitivity respectively increased by 4.11%, 5.81% and 12.69% compared with the method using dense connection block, and increased by 5.17%, 7.25% and 9.27% respectively compared with the method using vanilla convolution block. The reason of dense convolution block superior to vanilla convolution block in TC-Fuse is that skip connection suppresses the degradation of network and improves performance. TC-Fuse using residual convolution block is better than TC-Fuse using dense convolution block indicating that a large number of skip connections may not improve segmentation performance.



Figure 5: The type of CNN backbones: (A) residual convolution block. (B) vanilla convolution block. (C) dense convolution block
4.5 Comparison with Other Methods
In order to verify the effectiveness and progressiveness of our model, the proposed TC-Fuse model is compared with several methods including convolution-based segmentation network Segnet [41], U-Net [11], U-Net++ [17], and Attention U-Net [42], MLP-based segmentation network UNeXt [19], and self-attention-based segmentation network Axial Attention U-Net [25] and Medical Transformer [7] on GlaS and CRAG datasets. In addition, the proposed model is also compared with the above methods on COVID-19 CT Images Segmentation Dataset. The experimental results are shown in Tables 9–11.



It can be seen from Table 9 that the proposed TC-Fuse model ranks first on the GlaS dataset with F1 scores of 84.01%, IoU scores of 73.80% and PA scores of 71.21%. F1, IoU and PA by TC-Fuse model is 2.99%, 4.19% and 0.85% higher than those by the Medical Transformer model. The HD by the proposed TC-Fuse on the GlaS dataset is 8.95, ranks second in these several method, and is 0.32 higher than it by the UNeXt method. As shown in Fig. 6, the segmentation effect of our model is the best. In the first, second and fourth rows of Fig. 6, the glands predicted by TC-Fuse are the most intact. And in the third row of Fig. 6, TC-Fuse segments the small glands at the bottom of the image closest to the ground truth.

Figure 6: Segmentation results on sample test images from GlaS
It can be seen from Table 10 that our model ranks first on CRAG dataset with F1 scores of 83.13%, IoU scores of 71.13% and PA scores of 76.10%, and is 2.07%, 2.98% and 1.58% higher than Medical Transformer method which rank second. The HD by the proposed TC-Fuse on the CRAG dataset ranks first and is 2.54 lower than it by Axial Attention U-Net which ranks second. As shown in Fig. 7, the proposed TC-Fuse model has the best segmentation performance. The glands segmented by TC-Fuse in the first and third rows of Fig. 7 are the most intact. In the second row of Fig. 7, the glands segmented by Medical Transformer are fragmentary, and Attention U-Net, UNeXt and U-Net++ are not as effective as TC-Fuse in segmenting the glands at the bottom left part of the image. In the fourth row of Fig. 7, all of Attention U-Net, U-Net++, UNeXt and TC-Fuse predict the wrong segmentation. But the segmented image by TC-Fuse is most similar to the ground truth.

Figure 7: Segmentation results on sample test images from CRAG
U-Net++ adds more skip connections between the encoder and decoder of U-Net. Attention U-Net adds an attention mechanism on the basis of U-Net. UNeXt replaces the deepest two-layer convolution blocks of U-Net with MLP block. According to Tables 9 and 10, Figs. 6 and 7, the performance improvement by these variant structures based on U-Net is limited compared with U-Net. This shows that it is difficult for simple U-Net variants to extract long-range dependencies of images and achieve better performance improvement. For transformer-based baselines, both Axial Attention U-Net and Medical Transformer used Axial Attention, but Medical Transformer has a much better segmentation effect than Axial Attention U-Net, which shows the effectiveness of dual branch structure. The proposed TC-Fuse achieves relatively better performance on GlaS and CRAG datasets. The excellent prediction results can be attributed to modeling capability of long-range dependencies and powerful perception of location information of Mix-FFN and Axial Attention, which are necessary in medical image segmentation, as well as CNNs’ feature extraction capability on small-scale datasets and feature fusion capability of the fusion module.
It can be seen from Table 11 that the proposed TC-Fuse model ranks first on COVID-19 CT Images Segmentation Dataset in terms of F1 scores of 72.10%, IoU scores of 56.37% and PA scores of 93.57%. Compared with Medical Transformer, TC-Fuse increased by 3.95%, 4.68% and 0.41% in terms of F1, IoU and PA, respectively. The HD by the proposed TC-Fuse on the COVID-19 CT Images Segmentation Dataset ranks second and is 0.33 lower than it by Medical Transformer which ranks first.
For Segnet, the segmentation ability of COVID-19 CT images is far inferior to that of the gland. The reason should be that Segnet’s classification pixel by pixel makes it lack of spatial consistency, and it is difficult to extract COVID-19 CT images with high similarity. To sum up, small number of images in COVID-19 CT Images Segmentation Dataset also make the performance of transformer-based network Axial Attention U-Net and Medical Transformer inferior to that of U-Net and U-Net++. The comprehensive performance of the proposed TC-Fuse exceeds that of other models mentioned above, which indicates the proposed fusion model is effective on COVID-19 CT Image Segmentation. In the first row of Fig. 8, both Axial Attention U-Net and Medical Transformer fail to segment all targets on the right, while TC-Fuse segmented them correctly. The second row of Fig. 8 shows Medical Transformer is over-segmented and Axial Attention U-Net is under-segmented. The disease areas by TC-Fuse are correctly segmented and are most similar to the labels. In the third row of Fig. 8, the segmented image by TC-Fuse is more complete than those by Axial Attention U-Net and Medical Transformer.
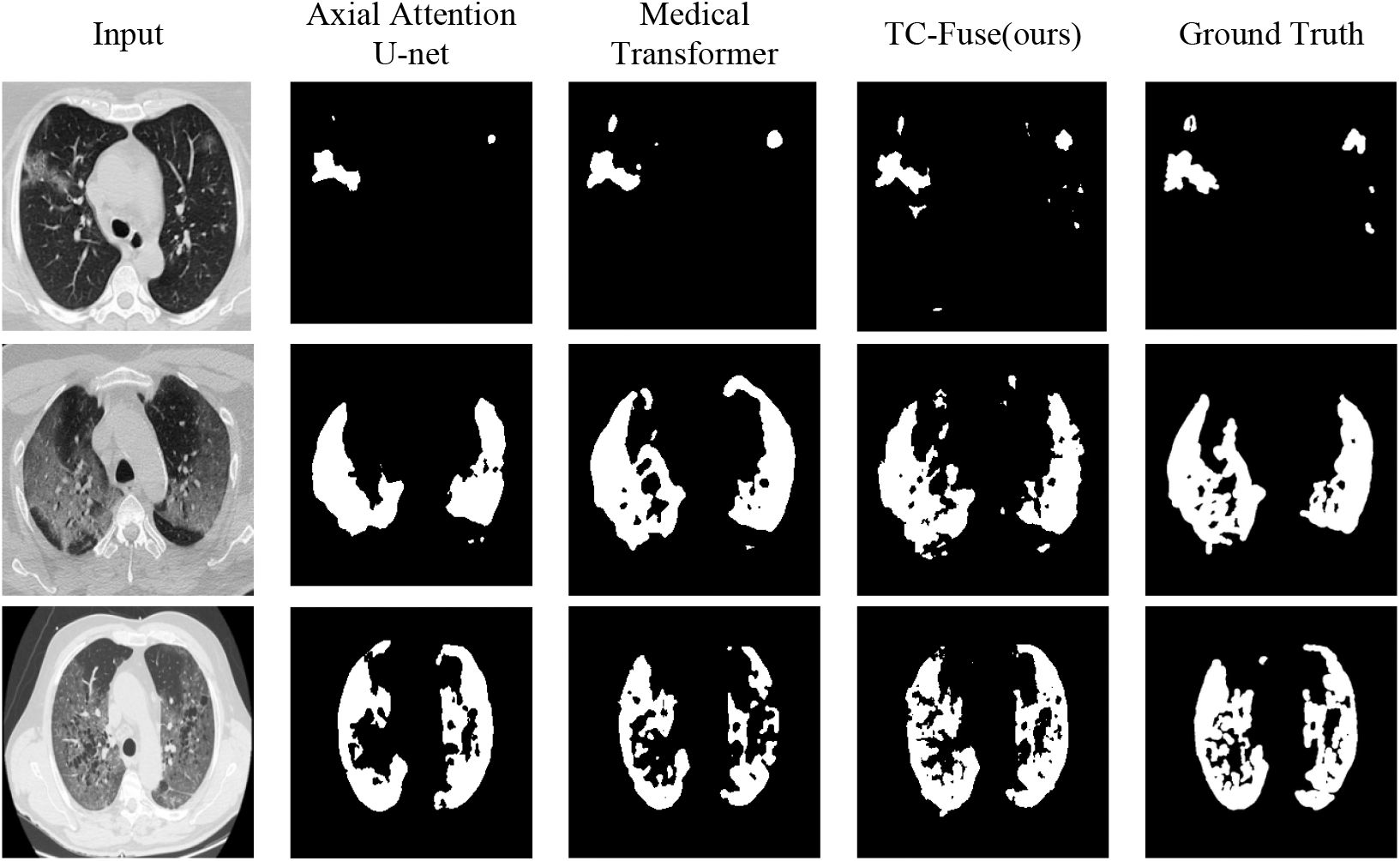
Figure 8: Segmentation results on sample test images from COVID-19 CT Images Segmentation dataset
The Receiver Operating Characteristic (ROC) curves of the different models on different datasets are shown in Fig. 9. It can be found from Fig. 9 that the proposed TC-Fuse has the largest area, which indicates that TC-Fuse can accurately distinguish the foreground from background of medical images. The Precision-Recall (PR) curves of different models on different datasets are shown in Fig. 10. In Figs. 10a and 10b, the areas of PR curves of the TC-Fuse on GlaS and CRAG datasets are the largest, which indicates that the performance of TC-Fuse is better than the other models on GlaS and CRAG datasets. In Fig. 10c, although the area of PR curves of the TC-Fuse is less than that of U-Net++ on COVID-19 CT Images Segmentation dataset, TC-Fuse has the largest area of ROC curves. Both indicators of PR curve focus on positive examples, but both positive and negative examples are needed to be considered in medical image segmentation. And ROC curve gives consideration to both positive and negative examples.

Figure 9: The ROC curves of different models: (a) GlaS dataset. (b) CRAG dataset. (c) COVID-19 CT Images Segmentation dataset

Figure 10: The PR curves of the different models: (a) GlaS dataset. (b) CRAG dataset. (c) COVID-19 CT Images Segmentation dataset
Table 12 shows the numbers of parameters of different model. The number of parameters of TC-Fuse is 1.69 M close to those of UNeXt, Axial Attention U-Net and Medical Transformer, but are significantly less than those of Segnet, U-Net, U-Net++ and Attention U-Net. The reason is that there are only two layers of encoders and decoders in the Transformer branch of TC-Fuse, and that there are only four ResBlocks in its CNN branch. Moreover, the operations in the BiFusion module are also very simple.

In this work, how to fuse transformers and CNNs for medical image segmentation is explored. Specifically, a dual branch structure composed of transformer branch and CNN branch, named as TC-Fuse, is proposed. The output features of the two branches are fused by the BiFusion module. The proposed TC-Fuse does not need to be pre-trained on large-scale datasets like other transformer-based models. Moreover, the proposed transformer branch is composed of axial attention and Mix-FFN, which can capture long-range dependencies without destroying the translation invariance of the model. Furthermore, the BiFusion module effectively captures the long-range dependencies extracted from the transformer branch and the details extracted from the CNN branch. A lot of experiments around TC-Fuse on GlaS and CRAG datasets have been done, and TC-Fuse achieves good performance. However, there are some limitations in TC-Fuse. The dependencies of the axial attention layer in the transformer branch are not enough to capture enough context information. And Mix-FFN may cause gradient exploding or gradient vanishing in deep networks. Besides, a lack of down-sampling operation could make the training speed slow. In the future, more powerful transformer branches can be introduced to replace the transformer branch of TC-Fuse to achieve better performance. And more skip connections and layer norm could be added into Mix-FFN to alleviate gradient exploding or vanishing caused by more powerful transformer branches. Besides, in order to better maintain the detailed information, the atrous convolution could be introduced to expand the receptive field. Due to the difficulty in labeling medical image, the proposed model did not be trained on large-scale datasets. In the future, semi-supervised learning algorithm can be considered to train with labeled and unlabeled images to further improve the performance.
Acknowledgement: The authors are thankful to the previous contributors of deep learning technology that used in this research.
Funding Statement: This work was supported in part by the National Natural Science Foundation of China under Grant 61972267, the National Natural Science Foundation of Hebei Province under Grant F2018210148, the University Science Research Project of Hebei Province under Grant ZD2021334, the Science and Technology Project of Hebei Education Department (ZD2022098).
Availability of Data and Materials: Publicly available datasets were analyzed in this study. The gland segmentation dataset (GlaS) for this study can be found in the Warwick, https://warwick.ac.uk/. The colorectal adenocarcinoma gland dataset (CRAG) for this study can be found in the Warwick, https://warwick.ac.uk/. COVID-19 CT Images Segmentation dataset for this study can be found in the MedSeg, https://medicalsegmentation.com/covid19/.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Wang, S. H., Govindaraj, V. V., Górriz, J. M., Zhang, X., Zhang, Y. D. (2021). COVID-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Information Fusion, 67, 208–229. https://doi.org/10.1016/j.inffus.2020.10.004 [Google Scholar] [PubMed] [CrossRef]
2. Wang, S. H., Nayak, D. R., Guttery, D. S., Zhang, X., Zhang, Y. D. (2021). COVID-19 classification by CCSHNet with deep fusion using transfer learning and discriminant correlation analysis. Information Fusion, 68, 131–148. https://doi.org/10.1016/j.inffus.2020.11.005 [Google Scholar] [PubMed] [CrossRef]
3. Zhu, Z., Wang, S., Zhang, Y. (2022). A survey of convolutional neural network in breast cancer. Computer Modeling in Engineering & Sciences, 136(3), 2127–2172. https://doi.org/10.32604/cmes.2023.025484 [Google Scholar] [PubMed] [CrossRef]
4. Chen, L. C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A. L. (2014). Semantic image segmentation with deep convolutional nets and fully connected CRFs. arXiv preprint arXiv:1412.7062. [Google Scholar]
5. Zhao, H., Shi, J., Qi, X., Wang, X., Jia, J. (2017). Pyramid scene parsing network. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2881–2890. Honolulu, USA. [Google Scholar]
6. Huang, Z., Wang, X., Huang, L., Huang, C., Wei, Y. et al. (2019). CCNet: Criss-cross attention for semantic segmentation. Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 603–612. Seoul, Korea. [Google Scholar]
7. Valanarasu, J. M. J., Oza, P., Hacihaliloglu, I., Patel, V. M. (2021). Medical transformer: Gated axial-attention for medical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 36–46. Strasbourg, France. [Google Scholar]
8. Devlin, J., Chang, M. W., Lee, K., Toutanova, K. (2018). BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805. [Google Scholar]
9. Lin, T., Wang, Y., Liu, X., Qiu, X. (2021). A survey of transformers. arXiv preprint arXiv:2106.04554. [Google Scholar]
10. Long, J., Shelhamer, E., Darrell, T. (2014). Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3431–3440. [Google Scholar]
11. Ronneberger, O., Fischer, P., Brox, T. (2015). U-Net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 234–241. Munich, Germany. [Google Scholar]
12. Chen, L. C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H. (2018). Encoder-decoder with atrous separable convolution for semantic image segmentation. Proceedings of the European Conference on Computer Vision (ECCV), pp. 801–818. Munich, Germany. [Google Scholar]
13. Xiao, X., Lian, S., Luo, Z., Li, S. (2018). Weighted Res-UNet for high-quality retina vessel segmentation. 2018 9th International Conference on Information Technology in Medicine and Education (ITME), pp. 327–331. Hangzhou, China. [Google Scholar]
14. He, K., Zhang, X., Ren, S., Sun, J. (2016). Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778. Las Vegas, USA. [Google Scholar]
15. Li, X., Chen, H., Qi, X., Dou, Q., Fu, C. et al. (2018). H-DenseUNet: Hybrid densely connected unet for liver and tumor segmentation from CT volumes. IEEE Transactions on Medical Imaging, 37(12), 2663–2674. https://doi.org/10.1109/TMI.2018.2845918 [Google Scholar] [PubMed] [CrossRef]
16. Huang, G., Liu, Z., van der Maaten, L., Weinberger, K. Q. (2017). Densely connected convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708. Honolulu, USA. [Google Scholar]
17. Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., Liang, J. (2018). UNet++: A Nested U-Net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, pp. 3–11. Cham: Springer. [Google Scholar]
18. Huang, H., Lin, L., Tong, R., Hu, H., Wu, J. (2020). UNET 3+: A full-scale connected unet for medical image segmentation. ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 1055–1059. Barcelona, Spain. [Google Scholar]
19. Valanarasu, J. M. J., Patel, V. M. (2022). UNeXt: MLP-based rapid medical image segmentation network. arXiv preprint arXiv:2203.04967. [Google Scholar]
20. Xie, Y., Zhang, J., Liao, Z., Shen, C., Verjans, J. et al. (2020). Pairwise relation learning for semi-supervised gland segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 417–427. Lima, Peru. [Google Scholar]
21. Graham, S., Chen, H., Gamper, J., Dou, Q., Heng, P. et al. (2019). MILD-Net: Minimal information loss dilated network for gland instance segmentation in colon histology images. Medical Image Analysis, 52, 199–211. https://doi.org/10.1016/j.media.2018.12.001 [Google Scholar] [PubMed] [CrossRef]
22. Yu, L., Qin, Z., Ding, Y., Qin, Z. (2021). MIA-UNet: Multi-scale iterative aggregation U-network for retinal vessel segmentation. Computer Modeling in Engineering & Sciences, 129(2), 805–828. https://doi.org/10.32604/cmes.2021.017332 [Google Scholar] [CrossRef]
23. Tie, J., Peng, H., Zhou, J. (2021). MRI brain tumor segmentation using 3D U-Net with dense encoder blocks and residual decoder blocks. Computer Modeling in Engineering & Sciences, 128(2), 427–445. https://doi.org/10.32604/cmes.2021.014107 [Google Scholar] [CrossRef]
24. Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Houlsby, N. (2020). An image is worth 16×16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929. [Google Scholar]
25. Wang, H., Zhu, Y., Green, B., Adam, H., Yuille, A. et al. (2020). Axial-deepLab: Stand-alone axial-attention for panoptic segmentation. ECCV 2020, pp. 108–126. Glasgow, UK. [Google Scholar]
26. Zheng, S., Lu, J., Zhao, H., Zhu, X., Zhang, L. (2020). Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. arXiv preprint arXiv:2012.15840. [Google Scholar]
27. Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y. et al. (2021). Swin transformer: Hierarchical vision transformer using shifted windows. Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10012–10022. Montreal, Canada. [Google Scholar]
28. Xie, E., Wang, W., Yu, Z., Anandkumar, A., Alvarez, J. M. et al. (2021). Segformer: Simple and efficient design for semantic segmentation with transformers. Advances in Neural Information Processing Systems, 34, 12077–12090. [Google Scholar]
29. Mahajan, S., Abualigah, L., Pandit, A. K., Altalhi, M. (2022). Hybrid aquila optimizer with arithmetic optimization algorithm for global optimization tasks. Soft Comput, 26, 4863–4881. https://doi.org/10.1007/s00500-022-06873-8 [Google Scholar] [CrossRef]
30. Chen, J., Lu, Y., Yu, Q., Luo, X., Zhou, Y. (2021). TransUNet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306. [Google Scholar]
31. Yao, C., Hu, M., Zhai, G., Zhang, X. P. (2021). TransClaw U-Net: Claw U-Net with transformers for medical image segmentation. arXiv preprint arXiv:2107.05188. [Google Scholar]
32. Zhang, Y., Liu, H., Hu, Q. (2021). TransFuse: Fusing transformers and cnns for medical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 14–24. Strasbourg, France. [Google Scholar]
33. Wu, H., Chen, S., Chen, G., Wang, W., Lei, B. et al. (2022). FAT-Net: Feature adaptive transformers for automated skin lesion segmentation. Medical Image Analysis, 76, 102327. https://doi.org/10.1016/j.media.2021.102327 [Google Scholar] [PubMed] [CrossRef]
34. Zhang, Z., Sun, B., Zhang, W. (2021). Pyramid medical transformer for medical image segmentation. arXiv preprint arXiv:2104.14702. [Google Scholar]
35. Chu, X., Tian, Z., Zhang, B., Wang, X., Wei, X. et al. (2021). Conditional positional encodings for vision transformers. arXiv preprint arXiv:2102.10882. [Google Scholar]
36. Zhang, Y. D., Dong, Z., Wang, S. H., Yu, X., Yao, X. et al. (2020). Advances in multimodal data fusion in neuroimaging: Overview, challenges, and novel orientation. Information Fusion, 64, 149–187. https://doi.org/10.1016/j.inffus.2020.07.006 [Google Scholar] [PubMed] [CrossRef]
37. Hu, J., Shen, L., Sun, G. (2018). Squeeze-and-excitation networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7132–7141. Salt Lake City, USA. [Google Scholar]
38. Woo, S., Park, J., Lee, J. Y., Kweon, I. S. (2018). CBAM: Convolutional block attention module. European Conference on Computer Vision, pp. 3–19. Munich, Germany. [Google Scholar]
39. Sirinukunwattana, K., Pluim, J., Hao, C., Qi, X., Rajpoot, N. M. (2016). Gland segmentation in colon histology images: The glas challenge contest. Medical Image Analysis, 35, 489–502. https://doi.org/10.1016/j.media.2016.08.008 [Google Scholar] [PubMed] [CrossRef]
40. Dlinradiology (2020). COVID-19 CT segmentation dataset. https://medicalsegmentation.com/COVID19/ [Google Scholar]
41. Badrinarayanan, V., Kendall, A., Cipolla, R. (2017). SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis & Machine Intelligence, 39(12), 2481–2495. https://doi.org/10.1109/TPAMI.34 [Google Scholar] [CrossRef]
42. Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich, M. et al. (2018). Attention U-Net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools