 Open Access
Open Access
ARTICLE
Dual-Branch-UNet: A Dual-Branch Convolutional Neural Network for Medical Image Segmentation
1 School of Information Science and Technology, Linyi University, Linyi, 276000, China
2 School of Computer Science and Technology, Shandong University of Finance and Economics, Jinan, 250014, China
3 School of Science & Engineering, University of Limerick, Limerick, V94 T9PX, Ireland
* Corresponding Author: Muwei Jian. Email:
# They contributed equally to this work and shared the first authorship
(This article belongs to the Special Issue: Advanced Intelligent Decision and Intelligent Control with Applications in Smart City)
Computer Modeling in Engineering & Sciences 2023, 137(1), 705-716. https://doi.org/10.32604/cmes.2023.027425
Received 28 October 2022; Accepted 30 December 2022; Issue published 23 April 2023
Abstract
In intelligent perception and diagnosis of medical equipment, the visual and morphological changes in retinal vessels are closely related to the severity of cardiovascular diseases (e.g., diabetes and hypertension). Intelligent auxiliary diagnosis of these diseases depends on the accuracy of the retinal vascular segmentation results. To address this challenge, we design a Dual-Branch-UNet framework, which comprises a Dual-Branch encoder structure for feature extraction based on the traditional U-Net model for medical image segmentation. To be more explicit, we utilize a novel parallel encoder made up of various convolutional modules to enhance the encoder portion of the original U-Net. Then, image features are combined at each layer to produce richer semantic data and the model’s capacity is adjusted to various input images. Meanwhile, in the lower sampling section, we give up pooling and conduct the lower sampling by convolution operation to control step size for information fusion. We also employ an attention module in the decoder stage to filter the image noises so as to lessen the response of irrelevant features. Experiments are verified and compared on the DRIVE and ARIA datasets for retinal vessels segmentation. The proposed Dual-Branch-UNet has proved to be superior to other five typical state-of-the-art methods.Keywords
The basic goal of medical image segmentation is to make pathological structural changes in the image more visible, allowing the imaging physician to make a diagnosis more quickly and objectively [1]. With the rise of deep learning, particularly the significant progress made by convolutional neural networks in recent years, computer-aided diagnosis has been greatly improved in terms of efficiency and accuracy. Among the well-known CNN approaches, U-Net [2] is unquestionably one of the most successful medical image segmentation models, requiring little training data while achieving good segmentation results. U-Net [2] originates from FCN (Fully Convolutional Networks) [3], and consists of two parts: encoder and decoder. After multiple convolutions and down-sampling, feature maps containing rich semantic information are obtained, and the feature maps are up-sampled and then spliced to obtain the output. Since U-Net [2] has efficient reasoning speed and excellent segmentation results, it has been widely concerned by researchers since it appears and has expanded many variant works [3]. For example, Attention U-Net [4] skillfully adopts the soft attention mechanism to replace the hard attention mechanism for pancreas segmentation, and obtains the attention weight by fusing the feature maps, then finally splices with the upsampling results to suppress irrelevant features and enhance relevant features. Another example is the connection-sensitive attention network CSAUNet [5], which achieves good segmentation results by combining the attention mechanism with its proposed connection-sensitive loss function. These studies showed that the attention mechanism is useful for lightweight medical picture segmentation models.
In recent years, the construction of innovative convolution modules has been a key research focus, and some research teams have produced dynamic convolution that is more suitable for lightweight neural networks [6,7], such as CondConv [8], DyNet [9] and Res-UNet [10]. Dynamic convolution, as opposed to traditional convolution, dynamically alters the convolution parameters based on the input image. This architecture can enhance the model’s expressiveness without expanding the network’s depth or width. Numerous investigations have shown that dynamic convolution improves performance more significantly than normal convolution for the same number of channels.
Semantic segmentation techniques in natural situations and related applications [11–14] have made significant progress with many outstanding segmentation models emerging, including the Deeplab series [15–18], FCN [19], FastFCN [20], and PsPNet [21]. However, there are still numerous challenges in the field of medical image segmentation. These challenges come from the intrinsic property of medical images themselves. The main reasons for segmentation difficulty are overlapping lesions with normal areas, diminutive variances between the anterior and posterior backdrops [22], small targets with various locations [23], and diverse noises generated by the medical imaging system.
In order to deal with these challenges, this paper proposes an improved network based on U-Net [2] and dynamic convolution as shown in Fig. 1, which aims to promote the accuracy rating of retinal blood vessel segmentation. The main contributions of this paper are condensed as follows:
1) We designed a two-branch encoder module composed of different convolutional blocks to replace the original single encoder to enhance the model’s processing capability for retinal image analysis.
2) We proposed a new attention module to guide the model to better identify vascular and non-vascular features.
3) Experiments implemented on both DRIVE and ARIA datasets show that the devised Dual-Branch-UNet outperforms the other typical models.
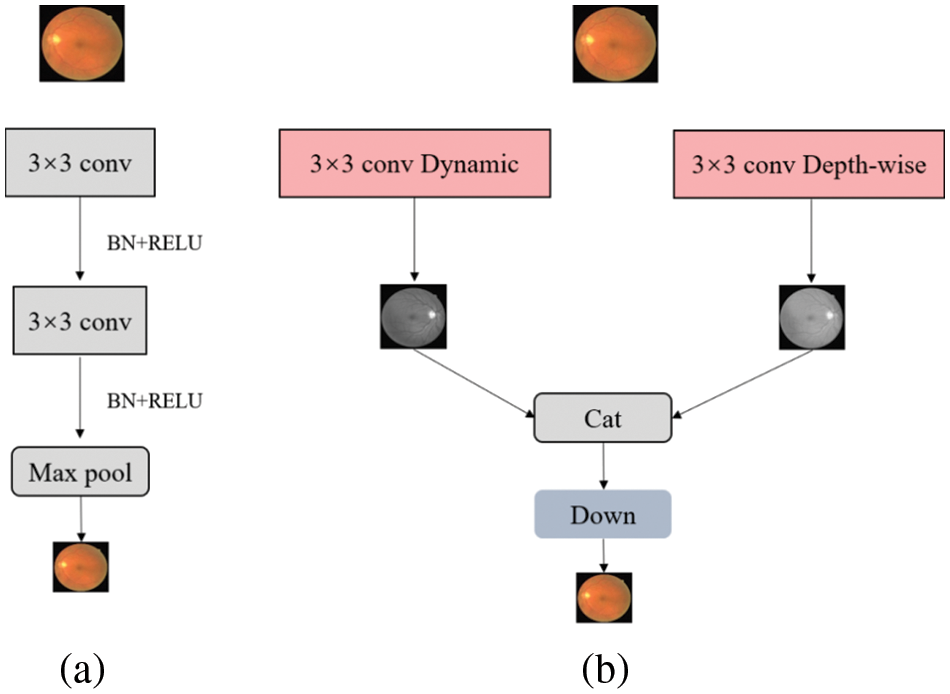
Figure 1: Encoder structure contrast of (a) UNet and (b) Dual-Branch-UNet. The down module is composed of conv, batch normalization and Relu
The remaining sections are described as below. Section 2 of this study provides a summary of current work on retinal vascular segmentation, and Section 3 shows how to extend U-Net to Dual-Branch-UNet. Section 4 presents our experimental results, and the last Section 5 summarizes and discusses the article.
Vessel segmentation is a challenging task for diagnosing diseases in medical image processing. Researchers have applied different methods, techniques and algorithms to extract retinal vascular structures to predict diseases such as laryngology, neurosurgery and ophthalmology [24]. This section presents some existing methods and models for retinal vascular segmentation from different technical perspectives.
2.1 Thresholds and Multiscale Segmentation
Thresholding is the most common and traditional technique for object region segmentation. This method is based on the feature that the gray values between neighboring pixels (e.g., background regions) and the target region is distinguished [25–27]. By setting a suitable threshold value, the pixels can be separated into two categories. Dash et al. [28] suggested a multi-stage framework for segmenting retinal vessels based on a threshold. This approach is divided into three steps. The first stage improves the retinal image using contrast-limited equalization and removes noise with median filtering, and the second stage is a Mean-C thresholding-based retinal vessel extraction segmentation procedure that uses a threshold to separate the input into black and white images, then the third stage is a morphology-based procedure that removes non-vessels and isolated pixels.
Additionally, the multi-scale approach adopts different convolution kernels to process the medical image, with the advantage of being able to take into account different ranges of spatial feature contexts. Michelson et al. [29] developed a multiscale-based approach for extracting retinal blood vessels. The method shortens the inference time by recognizing blood vessels of varying sizes with varying degrees of Gaussian kernel resolution. The Gaussian pyramid is separated into three tiers, which correspond to the original image’s largest resolution, the breadth of the retinal vessels, and the height of the retinal vessels. Similarly, Rattathanapad et al. [30] suggested a multiline detection-based vascular segmentation approach based on Gaussian linear detection of input images at different sizes. This technique is able to segment larger and smaller vessels in the retinal image. Comparative experiments are carried on the DRIVE dataset [31], which verify that both techniques can produce better segmentation results. However, despite the fact that the multiscale-based method can obtain different ranges of contextual information more effectively, the huge computing cost imposed by multiscale operations remains a problem to be considered.
2.2 Matching Filter-Based Segmentation
The matched filter segmentation is performed by convolving the kernel mask with the vessel structure of the retinal image, and the response image of the matched filter is generated by designing a kernel function on the basis that the filter can extract different features in each direction of the retinal vessels [32]. Using appropriate image threading, Fan et al. suggested a matched filter-based approach for the automatic extraction of retinal vasculature [33], which processes local entropy thresholds and filters misclassified pixels by eight contiguous regions around the target. Morphological refinement methods are also adopted to discover vessel trees and branch points in retinal pictures.
Dynamic convolution, a recent research hotspot, intends to overcome the challenges of limited depth of lightweight network models. The merit of dynamic convolution is that it is capable of enhancing feature extraction capability and diminishing the number of channels while preserving network performance. The usefulness of dynamic convolution is demonstrated by experimental verification on numerous publicly available datasets. Our research focuses on how to construct a deep learning model with dynamic convolution for medical image segmentation via extending the conventional U-Net [2] to Dual-Branch-UNet.
3.1 Structure of a Dual-Branch Encoder
In the current research, many algorithms use multi-branch network structures, such as Contextnet [34], Bisenet [35], Fast-SCNN [36], which have been successful in related medical image analysis tasks. This has triggered diverse thoughts about introducing multi-branch structures into U-Net [2] for medical image segmentation. Double U-Net [37], an improved U-Net based convolutional neural network with a composite structure, has been introduced in medical image segmentation. The encoder structure of Double U-Net [37] consists of a composite of VGG19 [38] and U-Net [2], and the input of U-Net [2] comes from the output of the VGG network, namely, it is two relatively independent networks that compose the codec module. Those models are different from our designed method, as our encoder is composed of two relatively independent convolutional modules, while we accomplish all feature fusion before decoding. Structurally, the devised model is simpler and lighter, as illustrated in Fig. 2.
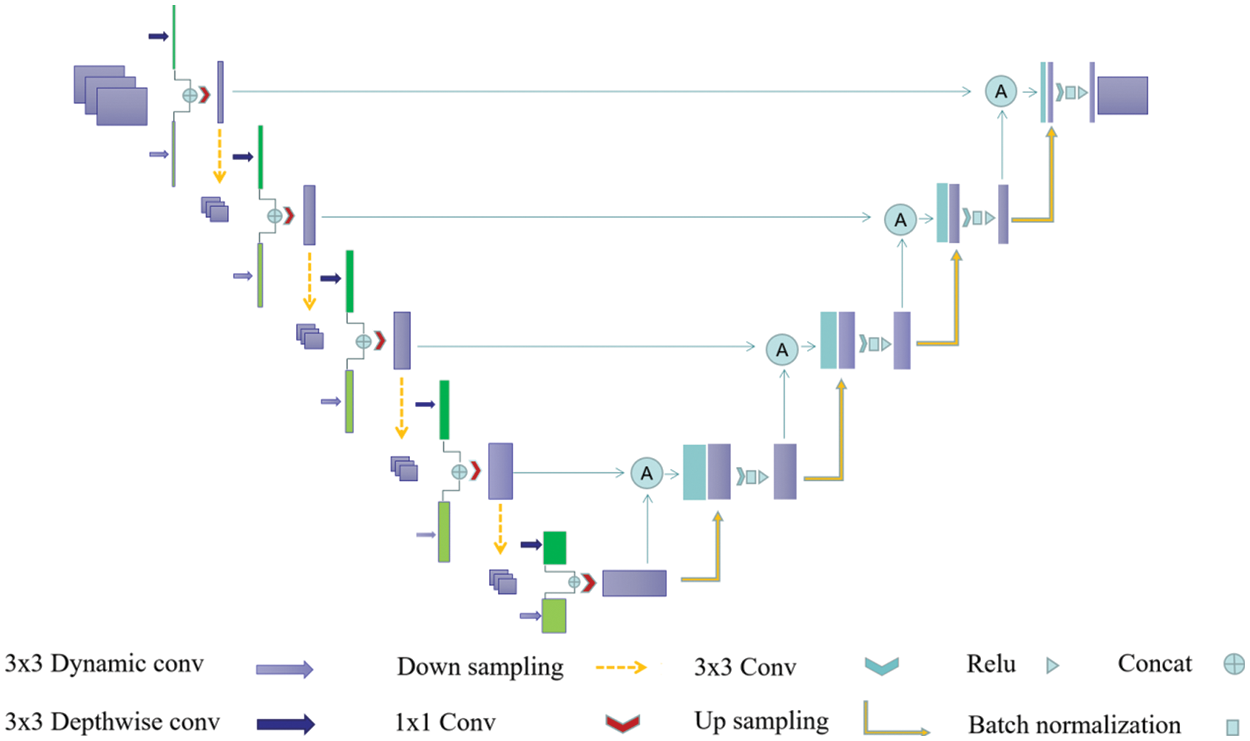
Figure 2: Diagram of the designed Dual-Branch-UNet model. Dark green and light green boxes are characteristic graphs generated by different branches, and the purple box represents the branch fusion result
In a multi-branch network structure, the deeper branches input low-resolution images with the aim of efficiently extracting global contextual features while ensuring less computational overhead. Meanwhile, the shallower network branches process high-resolution images with the aim of extracting spatial detail information. In our experiments, we found that the mutual combination of shallow networks can better control the computational overhead without losing accuracy so as to ensure the real-time performance of the algorithm. Therefore, we design a two-branch encoder structure, as depicted in Fig. 1, where each branch operates relatively independently and the intermediate results are fused to form the final semantic segmentation result.
3.2 Dynamic Convolution and Depth Separable Convolution
Compared with high-performance deep network, lightweight network has limited network depth and channel number, which leads to insufficient feature extraction ability and recognition performance of lightweight network. Dynamic convolution aims to solve these problems. In contrast to the standard convolution, the convolution kernel of dynamic convolution is associated with the input image, and different input images correspond to different convolution kernels. The exploitation of dynamic convolution not only enhances the robustness of the model to different datasets, but also brings a rapid rise in the total number of parameters. Considering this, it is desirable to seek out a solution to the large increase of the number of parameters caused by the introduction of dynamic convolution. Especially, we discovered that the combination of both dynamic convolution and depth-separable convolution has parallel segmentation accuracy to the combination of dynamic convolution and standard convolution, but the number of parameters is greatly decreased. Therefore, in the first branch, we replace the standard convolution with a deep separable convolution with a kernel of 3 × 3. In the designed architecture, the convolution kernel is set to 3 × 3 in its corresponding dynamic convolution branch, and the channel transform rate is set to 0.25, then the average pooling of its built-in attention module is replaced with maximum pooling.
3.3 Downsampling and Branch Fusion
In practical experiments, we found that the precision of down-sampling using convolution with step size of 2 and convolution kernel size of 3 × 3 is slightly higher than that of pooled down-sampling with convolution kernel size of 2 × 2. Therefore, we adopt convolution for down-sampling. Secondly, we fuse the feature information of two distinct branches in the channel dimension through the splicing operation, and then reduce the dimension through the standard convolution core of 1 × 1.
In order to better combine the low-level semantic information in the up-sampling process and suppress the responses of irrelevant features in the fusion process, we rearranged a new attention module by referring to Attention UNet [4] and the attention module in the full dimension dynamic convolution ODConv [39], as shown in Fig. 3. The input of the attention module is the sampling result
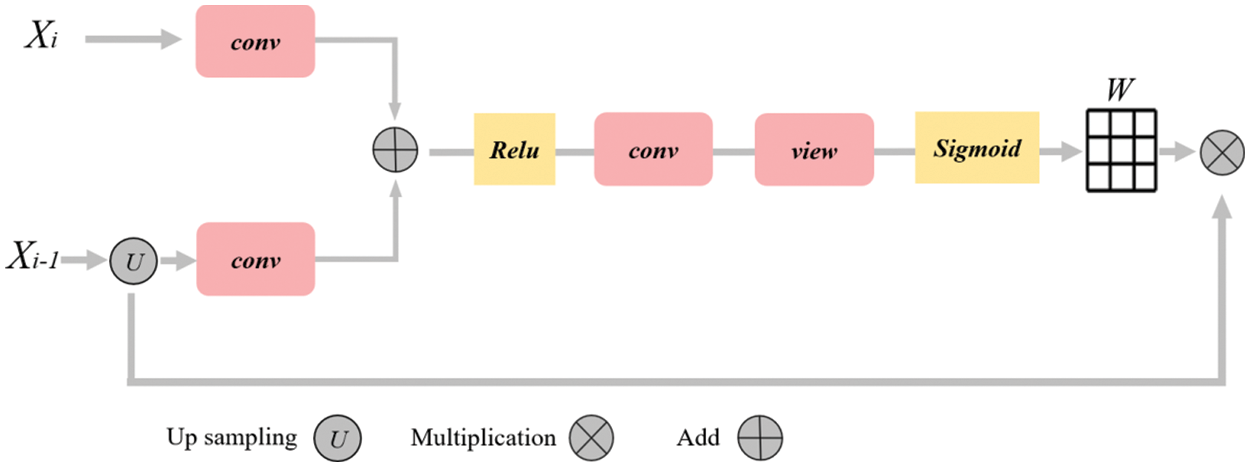
Figure 3: Diagram of attention mechanism. The rounded rectangle represents different operations, the yellow box represents the activation function, the arrow represents the direction of data flow, and the grid represents the weight matrix
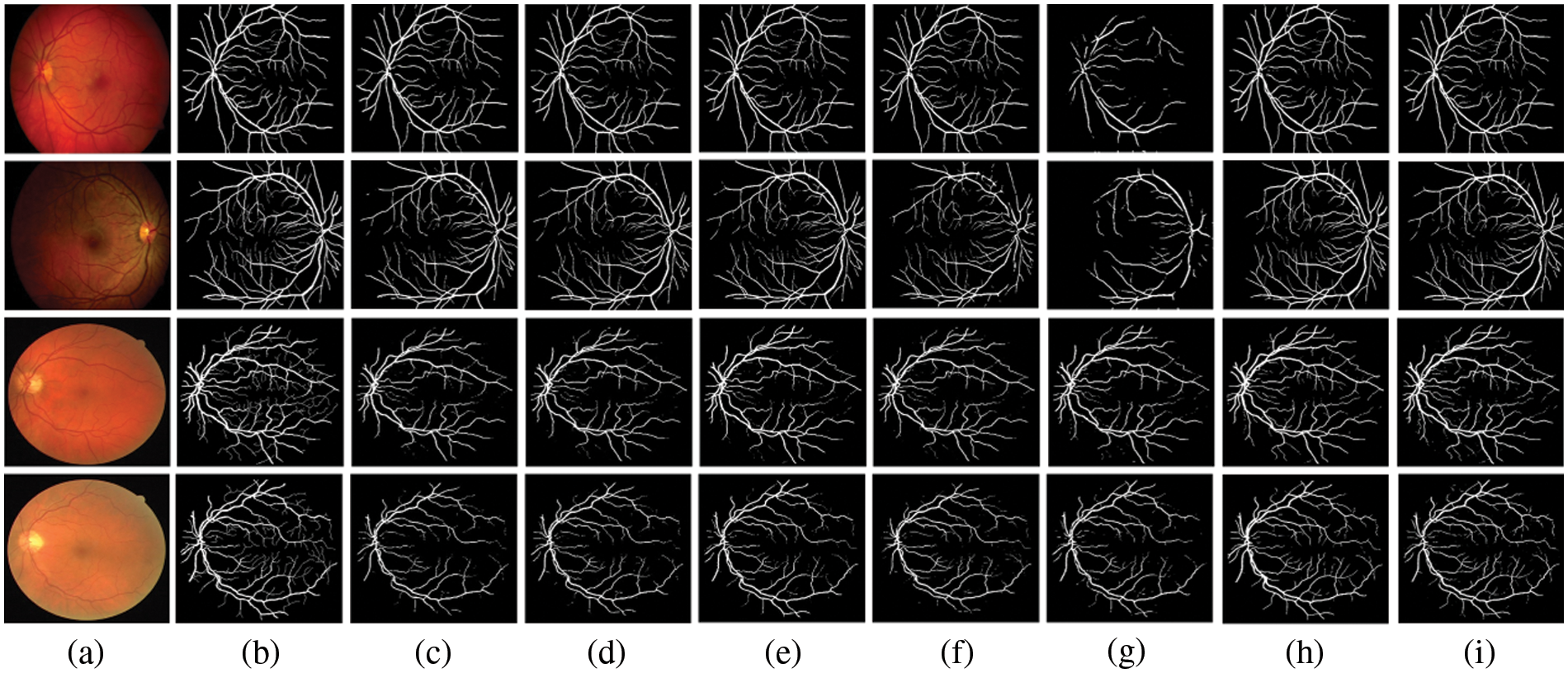
Figure 4: Comparison outputs of different methods for medical image segmentation. (a) the original image, (b) the ground truth, (c) U-Net [2], (d) Attention UNet [4], (e) UNet++ [40], (f) UNext [41], (g) R2UNet [42], (h) ConvUNext [22] and (i) the devised Dual-Branch-UNet
Immediately,
Finally, the weight matrix is multiplied with the up-sampling results to acquire the weighted features:
Experimental results showed that our attention module can suppress noise and increase the segmentation performance across diverse datasets and training sizes.
In this section, the utilized data set, evaluation indicators and parameter settings of the experiment are depicted. Through a series of experiments, the validity of the designed model is tested and compared with other advanced network models.
Data set: We verified the proposed model on two publicly accessible retinal vascular segmentation datasets, as depicted in Table 1. The DRIVE dataset is popular for retinal vessel segmentation [31]. This dataset consists of 20 training images and 20 testing images of size 565 × 565, including 7 cases of abnormal pathology. The Automated Retinal Image Analysis (ARIA) dataset [43] was provided by St. Paul’s Eye Unit, Liverpool, UK, which included a total of 142 RGB images of size 768 × 576 with three groups of subjects, containing the healthy, age-related macular degeneration, and diabetic groups. Among them, 120 were used for training and the rest of 22 were employed for testing.

Implementation details: To evaluate the devised model, we utilize the Dice coefficient and the average cross-merge ratio MIOU metrics for objective assessment. The early learning rate is set to 1e-4 for 200 training cycles using the Adam optimizer, the DRIVE dataset [31] is set to 2 for each batch size, and the learning rate is dynamically modified. The loss function is a combination of cross-entropy loss and dice loss. We also employ popular data preparation methods during experiments such as on-the-fly cropping, horizontal and vertical flipping, histogram equalization, and so on. Because the resolutions of the two datasets are different, we crop them into 480 × 480 for the DRIVE dataset and 600 × 600 for the ARIA dataset during training stage accordingly. Figs. 5 and 6 show the training loss curve on ARIA and DRIVE data sets.
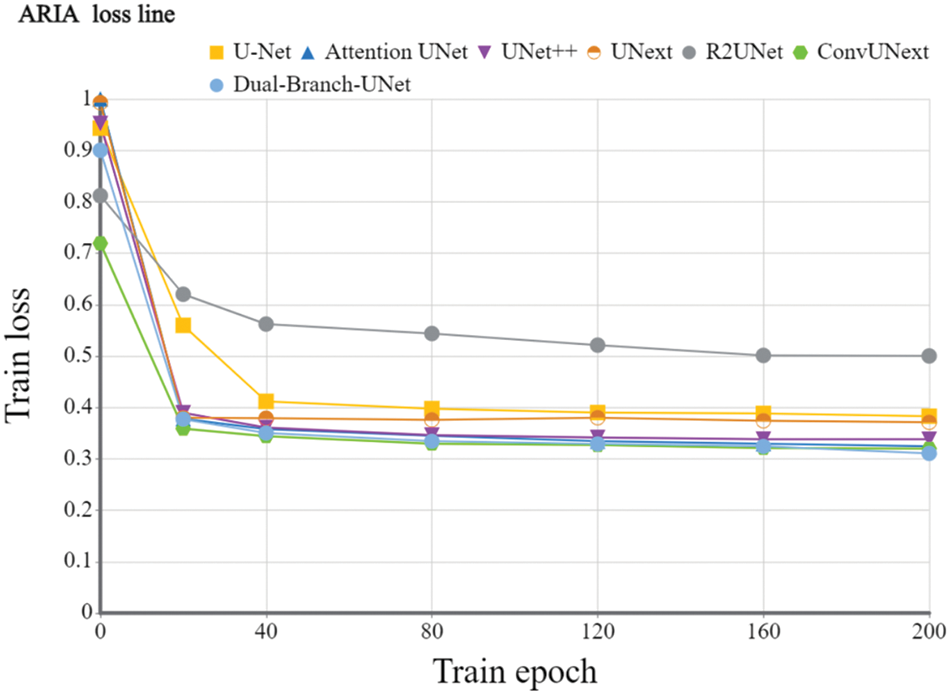
Figure 5: Training loss curve of the devised Dual-Branch-UNet on the ARIA dataset
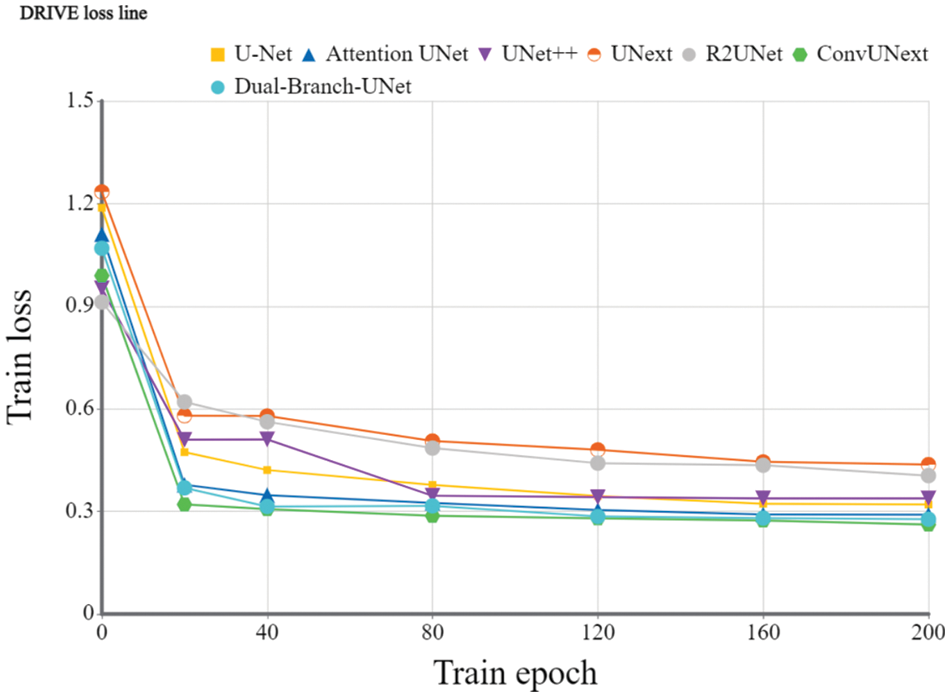
Figure 6: Training loss curve of the proposed Dual-Branch-UNet on the DRIVE dataset
The Dice coefficients and MIOU of U-Net [2], Attention UNet [4], UNet++ [40], UNext [41], R2UNet [42], ConvUNext [22] and Dual-Branch-UNet carried on the DRVIE and ARIA datasets are compared in Tables 2 and 3, respectively. The experimental results showed that our model achieves the best segmentation accuracy on DRIVE dataset. On ARIA dataset, the segmentation result of ConvUNext is also capable of generating promising results. We consider that ConvUNext adopted 7 × 7 large convolution kernel is more effective on ARIA data sets with objects of the larger original size.


The gap between UNext and the other models gradually closes as the training data increases, indicating that UNext is attentive to the amount of data volume and performs worse when the training data is minimal. In contrast, regardless of the amount of training data, our model delivers the best segmentation results. Fig. 4 depicts the visual outcomes anticipated by all models.
Some typical applications of computer-aided medical treatment for eye diseases are mainly aimed at some chronic eye diseases, such as early diabetic retinopathy and glaucoma [44,45]. This long-term chronic disease that will lead to blindness requires very experienced doctors to find signs from early scanning results. Computer-aided medical treatment can help doctors diagnose more objectively and improve the efficiency and accuracy of diagnosis [46,47].
In this paper, we improved the U-Net encoder structure by extending the classic single-branch encoder to a dual-branch structure in order to extract richer feature information for medical image segmentation. Furthermore, we substituted ordinary convolution with dynamic convolution and depth-separable convolution to ensure segmentation accuracy while dwindling the total number of parameters created by the multi-branch structure. Finally, we also developed an attention module to improve feature fusion further. Experiments based on the DRIVE, ARIA dataset show that the designed model outperforms the typical state-of-the-art approaches.
Since the number of parameters is increased compared to the original U-Net model due to the incorporation of the attention mechanism and dynamic convolution, how to accomplish the further lightweight design of the model will be our next study issue in future research. Besides, it is found that the segmentation results of the model have small fragmentation predictions. We know that blood vessels are continuous structures, and the fragmentation prediction obviously does not conform to this structure. In this case, the design of a new connectivity constraint function that is applied to solve the fragmentation prediction problem is also one of our concerned research directions in the future.
Funding Statement: This work was supported by National Natural Science Foundation of China (NSFC) (61976123, 62072213); Taishan Young Scholars Program of Shandong Province; and Key Development Program for Basic Research of Shandong Province (ZR2020ZD44).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Lei, T., Wang, R. S., Wan, Y., Du, X. G., Meng, H. Y. et al. (2022). Medical image segmentation using deep learning: A survey. IET Image Processing, 16(5), 1243–1267. https://doi.org/10.1049/ipr2.12419 [Google Scholar] [CrossRef]
2. Olaf, R., Philipp, F., Thomas, B. (2015). U-Net: Convolutional networks for biomedical image segmentation. Computing Research Repository, 234–241. [Google Scholar]
3. Wu, J., Liu, W., Li, C., Jiang, T., Shariful, I. M. et al. (2022). A state-of-the-art survey of U-Net in microscopic image analysis: From simple usage to structure mortification. arXiv preprint arXiv: 2202.06465. [Google Scholar]
4. Jo, S., Ozan, O., Michiel, S., Mattias P. H., Bernhard, K. et al. (2019). Attention gated networks: Learning to leverage salient regions in medical images. Medical Image Analysis, 53, 197–207. https://doi.org/10.1016/j.media.2019.01.012 [Google Scholar] [PubMed] [CrossRef]
5. Li, R. R., Li, M. M., Li, J. C. (2020). Connection sensitive attention U-Net for accurate retinal vessel segmentation. China Digital Medicine, 15(7), 125–129. [Google Scholar]
6. Chen, Y., Dai, X., Liu, M., Chen, D., Yuan, L. et al. (2020). Dynamic convolution: Attention over convolution kernels. Computer Vision and Pattern Recognition, 11027–11036. https://doi.org/10.1109/CVPR42600.2020 [Google Scholar] [CrossRef]
7. Felix, W., Angela, F., Alexei, B., Yann N. D., Michael, A. (2019). Pay less attention with lightweight and dynamic convolutions. International Conference on Learning Representations, New Orleans, USA. [Google Scholar]
8. Yang, B., Bender, G., Le, Quoc V., Ngiam, J. (2019). CondConv: Conditionally parameterized convolutions for efficient inference. Neural Information Processing Systems, 32, 1305–1316. [Google Scholar]
9. Zhang, Y., Zhang, J., Wang, Q., Zhong, Z. (2020). DyNet: Dynamic convolution for accelerating convolutional neural networks. arXiv preprint arXiv: 2004.10694. [Google Scholar]
10. Foivos, I. D., François, W., Peter, C., Chen, W. (2019). ResUNet-A: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS Journal of Photogrammetry and Remote Sensing, 162, 94–114. [Google Scholar]
11. Wu, Z. T., Karimi, H. R., Dang, C. Y. (2020). A deterministic annealing neural network algorithm for the minimum concave cost transportation problem. IEEE Transactions on Neural Networks and Learning Systems, 31(10), 4354–4366. https://doi.org/10.1109/TNNLS.5962385 [Google Scholar] [CrossRef]
12. Wu, Z. T., Karimi, H. R., Dang, C. G. (2019). An approximation algorithm for graph partitioning via deterministic annealing neural network. Neural Networks, (117), 191–200. https://doi.org/10.1016/j.neunet.2019.05.010 [Google Scholar] [PubMed] [CrossRef]
13. Wu, Z., Gao, Q., Jiang, B., Karimi, H. R. (2021). Solving the production transportation problem via a deterministic annealing neural network method. Applied Mathematics and Computation, 411, 126518. https://doi.org/10.1016/j.amc.2021.126518 [Google Scholar] [CrossRef]
14. Zhao, S., Zhu, S., Wu, Z., Jiang, B. (2022). Cooperative energy dispatch of smart building cluster based on smart contracts. International Journal of Electrical Power and Energy Systems, 138, 107896. https://doi.org/10.1016/j.ijepes.2021.107896 [Google Scholar] [CrossRef]
15. Chen, L. C., Papandreou, G., Kokkinos, L., Murphy, K., Yuille, A. L. (2018). Semantic image segmentation with deep convolutional nets and fully connected crfs. IEEE Transactions on Pattern Analysis and Machine Intelligence, 40, 834–848. https://doi.org/10.1109/TPAMI.2017.2699184 [Google Scholar] [PubMed] [CrossRef]
16. Chen, L. C., Papandreou, G., Kokkinos, L., Murphy, K., Yuille, A. L. (2014). Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. International Conference on Learning Representations, pp. 1412–7062. Banff, Canada. [Google Scholar]
17. Chen, L. C., Papandreou, G., Schroff, F., Adam, H. (2017). Rethinking atrous convolution for semantic image segmentation. arXiv preprint arXiv:1706.05587. [Google Scholar]
18. Chen, L. C., Zhu, Y. K., Papandreou, G., Schroff, F., Adam, H. (2018). Encoder-decoder with atrous separable convolution for semantic image segmentation. Proceedings of the European Conference on Computer Vision (ECCV), pp. 833–851. https://doi.org/10.1007/978-3-030-01234-2 [Google Scholar] [CrossRef]
19. Shelhamer, E., Long, J., Darrell, T. (2015). Fully convolutional networks for semantic segmentation. IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440. Boston, MA, USA. [Google Scholar]
20. Onim, S. H., Ehtesham, A. R., Anbar, A., Islam, N., Rahman, M. (2020). LULC classification by semantic segmentation of satellite images using FastFCN. International Conference on Advanced Information and Communication Technology (ICAICT), pp. 471–475. Macau, China. [Google Scholar]
21. Zhao, H. S., Shi, J. P., Qi, X. J., Wang, X. G., Jia, J. Y. (2017). Pyramid scene parsing network. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, no. 1612, pp. 6320–6329. Hawaii, USA. [Google Scholar]
22. Han, Z. M., Jian, M. W., Wang, G. G. (2022). ConvUNeXt: An efficient convolution neural network for medical image segmentation. Knowledge-Based Systems, 253, 109512–109522. https://doi.org/10.1016/j.knosys.2022.109512 [Google Scholar] [CrossRef]
23. Lu, X. W., Jian, M. W., Wang, X., Yu, H., Dong, J. Y. et al. (2022). Visual saliency detection via combining center prior and U-Net. Multimedia Systems, 28(5), 1689–1698. https://doi.org/10.1007/s00530-022-00940-8 [Google Scholar] [CrossRef]
24. Roseline, R. H., Priyadarsini, R. J. (2017). Survey on ocular blood vessel segmentation. Computer Science and Software Engineering, 7, 318–324. [Google Scholar]
25. Jian, M. W., Wang, J., Yu, H., Wang, G. G., Meng, X. et al. (2021). Visual saliency detection by integrating spatial position prior of object with background cues. Expert Systems with Applications, 168, 114219. https://doi.org/10.1016/j.eswa.2020.114219 [Google Scholar] [CrossRef]
26. Jian, M. W., Wang, J., Yu, H., Wang, G. G. (2021). Integrating object proposal with attention networks for video saliency detection. Information Sciences, 576, 819–830. https://doi.org/10.1016/j.ins.2021.08.069 [Google Scholar] [CrossRef]
27. Jian, M. W., Wang, J., Dong, J. Y., Cui, C., Nie, X. et al. (2020). Saliency detection using multiple low-level priors and A propagation mechanism. Multimedia Tools and Applications, 79(45–46), 33467–33482. https://doi.org/10.1007/s11042-019-07842-4 [Google Scholar] [CrossRef]
28. Dash, J., Bhoi, N. (2017). A thresholding based technique to extract retinal blood vessels from fundus images. Future Computing and Informatics Journal, 2, 103–109. https://doi.org/10.1016/j.fcij.2017.10.001 [Google Scholar] [CrossRef]
29. Michelson, G., Hornegger, J., Budai, A. (2010). Multiscale blood vessel segmentation in retinal fundus images. Bildverarbeitung für die Medizin, 574, 261–265. [Google Scholar]
30. Rattathanapad, S., Mittrapiyanuruk, P., Kaewtrakulpong, P., Uyyanonvara, B. (2012). Vessel extraction in retinal images using multilevel line detection. Biomedical and Health Informatics, 87, 345–349. https://doi.org/10.1109/BHI.2012.6211584 [Google Scholar] [CrossRef]
31. Joes, S., Michael D. A., Meindert, N., Max A. V., Bram van, G. (2004). Ridge-based vessel segmentation in color images of the retina. IEEE Transactions on Medical Imaging, 23(4), 501–509. https://doi.org/10.1109/TMI.2004.825627 [Google Scholar] [PubMed] [CrossRef]
32. Santosh Nagnath, R., Ranjan, K. S., Amol, D. R. (2019). A review on computer-aided recent developments for automatic detection of diabetic retinopathy. Journal of Medical Engineering & Technology, 43(2), 1–13. [Google Scholar]
33. Thitiporn, C., Guoliang, F. (2003). An efficient algorithm for extraction of anatomical structures in retinal images. International Conference on Image Processing, pp. 1093–1096. Barcelona, Spain. [Google Scholar]
34. Rudra P. K. P., Ujwal, B., Stephan, L., Christopher, Z. (2018). ContextNet: Exploring context and detail for semantic segmentation in real-time. British Machine Vision Conference, pp. 146–158. Newcastle, UK. [Google Scholar]
35. Yu, C. Q., Wang, J. B., Peng, C., Gao, C. G., Yu, G. et al. (2018). Bisenet: Bilateral segmentation network for real-time semantic segmentation. Proceedings of the European Conference on Computer Vision (ECCV), pp. 334–349. https://doi.org/10.1007/978-3-030-01261-8 [Google Scholar] [CrossRef]
36. Rudra P. K. P., Stephan, L., Roberto, C. (2019). Fast-SCNN: Fast semantic segmentation network. British Machine Vision Conference, no. 19, pp. 289–297. Cardiff, Wales, UK. [Google Scholar]
37. Jha, D., Riegler, M., Johansen, D., Halvorsen, P. (2020). Double U-Net: A deep convolutional neural network for medical image segmentation. 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), vol. 118, pp. 558–564. IEEE. [Google Scholar]
38. Karen, S., Andrew, Z. (2015). Very deep convolutional networks for large-scale image recognition. International Conference on Learning Representations (ICLR), no. 1409. San Diego, CA. [Google Scholar]
39. Li, C., Yao, A. B., Zhou, A. J. (2022). Omni-dimensional dynamic convolution. International Conference on Learning Representations, pp. 25–29. La Jolla, CA, USA. [Google Scholar]
40. Zhou, Z. W., Siddiquee, M. R., Tajbakhsh, N., Liang, J. M. (2018). UNet plus: A nested U-Net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, vol. 11045, pp. 3–11. DLMIA 2018, ML-CDS 2018. [Google Scholar] [PubMed]
41. Jose, J. M., Patel, V. M. (2022). UNext: MLP-based rapid medical image segmentation network. Medical Image Computing and Computer Assisted Intervention–MICCAI 2022: 25th International Conference, vol. 13435, pp. 23–33. Singapore. [Google Scholar]
42. Md Zahangir, A., Chris, Y., Mahmudul, H., Tarek, M. T., Vijayan, K. A. (2019). Recurrent residual U-Net for medical image segmentation. Journal of Medical Imaging, 6(1), 6–14. [Google Scholar]
43. Farnell, D. J. J., Hatfield, F. N., Knox, P., Peakes, M., Spencer, S. et al. (2008). Enhancement of blood vessels in digital fundus photographs via the application of multiscale line operators. Journal of the Franklin Institute, 345(7), 748–765. https://doi.org/10.1016/j.jfranklin.2008.04.009 [Google Scholar] [CrossRef]
44. Yao, F., Ding, Y., Hong, S., Yang, S. H. (2022). A survey on evolved LoRa-based communication technologies for emerging internet of things applications. International Journal of Network Dynamics and Intelligence, 1(1), 4–19. https://doi.org/10.53941/ijndi0101002 [Google Scholar] [CrossRef]
45. Szankin, M., Kwasniewska, A. (2022). Can AI see bias in X-ray images. International Journal of Network Dynamics and Intelligence, 1(1), 48–64. https://doi.org/10.53941/ijndi0101005 [Google Scholar] [CrossRef]
46. Yu, N., Yang, R., Huang, M. (2022). Deep common spatial pattern based motor imagery classification with improved objective function. International Journal of Network Dynamics and Intelligence, 1(1), 73–84. https://doi.org/10.53941/ijndi0101007 [Google Scholar] [CrossRef]
47. Wang, M., Wang, H., Zheng, H. (2022). A mini review of node centrality metrics in biological networks. International Journal of Network Dynamics and Intelligence, 1, 99–110. https://doi.org/10.53941/ijndi0101009 [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools