 Open Access
Open Access
REVIEW
Application of U-Net and Optimized Clustering in Medical Image Segmentation: A Review
1 School of Physics and Information Engineering, Jiangsu Second Normal University, Nanjing, 211200, China
2 State Key Laboratory of Millimeter Waves, Southeast University, Nanjing, 210096, China
3 Jiangsu Province Engineering Research Center of Basic Education Big Data Application, Nanjing, 211200, China
4 School of Data Science, Perdana University, Serdang, Selangor, 43400, Malaysia
* Corresponding Authors: Shuwen Chen. Email: ; Mackenzie Brown. Email:
# These authors contributed equally to this work. Jiaqi Shao, Shuwen Chen and Jin Zhou are regarded as co-first authors
(This article belongs to the Special Issue: Computer Modeling of Artificial Intelligence and Medical Imaging)
Computer Modeling in Engineering & Sciences 2023, 136(3), 2173-2219. https://doi.org/10.32604/cmes.2023.025499
Received 17 July 2022; Accepted 08 November 2022; Issue published 09 March 2023
Abstract
As a mainstream research direction in the field of image segmentation, medical image segmentation plays a key role in the quantification of lesions, three-dimensional reconstruction, region of interest extraction and so on. Compared with natural images, medical images have a variety of modes. Besides, the emphasis of information which is conveyed by images of different modes is quite different. Because it is time-consuming and inefficient to manually segment medical images only by professional and experienced doctors. Therefore, large quantities of automated medical image segmentation methods have been developed. However, until now, researchers have not developed a universal method for all types of medical image segmentation. This paper reviews the literature on segmentation techniques that have produced major breakthroughs in recent years. Among the large quantities of medical image segmentation methods, this paper mainly discusses two categories of medical image segmentation methods. One is the improved strategies based on traditional clustering method. The other is the research progress of the improved image segmentation network structure model based on U-Net. The power of technology proves that the performance of the deep learning-based method is significantly better than that of the traditional method. This paper discussed both advantages and disadvantages of different algorithms and detailed how these methods can be used for the segmentation of lesions or other organs and tissues, as well as possible technical trends for future work.Keywords
Medical imaging plays a leading role in the diagnosis, and treatment of diseases. It enables localization, qualitative and quantitative analysis of diseases through noninvasive imaging. As an important branch of computer vision in medical image processing, the goal of medical image segmentation is to realize the segmentation of complex medical images of special significance or interest. The precision and accuracy of segmentation will directly affect the subsequent diagnostic work. In addition, time is also a factor that cannot be ignored. When serious diseases are spreading, such as the novel coronavirus pneumonia, rapid automatic segmentation technologies such as deep learning and artificial intelligence technology, will be an alternative to something that more patients will see a doctor in time within the optimal treatment time [1]. Usually, medical images are affected by the image acquisition equipment, resulting in problems, such as low contrast and difficult identification of boundary areas. Different medical image imaging techniques, including computed tomography (CT), nuclear magnetic resonance imaging (MRI), positron emission tomography (PET), etc., have some problems in generating medical images. For example, CT images do not provide clear images of soft tissues and lesions, and MRI images do not produce images of the anatomical structure of bones. All these make the high-quality segmentation of medical images become an important and difficult problem for computer-aided diagnosis. In addition to its inherent disadvantages, medical image acquisition is so expensive and complex, that there are few available datasets [2]. In order to increase the number and diversity of training samples, the dataset can be augmented by synthesizing high quality adversarial images. As an important part of computer-aided diagnosis and treatment system, medical image segmentation has been the focus of attention at home and abroad. Due to the limitation of technical level, no general medical image segmentation method has been found yet [3]. Therefore, researchers need to take a commitment to study medical image segmentation in clinical diagnosis and case analysis.
Many researchers have devoted themselves to improving the accuracy as well as the speed of medical image segmentation [4], spending a lot of human and financial resources to solve some technical bottlenecks. After investigating many literatures, the clustering algorithm belonging to traditional algorithms and the U-Net automatic segmentation method based on deep learning accounted for a lot of articles. Therefore, this paper will explore and discuss the medical image segmentation method with universal applicability from these two perspectives.
Traditional medical image segmentation algorithms include threshold segmentation, edge detection, region growing and [5] clustering algorithms. Although the threshold method has the advantages of small computation and simple implementation, it only uses the gray value characteristics of pixels and does not consider their spatial characteristics. The key of the region growing method lies in the design of the criterion. The improper design will easily lead to the problem of over-segmentation or under-segmentation. Image segmentation based on edge detection solves the problem caused by detecting edges containing different regions. The common detection methods are the Roberts gradient operator, Canny operator, Wills operator and so on. The gray value of the boundary between different regions changes dramatically, and different operators cannot ensure the continuity and closure of the edge when extracting the edge. Among these traditional medical image segmentation algorithms, cluster analysis is more unsupervised, efficient, and adaptive than traditional medical image segmentation methods, so clustering algorithm is the most widely studied and applied method. In view of the problems of the traditional clustering algorithm, such as uncertain data, easy to be disturbed by noise, and unable to make full use of spatial information, many researchers have improved it. There are two kinds of clustering methods: hard clustering and soft clustering. K-means is a typical hard clustering algorithm. The basic idea is to assign each sample point only to a certain class. The problem is that the flexibility of segmentation results is poor, and the effect and effectiveness of segmentation are greatly affected by noise and spatial resolution. In order to cope with this problem, many scholars consider introducing concepts such as morphology. Agrawal et al. [6] combined K-means and morphology concepts and selected median filter to remove impulse noise in images. K-means is used to subdivide the image into different components, and finally extract the tumor region through morphological surgery. The combination of K-means and discrete wavelet transform is also a feasible method. Sumithra et al. [7] used this new method to segment tumor images with an accuracy of 87.8%. But unfortunately, the computational efficiency of the algorithm is not guaranteed. Therefore, soft clustering algorithm came into being. Researchers have done a lot of work and innovation on fuzzy clustering algorithm which belongs to soft clustering category. Puneet Kumar’s team proposed to refine the membership degree of noisy pixels through the information of adjacent pixels, which improved the problem of segmentation performance degradation in the case of noise interference. Spatial intuitionistic fuzzy clustering algorithm uses spatial function to update the membership function, but it is easy to cause the objective function not to converge. The fuzzy local C means increases the fuzzy local neighborhood factor [8] and integrates the local spatial information into the objective function. The distance between the pixel and the cluster center is calculated from the original and local information of the pixel. ACC reaches 0.9480, SEN reaches 0.9221, and SPE reaches 0.9610. But the acquisition of spatial information takes a long time. Kumar’s et al. team [9] proposed to refine the membership degree of noisy pixels through adjacent pixel information, which makes a difference to the segmentation performance degradation in the case of noise interference. Spatial intuitionistic fuzzy clustering algorithm applies spatial function to update the membership function, but it is easy to cause the objective function not to converge. In order to improve the segmentation accuracy, Almahfud et al. [10] proposed A method combining K-means and fuzzy C-means, which produced good results when detecting the optimal value and local outliers. The other is the popular deep learning algorithm based on artificial neural networks, which obtains features through the multi-layer network structure and classifies pixels by these features. A full convolutional network [11] can accept images of arbitrary size, and the segmentation result is close to manual segmentation. However, the simple up sampling operation still cannot achieve the expected accuracy, and it is easy to ignore the details. In view of these shortcomings, improved network models such as Deep Lab [12], SegNet [13] and U-Net [14] have emerged. Due to the complexity and cost of obtaining medical images, richer medical images cannot be provided. U-Net has the characteristics of residual connection and U-shaped structure, which can make full use of the global and local details of the image. In the absence of sufficient data sets, U-Net can achieve satisfactory segmentation performance and is the best choice among many network models. U-Net has a simple structure and is easy to train, and does not rely on large data sets like other networks [15]. From the perspective of protecting personal information, Yu’s team [16] proposed MIA-U-Net, a multi-scale iterative aggregation network for efficient segmentation of retinal blood vessels. Due to the low computational complexity of the network, there is no need to transfer data to the cloud for image processing and analysis, thus avoiding the leakage of personal information.
Although the underlying feature map can capture rich spatial information to locate the location of organs, it lacks the support of semantic information. On the other hand, although advanced feature maps acquire more semantic information, they lack the ability to perceive details. Therefore, a 2.5D image segmentation method based on U-Net is discussed in this paper [17,18]. This method achieves a balance between computation and training time, it must be pointed out that the segmentation accuracy is still insufficient. The automatic 3D model based on U-Net can be used for automatic segmentation of 3D magnetic resonance images. A context nested network containing two U-Net structures [19]. The use of dense blocks in the encoder can reduce the number of network parameters but deepen the network hierarchy and ease the gradient disappearance. The residual block used in the decoder instead of the convolutional nerve block in U-Net improves the segmentation effect [20]. Jia Chen et al. believed that the acquisition of more advanced information [21] could effectively improve the segmentation accuracy, so they proposed a multipath up-sampled convolution network MU-Net. Embedding spatial attention blocks in the segmentation process helps to extend the perceptual domain. To a large extent, it solves the problem caused by small tissue segmentation [22]. Influenced by the support vector Machine multi-classification model, Wang’s team [23] proposed a deep residual network based on U-Net, which achieved good results, but there was a serious gradient disappearance problem. To solve the problem of how to locate the target boundary more accurately, Leclerc et al. [24] provided the answer. They proposed an RU-Net model consisting of a U-Net segmentation network and a multi-level boundary detection network. How to compress the network structure model, further reduce the computing power of hardware devices, and ensure the accuracy and stability of image segmentation results is also a problem that researchers need to consider and solve.
In the first part of this paper, several methods commonly used in traditional medical image segmentation algorithms are introduced, including threshold segmentation, edge detection and region growing algorithms. The typical problems of traditional segmentation algorithms are discussed. The second part discusses the optimization and improvement based on clustering algorithm in detail, mainly around C-means and K-means. In terms of U-Net, taking the continuous improvement and innovation of the model as the main line, this paper clarifies the subdivision performance and existing problems of the network structure, which has certain significance for the development of U-Net [25,26]. These will be covered in Part three. Finally, the extension and network model of the clustering algorithm based on U-Net are summarized and prospected. From the perspective of observation and suggestion, the gaps, challenges, and future research trends of current technology are shown. With the continuous update and iteration of technology, medical image segmentation has been rapidly developed, which promotes the bright future of the whole medical and health career. Based on the research before August 23, 2022, this paper discusses and summarizes the improvement strategy based on clustering algorithm and deep learning algorithm led by U-Net. Our team aims to provide certain guidance for doctors and researchers, which has certain positive significance for the development of medical image segmentation technology.
2 Traditional Image Segmentation Technology
Traditional medical image segmentation algorithms include threshold segmentation, edge detection, region growth [14] and segmentation methods based on energy functional as well as graph theory. The idea of traditional segmentation algorithm is easy to understand, but each has its own shortcomings. For example, the threshold method is more suitable for the image with large gray difference between the background and the target, and it is sensitive to noise because it cannot consider the spatial characteristics. The region growing method is easy to calculate and has good segmentation effect on connected regions with the same characteristics, but the problem of over-segmentation still exists. Although the operator used in edge detection can extract the boundary, small edges are easy to appear in the high detail area, which is not suitable for the segmentation of lesions composed of small tissues in medical images. Table 1 lists the characteristics and disadvantages of several common traditional image segmentation methods. These traditional medical image segmentation algorithms are mainly based on various mathematical rules. With the continuous development of technology, researchers have improved and innovated the traditional segmentation methods which are based on much experimental work. Although they have improved segmentation performance, cluster analysis has more significant advantages, such as unsupervised, high efficiency and adaptability.

2.1 Image Segmentation Based on Threshold
As one of the segmentation algorithms, threshold segmentation technology divides the gray histogram of an image into several classes with one or more thresholds, and considers that pixels in the image with gray values in the same gray class have the same attributes. The selection of threshold value can be set manually or obtained by specific calculation. The segmentation of more than one threshold value is called multi-threshold segmentation. The pixel points of the image are divided into several classes according to the threshold value, and different sets are obtained through division. The pixels in each set have the same attributes. It is especially suitable for images with different levels of gray values between the target object and the background. The key point is to obtain the optimal threshold value, which directly affects the rationality and effect of image segmentation. There are three typical thresholding-based image segmentation methods, including iterative global threshold segmentation, Otsu global threshold segmentation [27–29] and local threshold segmentation [30]. However, thresholding-based image segmentation algorithm still has some problems in practical application. Since the spatial distribution of pixels in the image is not fully considered, it is easy to be affected by noise. On the other hand, this method is difficult to achieve image segmentation if the gray difference between the background and target area is very small. Considering these problems, researchers have improved the original thresholding-based image segmentation algorithm.
When the contrast between the object and the background is not the same everywhere in the image, different thresholds can be used for segmentation, according to the local features of the image. The study in [31] proposed an Otsu method which is called an adaptive threshold method. With this method, it can make insurance to extract lungs with minimal intraclass variability. In the paper [32], the authors put forward a kind of anisotropic diffusion filter which is based on the threshold method for brain tumor detection and segmentation. Wang’s team [33] proposed a new medical image segmentation method. The innovative point is the combination of threshold segmentation method and fast-moving method, which is called level set method based on threshold. The diffusion coefficient of the curve can be effectively controlled within the threshold set, while the fast-moving method aims to redefine the velocity function according to the similarity of the statistical characteristics of each region. The experimental data also show that this method greatly improves the segmentation efficiency and speed.
2.2 Medical Image Segmentation Based on Edge Detection
The basic idea of image segmentation method based on edge detection is to determine the edge pixels in the image first, and then connect these pixels together to form the desired region boundary [34–36]. Image edge can be understood as a collection of pixels with spatial mutation of image gray scale. The first and second derivatives are usually used to describe and detect edges. Roberts, Prewitt and Sobel commonly be chosen as the first-order differential operators, and Laplace and Kirsh usually be the second-order differential operators. The image segmentation algorithm based on edge detection tries to solve the problem of segmentation by detecting the edges containing different regions [37]. It provides a guiding method for extracting the weak edges of uneven gray scale in images such as coronavirus. The author verified the effectiveness of the improved multi-scale morphology and the adaptive threshold binarization of the weak edge detection method [38]. Using COVID-19 images as the data set, the entropy is 1.8539 and the accuracy is 0.9992.
Canny applies non-maximum suppression to remove unwanted responses. Under the inspiration of Canny edge detection technology, the authors proposed a gaussian filtering fuzzy C-mean threshold segmentation method for edge detection technology [39]. Firstly, the cluster was created, and then 5% Gaussian fuzzy noise was added to the cluster image. After removing the noise, edge detection was carried out on the cluster image, and the detected edge was segmented and extracted. However, it still cannot guarantee the continuity and closure of the edges, especially in the high detail area, there will still be broken edges.
2.3 Medical Image Segmentation Based on Region Growth
The region growth method belongs to the region-based segmentation method. The region-based segmentation method [40] mainly combines the similarity of pixels into segmentation results by using local spatial information of the image. The basic idea of the region growing method is to divide the seed points and successively add adjacent pixels or small regions that meet certain similarity criteria to the same region to obtain the target region [15]. The seed point is the starting point of regional growth, and the selection of its location has a great influence on the segmentation result.
In a word, the key of seed region growing method is to select the initial seed pixel and the growth criterion. Vyavahare’s team [41] proposed a seed region growth algorithm based on the concept of “affinity”. It is mainly to achieve the separation of different parts of the image according to the function definition. The user will specify the seed pixel which can be a single pixel or a group of pixels in the region to be divided, and then calculate not only the similarity between the adjacent pixels but the seed pixel. When the calculated similarity compared to a threshold, once the outcome is greater than the threshold, then the group operation will be performed.
However, there is no way to avoid the problem of noise and uneven gray level. The idea often adopted is to combine the region method with other segmentation methods, Angelina et al. [42] used genetic algorithm based on region growth to achieve. The advantage of combining genetic algorithm and region growth algorithm is that it avoids the problem of using a single method.
2.4 Medical Image Segmentation Based on Energy Functional
This method is based on the active contour model, which uses continuous curves to express the target edge. According to the different curve evolution ways, the active contour model can be divided into boundary based, region based and mixed active contour model. The basic idea of image segmentation based on energy functional is to define an energy functional so that its independent variables include edge curves. Generally, the minimum value of the energy functional is obtained by solving the Euler equation corresponding to the function, and the curve with the minimum energy is the position of the target contour [43].
Active Appearance Models (ASM) and Active Appearance Models (AAM) are two of the parametric Active contour Models. Both are based on point distribution Models [44]. The difference is that ASM is based on statistical shape model, while AAM is based on ASM for statistical modeling of texture. AAM has two main improvements over ASM model. One is that AAM uses two statistical models to fuse instead of ASM gray model. The other is to improve the feature descriptor of feature points. Increased the complexity and robustness of descriptors [45].
After parametric active contours, geometric active contours model is another great development based on curved fireworks theory and level set method. In contrast to parametric active profile models, geometrically active profile models can handle topological changes of curves, insensitivity to initial positions and stability of numerical solutions [46].
2.5 Medical Image Segmentation Based on Graph Theory
Image segmentation methods based on graph theory are often associated with the problem of minimum cut of graph. Each node in the graph corresponds to each pixel in the image, and each edge is connected to a pair of adjacent pixels. The weight of the edge represents the non-negative similarity between adjacent pixels in terms of gray level, color, or texture. The segmentation problem is transformed into a label problem. The optimal principle of segmentation is to make the divided subgraph keep the maximum similarity inside. The similarity between subgraphs is kept to A minimum [47]. Graph cut [48] and grab cut [49] are two basic graph theory methods. One is a one-time energy minimization, and the other is an interactive iterative process of continuous segmentation evaluation and model parameter learning. Graph cut requires the user to specify some seed points [50] of target and background, while grab cut only needs to provide pixel set of background area. It can be thought of just selecting the target, then pixels outside the box will be considered as backgrounds [49,51]. New research shows that a super pixelated based algorithm can be used to avoid as figure node area of the image texture segmentation problem again [52]. In the article [53], it is mentioned that the complex assignment of digraph to Images is carried out. And then minimizing the edge weights using the minimum spanning tree.
3 Medical Image Segmentation Method Based on Mean Clustering
Medical image segmentation can be popularly understood as clustering by dividing pixels into homogeneous regions. Clustering methods belong to unsupervised machine learning methods. Different medical image segmentation has different requirements [54]. Cluster analysis has the characteristics of unsupervised, efficient, and self-adaptive. The image segmentation of clustering can be roughly classified into hard clustering and fuzzy clustering. A typical example of hard clustering is the K-means clustering image segmentation algorithm. And fuzzy clustering such as fuzzy C-means clustering. The improvement of the traditional K-means algorithm usually starts with how to select the optimal clustering center point, or combines with other medical treatment methods. Usually, morphological image processing technology or filtering and other operations are used to reduce the noise of the initial segmented image, and then K-means algorithm is used to segment the labeled area. Such as the optimized K-means clustering algorithm (FGO) based on football match, the four-stage fuzzy K-means algorithm based on deep learning, the intuition-based fuzzy clustering algorithm, the IFLICM clustering algorithm combining local space and local gray information, and other new technologies. These algorithms and methods are extensions of traditional clustering algorithms. The fuzzy C-means clustering method is more extensive and perfect in daily use by virtue of its simplicity and flexibility. For example, intuitionistic fuzzy C-means combines fuzzy entropy and hesitancy factor. Based on the original fuzzy C-means, local information with intuitionistic and non-central characteristics is added to the objective function term. These new algorithms, k-means, solve the problems that traditional clustering algorithms cannot solve, such as the pixel arrangement in the clustering space and the uncertainty of the preprocessed image boundary, the quality index of the segmented image and the processing time.
3.1 Methods Based on K-Means Clustering
The choice of K value in K-means algorithm will greatly affect the segmentation effect. Improper selection of K value will lead to over-segmentation or under-segmentation. Many scholars and researchers put forward different optimization methods after many experiments. Combined with spatial region information or other segmentation algorithms, it can solve the problems of fuzzy edges, inaccurate gray distribution, and local optimal solutions. Table 2 shows the differences between the five optimized K-means algorithms discussed in this article and the traditional K-means.

As the simplest unsupervised learning method, K-means clusters the data by calculating the average intensity of each category [6]. K-means clustering is popular due to its simplicity and minimal execution time.
Inspired by deep learning and natural simulation algorithms for football game optimization, in the paper [55], the author applied the optimized K-means clustering algorithm based on football match (FGO) to multi-stage segmentation of medical images. FGO is a crowd-based optimization method that mimics the behavior of soccer players. Firstly, the median filter is used to remove the noise points from medical images. It is transformed into the color space formed by convolution of L, A and B, and the last two layers are segmented by the optimized FGO algorithm to obtain the global optimal clustering, which solves the problem of classifying the pixels in the preprocessed image into the clustering space. Four indicators, namely Sensitivity, Specificity, Jaccard index and Dice Coefficients, were used for evaluation. Experimental studies clearly show that the proposed method for medical image segmentation is more effective than existing methods, especially in noisy environments. In order to avoid the problem of local optimality in K-means, the cuckoo search optimization (CSO) algorithm can be used to initialize the centroid [57].
Mix different segmentation techniques provides more possibilities. The author combined median filtering, K-means clustering, Sobel edge detection and morphological write operation to segment medical images from magnetic resonance imaging (MRI) and computed tomography (CT) under different imaging modes [6]. Firstly, a popular nonlinear filter, median filter, is selected as an important preprocessing step. Its function is to remove the pulse noise in the image while preserving the edge of the image, to improve the quality of image. Secondly, K-means clustering subdivides the image into various components. Sobel edge detection method can obtain high contrast binary mask of segmented image. Morphologic operations were performed to extract brain tumor regions. Finally, in the performance evaluation phase, detailed experiments were carried out on two brain image datasets with different modes through an OVA test, correlation coefficient, segmentation accuracy and execution time. The segmentation accuracy of CT scan imaging method is 95%, and the minimum accuracy of MRI brain image method is 64%. In addition, the shortest execution time of CT scan image reached 0.976 s. But there is still room for improvement, mainly on two fronts:
1) Research methods can be applied to a larger database.
2) Expand future research methods.
K-means clustering and discrete wavelet transform are also a feasible method for medical image segmentation. After the image is acquired and normalized, the gray image or color image after binarization is further eliminated by the color threshold technology. The region of interest is separated from the specified background by K-means algorithm, and then the signal is decomposed by a pair of low-pass and high-pass filters in discrete wavelet transform [51]. Sumithra et al. [7] developed the fuzzy K-means algorithm combining wavelet and ROI for tumor image Segmentation which achieved satisfactory results, with accuracy up to 87.8%. Based on K-means segmentation, detailed segmentation is carried out on the image. Islam et al., in article [58,59], gave the answer on how to accurately segment the image, and combined K-means, denoising factor and Canny edge detector to distinguish the edges. The probability rand index was used to qualitatively evaluate the image index of 10 different grades. When the image index was I8, the best result of 0.8556 was obtained. In addition to this method, another method which was mentioned in [60] is the fusion of the commonly used Sobel edge operator and Canny edge operator to extract the boundary. Similarly, the authors proposed the combination of K-means, threshold, and morphology [61]. Different from what was mentioned above, the method listed in this paper [62] uses K-means as the intermediate step before threshold, taking steps to remove non-brain tissue. Experimental tests on BRATS dataset with high-resolution and low-resolution images show that good Dice scores are achieved on both high-resolution and low-resolution images.
In order to overcome the limitations of the standard K-means algorithm, such as random initialization of cluster centroid and noise sensitivity, Mehidi et al. proved through experiments that the combination of Darwinian particle swarm optimization technology, K-means algorithm and morphological reconstruction operation could segment images more effectively [56]. The main idea of particle optimization algorithm is that the first particle is generated from the best position in the parameter space of itself, and the second is generated from the whole population. Running multiple parallel PSO algorithms on the same optimization problem is the main idea of DPSO algorithm. The initial clustering center was obtained by DPSO algorithm, and then the image was segmented by K-means algorithm. Finally, the tumor shape was separated by magnetic resonance imaging on the previous step image. Finally, the tumor shape was separated by magnetic resonance imaging of the previous image. Through experimental calculation, accuracy, specificity, precision, sensitivity reached 99.88, 0.9992, 0.9123, and 0.9502, which also indicated that the method could produce a more compact cluster.
Deep learning moves forward in current medical research. Using computers to undertake a large quantity of work of calculation greatly improved the work efficiency. A four-stage fuzzy K-means clustering method based on automatic deep learning is proposed for brain tumor segmentation [63]. In the pretreatment stage, wiener filter was used to eliminate the noise of MRI images. The preprocessed images are used as input to extract important features through CSOA algorithm. Artificial neural network is applied to classify the images, and fuzzy K-means algorithm makes an approach to segment the abnormal images. Experimental results clearly show that the maximum accuracy of the proposed method is up to 94%. Different algorithms are prospects for segmentation of various types of disease images in the future. The performance of the model is evaluated by using a confusion matrix that can visually show the real category and the category judgment predicted by the classification model, where the rows of the matrix represent the real value and the columns of the matrix represent the predicted value. TN indicates that the negative class is predicted as the number of negative classes, that is when the truth is 1 and the prediction is also 1. FN denotes the number of positive classes predicted to be negative, that is, the case where the truth is 0 and the prediction is 0. TP indicates the positive class is predicted as the number of positive classes, that is, the true is 0 and the prediction is also 0. FP represents the case where the negative class is predicted as the number of positive classes, that is, the true is 1 and the prediction is 0 [64]. Because the performance of a model cannot be evaluated by only one or two aspects, especially when the data category is not balanced; The authors’ team binarized the predicted values of the model into masks to evaluate the performance of the segmentation algorithm. There are three methods used: recall, specificity, and accuracy. Among them, recall represents the proportion of correct prediction among all results whose true value is positive. Specificity indicates the specific proportion of a specific result whose true value is negative. Accuracy indicates the percentage of the total predicted value in which all predictions are correct.
The method mentioned in article [65] has similar works, which combines fully convolutional neural network, fully connected conditional random field (DenseCRF) and K-means algorithm to optimize the existing medical image segmentation technology. In general, CNN is first segmented by fully convolutional neural network, and then the cascaded FCN fusion model composed of DenseCRF and K-means is used to refine and supplement the segmentation results. How does DenseCRF refine the results? The team worked out that the space could be smoothed by combining unitary potential and pairwise potential, and more segmentation edge details could be obtained by removing some isolated small regions. Consciously, the results obtained by FCN algorithm are after several iterations of training and learning, while the results obtained by K-means are those with traditional features. The researchers thought that better segmentation results could be obtained by comparing and fusing the two, so they used the bounding box based clipped area of the input image as the input of K-means, and predicted and voted the category of the pixel of the corresponding position in the segmentation results of FCN and K-means.
3.2 Methods Based on Intuitionistic Fuzzy C-Means Clustering
Intuition-based fuzzy clustering is an extension of fuzzy C-mean [40], one of whose goals is to suppress the noise in image segmentation. Intuitionistic fuzzy C-mean (IFCM) is a clustering technique combining fuzzy entropy and hesitation factor. As listed in the paper [66], the authors propose a method called the credibilistic intuitionistic fuzzy C-means (CIFCM) based on intuitionistic fuzzy sets. The proposed new clustering technique was validated on simulated and real MRI and CT brain images. In order to test its performance, three simulation experiments are done. The first was to test the segmentation of images from Brain Web into white matter, gray matter, background, and cerebrospinal fluid. The proposed CIFCM is compared with IFCM and FCM, and the segmentation result of CIFCM is closer to the real value. The second experiment is to segment the same image which is interfered by mixed noise. CIFCM shows good performance of suppressing noise. In the last simulation experiment, CIFCM not only detected the tumor area but also was close to the area existing in the ground truth image. In the paper [67], considering the fuzziness of MR brain image and the intuitive property of MR image——complex data classification, the author’s research team not only added spatial constraints in the defined pixel-cluster similarity which replaced the Euclidean distance between pixel and cluster center, but also endowed it with intuitionistic fuzzy property. On this basis, by adding a local information term to the objective function term, which is intuitive and has no center, the robustness to MR image noise gets improved. In a word, the local information of each pixel is fused in the whole clustering process, which effectively realizes the segmentation of medical images. Finally, they proposed an intuitive acentric fuzzy C-means clustering method (ICFFCM) for MR brain image segmentation. In the study, they use the average weighted pixel-to-pixel similarity to construct new pixel-to-cluster similarity measures. Intuitionistic fuzzy attributes are embedded in the defined pixel-clustering similarity to deal with the fuzziness in MR brain images and the uncertainty in clustering process. Spatial constraints were added to suppress the effect of noise in MR brain images. In order to verify the effectiveness and robustness of the proposed method, qualitative and quantitative tests were performed on Brain Web simulated Brain database and real MR Brain image dataset respectively. Although smooth, fuzzy, and intuitionistic fuzzy clustering methods are mostly adopted in the literature to deal with noise during segmentation, important structural information, such as edges or other details, is ignored. The image affected by noise is difficult to be segmented correctly, so one way to solve this problem is to add spatial information coding in the process of segmentation. The authors adopted IFCM (IFCMSNI) method [68] based on spatial domain information, new spatial regularization term based on domain membership value and intuitionistic fuzzy set theory to deal with the noise problem in medical image segmentation, and retain the structural information which is lost because of smoothing in the image.
In order to deal with the problems such as noise and bias field effect in the process of MRI image segmentation, the spatial domain information is applied to the bias corrected intuitionistic fuzzy C-mean based on intuitionistic fuzzy set theory [69]. The direct fuzzy set can be used to represent the non-membership value, blur, and membership value of the image [70–72]. In both papers, Sugeno’s negation function is used to visually blur the image. Experiments were conducted on publicly available MRI image datasets, and quantitative performance indexes such as dice score (DS) and average Segmentation accuracy (ASA) were used to compare the proposed method with existing methods. In addition to the above methods for improving the performance of medical image segmentation, it is mentioned in paper [73] that the genetic algorithm can be used to select the optimal parameters in the clustering algorithm. In many studies, Euclidean distance is usually used as an intuitive fuzzy C-means clustering distance metric, but in paper [74], the authors added kernel function based on IFCM algorithm and replaced Euclidean distance with kernel distance. This reduces the number of iterations in the experiment but at the expense of the length of each iteration. This is also the direction of future research efforts.
As the performance of classical algorithms in medical image segmentation cannot meet the ideal requirements [75], FCM is a very common medical image segmentation method, but for images with noise interference, excessive segmentation will cause [76].
When medical images obtained by different scanners or different scanning protocols are often polluted by noise or interfered by outliers, which increases the uncertainty of the boundary among different tissues. The quality of medical images may vary greatly, leading to unsatisfactory segmentation results and bringing great challenges to medical image segmentation [54]. In order to solve this problem, in the paper [54], the author studied the segmentation of medical images under noise scenes. The novelty of the study lies in introducing the idea of migration into the clustering method. They proposed a negate-transfer-resistant Fuzzy clustering model with a shared cross-domain transfer Latent space (called) NTR-FC-SCT), which integrates anti-negative transfer and maximum mean difference (MMD). The focus of the researchers’ study is how to adopt the transfer matching scheme based on cluster center.
To solve the problem of inconsistent cluster number between source domain and target domain. Transfer learning plays an important role in fuzzy clustering framework to deal with the distribution, feature space or task difference between source domain and target domain. The maximum mean difference (MMD) inspired an in-depth study of the transmission capability of each cluster belonging to the source domain in the shared potential space, which contributes to transmit knowledge across different domains. Motivation is shown in Fig. 1. The black triangle and black circle presented by the two cluster centers shown in the figure have a positive shift, and the black rectangle presented by the cluster center has a negative impact on the clustering in the target domain. The negative transfer resistance mechanism is used to reject the black rectangles in the target domain automatically. In Fig. 2, the schematic diagram of NTR-FC-SCT in a migration learning scenario with noise interference, compute cluster centers with sound theory and rigorous procedures, using cluster centers in the source domain as auxiliary knowledge. This model has achieved the best performance in all brain CT data sets, but how to improve the model processing speed remains to be further studied.

Figure 1: The motivation of the proposed method

Figure 2: The schematic diagram of NTR-FC-SCT
Since traditional fuzzy C-means clustering cannot abstain ideal segmentation results. In the paper [77], the authors proposed an improved brain image segmentation method based on both Gaussian filtering and FCM clustering algorithm. Gaussian filter, which is a linear smoothing filter, is to overcome the influence of noise. The initial clustering center is obtained from gray histogram. T1-weighted MR images with slice thickness of 1 mm and dimension of 181 × 217 × 181 were used in the study. In the experiment, by comparing the traditional FCM and the improved new method, it is intuitively shown that the improved new clustering method can distinguish the details of the noisy image. With the increase of the noise level, the segmentation result of FCM decreases sharply. Two functions were used to evaluate the clustering performance: the area overlap measure AOM and the misclassification error ME. The criteria for the best cluster are the maximum AOM value or the minimum ME value. After comparison with FCM, the AOM value of IFCM is 0.95295, the ME value is 0.09595, and the FCM is 0.87771 and 0.23873, respectively.
Dynamic correlation analysis can better control the precise correlation between pixels. Membership is used to measure correlation rather than image features, thus avoiding segmentation difficulties caused by noise pollution. In the paper [78], the authors propose a dynamic correlation model. Compared with the traditional fuzzy clustering algorithm, the proposed model measures the dynamic pixel correlation model and avoids the influence caused by inaccurate features in noisy images. In this model, pixel correlation degree is determined by the membership degree of related pixels to the corresponding cluster, and has nothing to do with the image features. In addition, the authors proposed an algorithm, DRFLICM (FLICM with Dynamic Relatedness), which was not affected by noise characteristics and used similarity among members to measure damping.
When the classical FCM algorithm cannot directly process all the data sets in the multi-task scenario simultaneously, it is usually difficult to find the common information of related tasks by clustering the data of different tasks independently. Zhao et al. [75] proposed a distributed multi-task fuzzy C-means (MT-FCM) clustering algorithm for MR brain image segmentation based on public and personal information of different tasks. The workflow of distributed MT-FCM algorithm is shown in Fig. 3. The negative effects of noise data in MR brain images can be avoided by effectively utilizing the common information in different but related MR brain image segmentation tasks. Each MRI data set consisting of 256 image slices was used as the input of the clustering algorithm. The consumption of clustering time is reduced and the common information of related tasks is effectively used to improve the performance of clustering. But one of the limitations is that it takes more run time to adjust regularization parameters.

Figure 3: Workflow of distributed MT-FCM algorithm
The main challenges in the segmentation of different brain tissue regions in brain magnetic resonance (MR) images are limited spatial resolution, signal-to-noise ratio (SNR) and rf coil heterogeneity. In the paper [79], the authors proposed an entropy-based FCM segmentation method. The non-Euclidean distance is defined based on gaussian probability density function, and the uncertainty of single pixel classification is combined with the classical FCM framework. Specifically, the fuzzy C-mean objective function uses Shannon entropy measure to correct the uncertainty between pixels. And a square window is used to calculate pixel reflexes and averages. Fig. 4 shows the process of the whole algorithm. The algorithm was validated on both simulated and real patient MRI images and required 30–35 iterations to converge.

Figure 4: Proposed framework for entropy-based fuzzy C-means clustering algorithm
Kollem et al. [80] designed an efficient total variational denoising method based on partial differential equations and a probabilistic fuzzy C-mean clustering segmentation algorithm. In order to better analyze the proposed method, the authors accessed MRI brain tumor medical images from the BRT TS2018 database. This method includes two stages. In the pretreatment stage, as shown in Fig. 5, the wavelet transform was used for image noise and sampling contour decomposition. Besides, the improved method was applied for noise image enhancement processing. In the segmentation stage, the improved possibility mean fuzzy C-mean algorithm is used to segment the preprocessed image. The final experimental data showed that the peak SNR reached 78.0001 dB, the mean square error reached 0.010336%, the sensitivity reached 99.999%, the specificity reached 100%, and the accuracy reached 99.999%.

Figure 5: Pipeline of the proposed denoising and segmentation methods
An optimized fuzzy clustering image segmentation algorithm is proposed [81], which adopts graph decomposition and region combination. The optimized clustering method is verified from two aspects of algorithm and objective index, and it is obvious that the algorithm has great advantages over F_stl algorithm and SCG algorithm in terms of running time and image segmentation accuracy. While gaining advantages, defects are also introduced, for example, the determination of threshold value increases the space of distance.
The study in [82] tried to quicken fuzzy C-means clustering based on super pixels (SPFFCM) segmentation method which is used for skin lesion segmentation in image processing. The input color image is segmented into super pixel regions, and the gradient image is generated by Sobel operator by calculating the histogram of the image. The results of the preliminary study were evaluated using six indicators, namely accuracy, precision, F-measure, Matthews correlation coefficient, sensitivity, specificity, Dice index, and Jaccard index. Compared with super pixel segmentation (SPS), the experimental results show that the fast fuzzy C-means method based on super pixel has the advantages of fast calculation speed, weak sensitivity to parameter changes, and strong adaptability to obtain local and global spatial information.
Halder’s team proposed a new clustering technique combining rough set and spatially oriented fuzzy C-means clustering (SKFCM) [83]. One of the advantages of this algorithm is that the higher and lower bounds ensure that the uncertainty of data belonging to multiple clusters is eliminated.
Semi-supervised clustering is the incorporation of a small amount of prior knowledge into the clustering process, in the paper [84], the authors proposed a semi-supervised clustering algorithm called seed fuzzy C-means (SFC-means), which enables elements to be placed in a group with a higher degree of certainty. It uses the correlation and clustering threshold derived from fuzzy logic to group image pixels, and ensures as far as possible that samples previously labeled as different categories are not assigned to the same cluster. In the clustering process, SFC-means arranged the membership degree of input samples of each seed calculated in descending order in the membership degree vector. The performance of the proposed algorithm is verified on 2200 images of different types, and the results show that the proposed algorithm can maintain good segmentation accuracy even when different types of images are used and thresholds are changed.
On the other hand, the authors believed that image intensity and spatial characteristic information should be fully considered in MRI image segmentation [85], so a new FCM formula is designed and regularization method is introduced to deal with images with uneven intensity and noise. Compared with the traditional FCM algorithm, the similarity coefficient of dice (DSC) reaches 92.56%. In the paper [86], the author proposes a fuzzy C-mean (RFRBSFCM) clustering method based on bounded support degree in rough fuzzy region. The rough fuzzy regions are determined according to local and global spatial information, and the membership degree is balanced by estimating adaptive parameters to minimize the objective function. The proposed RFRBSFCM algorithm can estimate the spatial constraints adaptively, and eliminate the extreme light and darkness at the boundary of the region, and achieve accurate separation of gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) in MRI brain images.
Image segmentation is not limited to local information. The author proposes an improved no-parameter fuzzy clustering algorithm NLFCM. In the fuzzy factor, the influence of adjacent pixels on the center pixel is called the damping range [87]. The algorithm measures the damping degree by introducing pixel correlation into the fuzzy factor, and uses non-local information to guide the image segmentation process. The brain images were divided into background, cerebrospinal fluid (CSF), gray matter (GRY) and white matter (WHT). Experimental results show that NLFCM segmentation algorithm can accurately segment the corresponding tissue. Specifically, the segmentation accuracy of CSF, GRY, and WHT in MRI brain images damaged by 30% Rician noise was 94.59%, 92.88%, and 94.88%, respectively. On the other hand, in the paper [88], the authors also proposed an improved algorithm based on non-local information and fuzzy clustering to study pixel correlation based on facet similarity, aiming to use more information to guide image segmentation. Based on a given pixel, an image block centered on the given pixel is constructed, and the correlation of pixels is measured by the similarity of image blocks. Experiments on medical images show that this algorithm is better than traditional FCM and FLICM.
3.2.2 Fuzzy Local Intensity Clustering
Fuzzy local information C-means clustering algorithm (FLICM) introduced a fuzzy factor as fuzzy local similarity measure. Although the damping degree of adjacent pixels on the central pixel achieves convergence of the membership degree of pixels in local Windows to similar values, the accuracy of estimating the damping degree of adjacent pixels is relatively low, and the FLICM algorithm only considers local information, which leads to unsatisfactory performance in high-noise images [86]. If non-local information is embedded in FLICM algorithm, a new algorithm will be generated. The new algorithm has both local information and non-local information, but it wrongly amplifies the importance of domain information when calculating the distance between pixel and cluster center. In order to solve this problem, the authors [8,89] believed that the distance measure of weights obtained through continuous iterative self-learning can be embedded into the objective function of fuzzy clustering algorithm. When the correlation between pixels is calculated, the non-local information of pixels is used, and the original information and local information of pixels are used to calculate the distance between pixels and the cluster center. Experiments also prove that this method is effective in complex noise images. Compared with FLICM algorithm, ACC reached 0.9480, SEN reached 0.9221, SPE reached 0.9610. It can be said that after the improvement, the segmentation performance of the original image has made greater progress.
In order to overcome the shortcomings of the existing fast generalized fuzzy C-means clustering algorithm, the robustness of image segmentation is improved. In the paper [88], the author used the improved IFLICM clustering algorithm which combined local space and local gray information to automatically detect and segment tuberculosis bacilli in images. IFLICM clustering directly uses the original sputum image for processing, thus avoiding the loss of details that may result from image preprocessing. The flow chart of IFLICM clustering algorithm in the process of image segmentation is shown in Fig. 6. Remove noise data by developing an appropriate objective function to capture bacterial features more accurately. According to the data of experimental results, IFLICM algorithm has high efficiency, anti-noise and feasibility, and the average segmentation accuracy of sputum image data set is 96.05. The data set used in this experiment is ZNSM iDB which is a repository of 350 sputum images. The results of the experiment used peak signal-to-noise ratio (PSNR) and precision value to measure the performance of different clustering algorithms and IFLICM clustering algorithms. Compared with other background blur-based systems, parameters such as noise are reduced to the maximum extent. The FLIC model performed tissue segmentation on noise images with Dice similarity coefficients of 0%, 3%, and 5%, respectively. When the number of slices was fixed to 90, the corresponding optimal results were 0.868, 0.864, and 0.837, respectively. Similarly, in order to improve the robustness of the segmentation algorithm to images with noise and intensity inhomogeneity, the global membership function is kept unchanged based on fuzzy C-means, and the local membership function with spatial constraints is introduced [90]. Qualitative and quantitative analyses of simulated 3D brain images with high noise and intensity inhomogeneity and real patient brain images have been carried out and satisfactory results have been obtained.

Figure 6: The flow chart of IFLICM clustering algorithm
In order to alleviate the influence of gray level inhomogeneity and noise pollution, in the paper [91], the authors proposed a fuzzy local intensity clustering algorithm (FLIC) based on the combination of level set algorithm and fuzzy clustering. Automatic and simultaneous segmentation and deviation correction are realized by using local clustering and local entropy. The flow chart of medical image segmentation method is shown in Fig. 7. It is mainly divided into two stages. In the first stage, various medical images with noise pollution are segmented as input. In the second stage, the output obtained in the first stage is taken as the initial contour, and the approximate fuzzy segmentation is completed through the level set method to further realize the automatic and accurate segmentation of the image. Experimental results show that the proposed algorithm is superior to other advanced segmentation algorithms in terms of accuracy, robustness, and computation time.

Figure 7: The proposed method flowchart
3.2.3 Fuzzy Clustering Based on Spatial Information
Introducing spatial information to optimize the objective function is beneficial to improve the noise resistance of the model. The introduction of non-local spatial information and local spatial information provides more information for image segmentation. Based on many experiments, many researchers have improved the segmentation effect by introducing prior probability and membership penalty terms. As listed in the paper [92], the author integrated different forms of spatial information into the traditional clustering algorithm. In order to consider the distance between adjacent pixels, the authors sum the standard FCM function and the optimized FCM function together to improve the objective function. This method is implemented by weighting the membership degree in the objective function to the residual of the complement of the local membership function. In paper [93], the spatial weight information is calculated by fusing spatial adjacent and similar super-pixels, and the crow search algorithm is used to optimize the performance. This provides a better solution for suspicious lesions or organ segmentation. Firstly, Daubechies wavelet transform with perfect reconstruction characteristics and symmetry is applied to each input image, and the phase information of the input image is saved, and the wavelet characteristics are obtained. Then, the super-pixels corresponding to the medical image is used to search its adjacent super-pixel. After that, the membership degree of each super-pixel and cluster center is iteratively updated until the maximum evolution period is met.
The authors put forward a kind of context based on the reliability of the space of fuzzy C-means (RSFCM) used for image segmentation [94]. In order to make up for the insufficient noise sensitivity of the traditional FCM algorithm, RSFCM combines neighborhood correlation model [95] with reliability measure to reduce edge blur. The advantage of this algorithm is that it can automatically determine the local similarity measure according to the gray relation between the local space and the central pixel and make full use of the local information in the neighborhood window, which can effectively suppress the heterogeneity of the objects within the class and improve the accuracy of image segmentation.
Introducing spatial information into fuzzy clustering algorithm can further improve the accuracy of segmentation of cerebral hematoma. In papers [96,97], authors proposed a kernel clustering segmentation algorithm based on spatial information intuitive fuzzy algorithm and kernel function algorithm. The neighborhood mean of the image is introduced into the objective function, the weight factor is introduced according to the spatial information, and the membership matrix [98] is modified by the location information of the hematoma. The algorithm is used to segment cerebral hemorrhage in brain CT images with skull structure removed. The data set is the brain CT images of 17 patients from Yuncheng Central Hospital, Shanxi Province, China. The algorithm not only improves the segmentation accuracy, but also avoids the error segmentation of the non-hematoma in the image whose gray value is similar to that of the hematoma into the cerebral hemorrhage region.
Due to the complexity of left ventricular geometry and heart movement, automatic segmentation of clinical cardiac PET images is challenging. The author proposes a novel approach to segmentation of left ventricular medical images [99]. They experimentally demonstrated that fitting the ellipsoid model into a three-dimensional cluster produced some effects by creating personalized myocardial models and segmental myocardium. In this study, the premise was that four clusters were assumed, including myocardium, left ventricle, non-cardiac region, and background. In the study, K-means (K-MS4) was adopted as well as in the fuzzy C-means (FCM4). The binary images of the myocardial clusters transformed by clustering were combined with the semi-ellipsoid model to generate personalized myocardial model. The segmentation results were evaluated in a normal group of 17 patients with normal cardiac perfusion and in an abnormal group of 20 patients with abnormal myocardial perfusion. The results showed that the difference in myocardial volume calculated by K-means and FCM4 was not very significant, while the difference in myocardial volume of abnormal group was larger when three-dimensional clustering was used. However, the only limitation is that the heart may be limited by the volume effect of the small LV cavity.
Computer-aided diagnostic automation, such as automatically finding the location of a tumor by automatically clicking on any MRI image, has been a huge help for neurosurgeons, and for this reason, the article [100] described a system for diagnosing brain tumors that Anina Miranda’s team successfully developed to diagnose brain tumors. On this basis, the MRI image is filtered by morphological related techniques. Compared to K-means, the newly developed system is a big step forward in terms of accuracy. The development of the system can be divided into five stages, including input brain MRI images, image preprocessing, image segmentation, execution of FCSPC algorithms to segment MRI brain images, output detected tumor regions. Systematically tested by a dataset from Kaggle, which covered 40 images with tumors, 7 images without tumors and 3 images with some noise. The test results well, the time complexity of the K-means algorithm is
As the plane-based soft clustering method can effectively process the data of non-spherical shape, for example, the material in human brain is non-spherical and the overlapping structure of brain tissue is uncertain [9]. Puneet Kumar’s team found that by adding local spatial information to Fuzzy K-plane clustering (FKPC), which belongs to one of the soft plane-based clustering algorithms, the noise in the image can be well processed. On the other hand, the core of the FKPC_S method proposed by the authors is to refine the membership values of noisy pixels in the way of neighboring pixel information, and the spatial regularization term added to the objective function of FKPC can effectively preserve information in the noisy environment. Through experiments on brain images with different levels of noise pollution (3%, 5%, 7% and 9%), the segmentation performance of FKPC_S proposed by the team decreases less than that of the existing 10 related methods under noise interference. The results were quantitatively analyzed by means of the average segmentation accuracy and dice score. The average segmentation accuracy of FKPC_S reached 0.9328, 0.7360, 0.6351, and 0.7680 when the image noise level was 3%, 5%, 7%, and 9%, respectively. The Dice score which is the evaluation of the quantitative segmentation performance of the synthetic image dataset damaged by Gaussian noise reaches the highest of 0.9738 and the lowest of 0.3256. Despite the advantages mentioned above, the shortcoming of this method is that it cannot automatically adjust the spatial regularization parameters. Manually adjusting the parameters means that more accurate segmentation results may not be obtained, and the whole calculation process is longer than FKPC. In addition, the application scope of FKPC_S method is limited, and it is only applicable to imaging data.
Due to the high noise environment, the existing intuitionistic fuzzy clustering algorithm cannot perform accurate segmentation of medical images. The authors explored a full Bregman divergence fuzzy clustering algorithm based on multi-local information constraints driven by intuitive fuzzy information, aiming to improve the anti-noise robustness and segmentation accuracy of intuitive fuzzy clustering correlation algorithm [101]. In order to further enhance the noise suppression, the algorithm constructs the total Bragg intuition of the divergence between clustering the center, rather than the current pixel and the square of the Euclidean distance, through the pixels in the field of normalized variance of the current pixel and clustering center Bregman divergence and discipline. The authors also proved the convergence of the algorithm strictly using Zangwill’s theorem. Fig. 8 shows the main framework of the algorithm. A local weighting coefficient is constructed and embedded into the objective function of intuitively fuzzy local information clustering. Then the segmentation accuracy and robustness of fuzzy clustering to noisy images are improved by adaptive modification of distance measure. The authors have carried out a lot of experiments on medical images, and the experimental results show that the algorithm can maintain strong anti-noise robustness in a high-noise environment. The segmentation performance of the proposed algorithm was compared with that of the existing intuitive fuzzy clustering correlation segmentation algorithm by ME, PSNR, ACC and P indexes.

Figure 8: Framework of the proposed method
Traditional medical image segmentation methods are difficult to accurately process images with weak edges and complex iterative processes. BIRCH (Balanced Iterative Reduction and Clustering Using Hierarchies) is a multi-stage clustering method using the clustering feature tree [102]. In the paper [102], the author proposed BIRCH (Balanced Iterative Reduction and Clustering Using Hierarchies) clustering method, and they believe that introducing a new energy function into the active contour model can make the contour curve complete target segmentation at the edge of the image. The flow chart of the specific method is shown in Fig. 9, which mainly covers four stages. Pixel gray values of medical images were input as sample data. Considering that adjacent pixels may belong to the same category; a new image was reconstructed according to the gray mean value of a small neighborhood of each pixel. In the study, different retinal vascular images were selected to validate and evaluate the performance of the proposed method. Experimental results show that the SEC value reaches 2687.3, the Entropy reaches 4.97, and the segmentation time reaches 2.7 s.

Figure 9: Flowchart of the proposed segment method
Non-local information and low-rank prior knowledge [103] are added to the framework of fuzzy clustering, as shown in Fig. 10, which roughly includes initialization of clustering centers, retrieval of relevant pixels and image segmentation. The predefined cluster number is very important for the fuzzy clustering algorithm. In the traditional fuzzy clustering algorithm, the cluster center is calculated by the intensity value and randomly initialized membership degree. But in the proposed framework, peak detection is used to initialize the cluster center. The authors considered it unreasonable to measure pixel correlation by the distance between image blocks, so they introduced weights in different directions into the model. When there were only a few pixels, the number of pixels in a cluster can be closely related to the grade of the image block. Low rank indicates many pixels in the same cluster, improving segmentation efficiency without degrading performance. Compared with FLICM algorithm, the algorithm which is called LRFCM proposed by the author replaced Euclidean distance measure by pixel correlation in the inter-pixel link. In the experiment, LRFCM is applied to medical image segmentation, including lung computed tomography (CT) image and brain magnetic resonance (MR) image. And compared with a typical FCM correlation algorithm, both VPC and VPE are indicators representing the fuzziness of segmentation results. A larger VPC and a larger VPE indicate the better segmentation. Segmentation accuracy is, in some ways, the ratio of correctly classified pixels. Therefore, accuracy (Acc), sensitivity (Sen) and specificity (Spe) were also used. Data show that LRFCM algorithm performs best in lung CT images with lobulation or burr. In order to test the robustness of the algorithm, 5% Rice noise was added to the images waiting to be segmented. The test results show that LRFCM algorithm is not only insensitive to medical images, but also retains the details of the images.
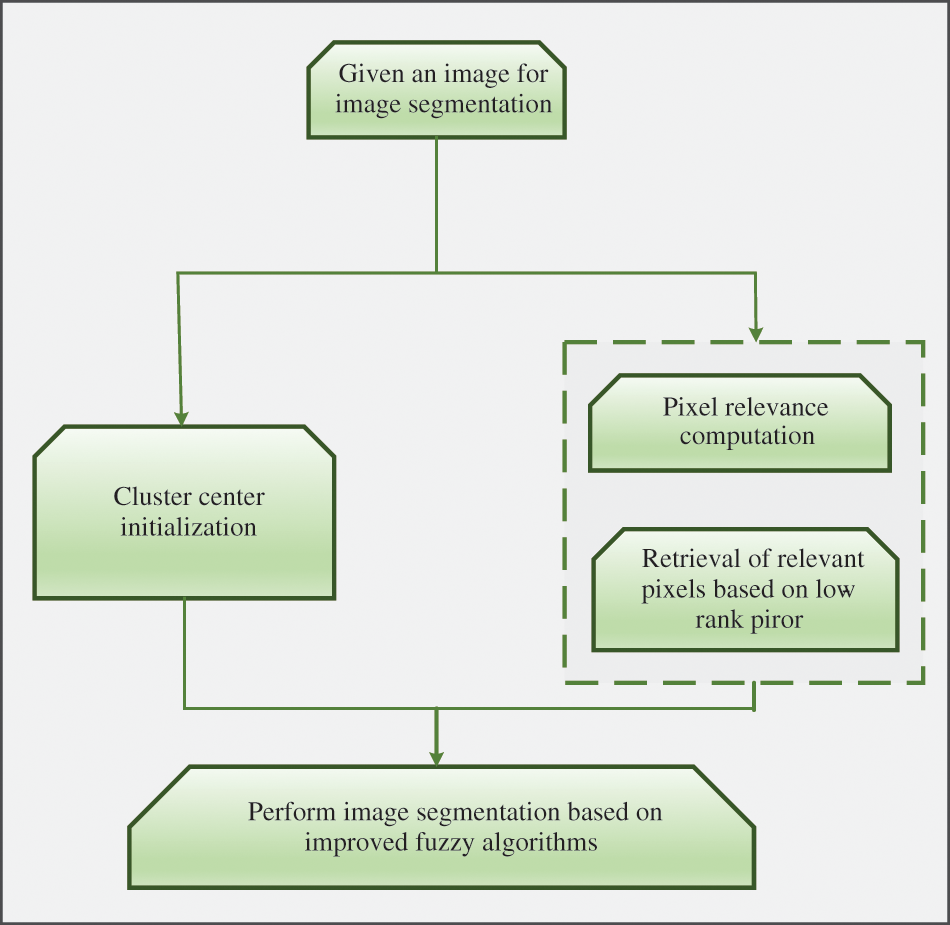
Figure 10: Pipeline of the proposed approach
In view of how to segment human MRI images more effectively, a grouping method combining two types of K-means and fuzzy C-means (FCM) is proposed. It uses FCM to cluster the K-means clustering results which were generated by four categories into three categories again, achieving the requirements of improved accuracy. In the paper [10], because K-means is more sensitive to color difference, it has a better effect in detecting optimal value and local outliers. However, the fly in the ointment is that different K-means clustering results may be obtained when the program is started at different times, and the accuracy rate is only about 56.45%. The four-stage experiment was based on 17 normal BRAIN MRI images and 45 brain images diagnosed with tumors. The brain MRI images were transformed into gray scale images and the images were clustered according to the color space. Considering that the intermediate value-based substitution process can change the pixel noise through the surrounding pixel values, the author chooses the median filter to merge the noise into the preestablished class population. And using morphological treatment to eliminate and detect brain tumors. The clustering process of FCM is to group the data according to the weight calculation from the data to the center of mass, so that the weight of the search process on FCM becomes lighter and the number of iterations decreases. The final experimental results also confirmed the author’s hypothesis that the clustering effect is better, the calculation process is lighter, and the accuracy of brain tumors detected by this method is 91.935%.
The above is the segmentation of unimodal images. In paper [104], the authors showed that hybrid positron emission tomography and computed tomography as complementary imaging methods combined with co-clustering algorithm can effectively achieve the segmentation of medical images containing 3D tumors. In order to achieve a consistent segmentation between two different single patterns, on the one hand, the research team added a specific contextual term to the framework of belief functions. On the other hand, Dempster’s combination rule is used to fuse the clustering results of the two single modes. The authors conducted experiments on images with and without texture features, and the Dice coefficient of the experimental results in the group with texture features increased from 0.83 ± 0.06 in the other group to 0.87 ± 0.04.
In addition to the mentioned method of combining K-means and C-means, Abraham’s research team proposed a region segmentation and clustering technology of K-region clustering (KRC) image segmentation method [105]. Initially, the image is divided into four regions, and the pixels with the same intensity value in each region are grouped into a cluster. The results in each adjacent region are then combined to form a new cluster according to the threshold. Fifty medical images were segmented, and the performance of the experimental results was tested with the help of quality indicators and processing time. Compared with the existing K-means clustering algorithm, watershed algorithm and regional growth algorithm, it was obvious that the image segmentation effect under KRC technology was better.
4 Medical Image Segmentation Method Based on U-Net Network
This section mainly focuses on the continuous optimization and improvement of medical image segmentation algorithms based on U-Net [106–108]. The symmetric structure of U-Net can make full use of the global and local details of the image. Compared with other networks used for medical image segmentation, the biggest advantage of U-Net is that it obtains better segmentation results when the training dataset is small. Therefore, U-Net has become the most popular network structure for medical image segmentation. Firstly, a 2.5D image segmentation method based on U-Net is discussed to achieve the balance of segmentation accuracy and computation. The automatic 3D model based on U-Net can realize automatic segmentation of 3D magnetic resonance images. A context nested network containing two U-Net structures. There are also full-size connections U-Net, RU-Net, VGG16 U-Net. Most of these algorithms rely on complex neural network architecture, and their performance has been greatly improved compared with the old algorithms, but there are still shortcomings in calculation cost, segmentation efficiency, application scope and other areas. Table 3 summarizes the benefits and performance of the main algorithms mentioned in this section.

The accuracy of U-Net in image segmentation under the framework of two-dimensional CNN needs to be improved, while 3D CNN requires a lot of computational costs and the convergence speed is slower than that of Two-dimensional U-Net. However, as listed in the paper [17], the authors proposed a 2.5D image segmentation method based on U-Net, achieving a balance between segmentation accuracy and computation. If 2D U-Net is used, it is easy to ignore the relationship between layers, while 3D U-Net requires more extra computation cost. This results in a compromise——2.5D U-Net, which is both efficient and makes full use of the spatial information in the data. The 2.5D model can be regarded as a special 2D model. In other words, the 2.5D model can be regarded as a 3-channel 2D convolution. In the preprocessing stage, in order to reduce the computational complexity, the 3D MRI image data are converted into two-dimensional surfaces in the three orthogonal directions of depth, width and height, which overcomes the limitations of 2D model in image segmentation. In contrast, the input to a 3D model is a 3D matrix. Then, the three two-dimensional U-shaped networks are used to correlate the two-dimensional surfaces. The U-Net framework consists of a contraction path which is used to capture context consisting of two 3 × 3 convolution and a symmetric expansion path where each step involves up-sampling the feature graph. Each convolution is followed by a ReLU function and a 2 × 2 maximum pooling operation. Although the experimental results show that the segmentation accuracy of 2.5D method is not as good as that of 3D U-Net, it is satisfactory in terms of computational efficiency. The 2.5D method outperforms the current 2D method on the test set. 2.5D U-Net is more suitable when there is little demand for segmentation accuracy and the calculation cost is limited. How to improve the segmentation accuracy will be the next research focus of the author. Kittipingdaja and his team [18] used a 2.5D ResU-Net that can integrate partial 3D information, balancing both accuracy and computational resources. This kind of network structure has obvious advantages, which can not only simplify the training but also increase the complexity of the model by adding more layers or feature channels. Taking the stack of an adjacent CT image slices as input, not only more content information can be extracted in the axial plane, but also additional context information can be provided in the orthogonal direction. In the model, all the convolutional layers share the same 3 × 3 kernel size, and the nonlinear activation function adopts the parametric rectified linear unit. As shown in Fig. 11, the problem of vanishing gradients is alleviated in the form of dense connections. To test and evaluate the performance of the model, the team selected slices of abdominal CT images at the time of intravenous contrast medium injection, where the kidneys were clearly visible. As the dataset, it achieves a high mean renal Dice score of at least 95%. In addition, the team specifically tested the model on abdominal CT images of four Thai patients, but the optimal average was only 87.60%.

Figure 11: The dense block structure
The rapid development of deep learning (DL) has been widely used in medical image segmentation. The author studied the automatic segmentation of human brain by Klaus Strum (CL) [109]. Although CL’s unique thin sheet structure, incomplete label and unbalanced data have brought challenges to the research, the author still proposed an automatic 3D segmentation model based on U-Net [19], which is called AM U-Net, and realized automatic segmentation of images in 3D-magnetic resonance images. In order to reduce the training data set, the three parts including pre-processing, segmentation and post-processing are optimized under the premise of ensuring the accuracy of the model. In this study, MRI data of 30 healthy adults aged between 21 and 35 were used for CL segmentation. In addition to preparing the data set, the pretreatment stage also included three sub-parts:
1) Sections were processed from 3DMRI.
2) Select the section of interest in the slice (ROI).
3) Use machine learning technology to normalize and enhance data operations.
Each MRI contains about 260 slices which are adjusted to have the same 256 × 256 pixel size. In order to ensure that only the slices containing CL are trained in the model, only 36 slices out of the 260 slices need to be left. The filtered slices are superimposed on the slices previously marked as areas of interest. The core of post-processing stage is to improve the accuracy of image segmentation.
Specifically, by removing false positives and again filtering out pixels that fell off from the region, annotated slices used for CL segmentation were finally stacked for 3D reconstruction. Dice scores are used to measure the similarity between the two samples on a scale of 0 to 1, with higher scores indicating better accuracy of the model. Joint crossing (IoU) [110] is an evaluation index used to measure the accuracy of image segmentation models. The MRI data set of 30 subjects was cross-validated by K-fold. During each run, the data set was divided into K equal parts and the model was trained. The process of which training was repeated for k times. The average IoU was 69%, 68% and 70%, respectively.
Three consecutive images were combined into a 2.5D slice image by means of spatial constraint information between adjacent slices of MRI brain image sequence. 2.5D section can not only make full use of the information of 3D brain section, but also directly reduce the training time. The authors proposed a Tripple U-Net based whole-brain segmentation method, which consists of a primary U-Net and two secondary U-Nets, with each of which consisting of encoding path and decoding path [111]. When training the network model, ShakeDrop regularization can be added to alleviate the overfitting problem [111]. As shown in Fig. 12, the multi-auxiliary mixed loss function based on binary cross entropy loss and dice loss is used to optimize the training model. In addition to this approach, the model can be optimized using Fourier entropy with a native RAV network and a stacked sparse autoencoder [112]. In order to overcome the dependency between sequential image slices, all images need to be input into the model independently. The network is about 0.55% higher than the two-dimensional U-Net model and 5.63% higher than the 3D Skull Strip model.

Figure 12: AM U-Net architecture
Many researchers have focused on how to accurately segment lesions or regions of interest from CT images, such as liver tumors, kidney tumors, cardiovascular diseases, choroid plexus [113] lymph nodes [114], etc. Wu et al. [115] designed an end-to-end [116,117] 3D U-Net model with a cascade structure. To ensure that the network retains more detailed features, the model increases the number of shrinking and expanding paths. It can effectively improve the segmentation accuracy of liver tumors from the aspects of feature fusion, multi-scale features and attention mechanism. To some extent, the key segmentation information can be grasped and the influence of useless information on subsequent segmentation can be reduced as much as possible [118]. The Dice Per case index was 0.665 and 0.719 after testing on LiTS and 3DIRCADb datasets. Accurate segmentation of the kidney in CT images can give medical professionals information about the shape and size of the kidney deformity. In paper [119], an automatic kidney segmentation method based on active contour and deep learning is proposed. The segmented regions of the kidney were extracted by active contour technology, followed by accurate segmentation of the tumor kidney by 3D U-Net. Similarly, a two-stage segmentation model [120,121] based on deep learning with attention mechanism can achieve the generation of the most accurate segmentation contour [122] in a short time (8.61 s). Accurate and automatic segmentation technology can make preoperative planning more convenient and reduce medical costs for doctors. Safer and more digital treatment options are available to patients [123].
In paper [124], the authors and their team proposed to complete tumor segmentation by using parts of images with similar values. The input medical images were standardized. In order to improve computational efficiency, the size of 3D MRI images was uniformly adjusted from 240 × 240 × 155 to 32 × 32 × 32. Using the hop architecture based on the U-Net architecture, the filter size is defined as 3 × 3 × 3, and ReLU is used as the activation function [125]. The accuracy of the model on the training data is 93.08%. The deep residual module can effectively solve the gradient degradation problem in the model training process [126]. The generalization ability of the model is enhanced by alleviating the imbalance of positive and negative samples in the data [127]. Although the construction of neural network structure is of great help for medical image segmentation, Liang et al. [128] believed that Vision Transformer with powerful remote information interaction capability can make up for the deficiency of convolutional network in modeling remote context interaction and spatial dependence. They construct a self-supervised learning framework to train the model encoder through the reconstruction task.
The segmentation of risk organs is very important in guiding radiotherapy treatment planning. The segmentation of organs at risk is decomposed into two stages or two sub-tasks. The boundary needs to be located first, and the organs within the bounding box need to be segmented second. Each task uses a 3D U-Net [129].
Full use of context information and feature representation can achieve more accurate segmentation of lesions. Hu et al. proposed a three-dimensional attention context U-net algorithm for MS lesion segmentation in paper [130]. The introduction of 3D context guidance module is helpful to expand the perceptual field and guidance context information. In the process of segmentation, the embedding of spatial attention blocks is needed. Leng et al. designed a context transmission module to make full use of context information. The context information is transmitted from four directions and the corresponding four context feature maps are generated, and then the context feature map containing rich information is finally obtained by combining with the input convolutional feature map. The experimental results of ISBI model of neuron structure segmentation show that the RAND error is only 0.0212 and the pixel error is only 0.0346 [131]. Fu et al. [132] proved through experiments that Attention gate block could be added to download jump connection part without adding a lot of calculations to increase the spatial extraction ability of vascular features. Furthermore, the combination of 1 × 1 × 1 convolution and softmax activation function in the decoding stage of the mapping at the output side ensures the ability of attention blocks in 3D space. The model was evaluated using the publicly available ISBI 2015 longitudinal MS lesion segmentation dataset. The model output was evaluated on the image mask manually delimited by the experts themselves, and the performance score reached 92.6.
In the paper [133], inspired by U-Net and its variants in medical image segmentation, the authors proposed a new architecture called Double U-Net which contains two U-Net architectures, hoping to achieve the goal of improving the overall segmentation performance of the network. This new architecture is a combination of improved U-Net and VGG-19. As a lightweight pre-training model, VGG-19 is easy to connect with U-Net due to its similar architecture, resulting in better output segmentation mask. Fig. 13 shows a brief overview of the architecture. Double U-Net selects VGG-19 as the encoder subnetwork, and the prediction mask (Output1) is generated after the image to be segmented input into the network. The input of the second network is multiplied by the prediction mask and the input image, finally generating another mask (Output2). The addition of ASPP helps to extract high-resolution feature images during image segmentation. Compared with U-Net, each piece in the decoder performs bilinear up-sampling of the input features, and finally generates a double U-Net mask of a convolution layer with a sigmoid activation function. The advantage of this method is that squeezing and excitation modules in the network can reduce redundant information, but there is still room for improvement in the output feature mapping of network 1. Validated on four different publicly available medical image datasets, Sørensen–dice Coefficient (DSC), mean Intersection over Union (mIoU), Precision, and Recall were used for evaluation. One of the major limitations of Double U-Net networks is the increased training time due to more parameters being used.

Figure 13: Overview diagram of the architecture
Rich multi-scale semantic information can generate more representative feature maps. It is crucial for segmenting the tissue structure of small images, such as blood vessel segmentation and eye diseases [134]. A two-channel encoder can not only capture more receiving fields, but also retain spatial information, increasing the ability of U-Net to acquire spatial and contextual information end-to-end [22]. In order to obtain semantically similar feature maps and obtain richer semantic information. The authors proposed a multi-scale nested U-Net (MSN U-Net), which aims to alleviate the semantic gap caused by simple jump join and improve the segmentation accuracy [135]. MSN network is mainly composed of Res block composed of dual convolutional layer and 1 × 1 bottleneck layer and multi-scale context fusion block combining the bottom layer and the top layer. To obtain more semantically similar feature maps without adding more parameters, they redesigned a new jump connection, which is composed of MSCF and dense connection. The 14352 selected CT images were divided into training dataset and validation dataset according to the ratio of 8:2. MSN network model achieved better evaluation index values than U-Net model. From the data point of view, the Dice value of U-Net is 0.9124
U-Net is a full convolutional network with simple structure and it is easy to study, as shown in Fig. 14. In the paper [136], the author proposed a method to segment the abdominal organ region using U-Net. 11824 full-cross-section CT images were collected from 99 patients, with 89 randomly selected for training and 10 U-Net tests. By analyzing the image dice coefficients obtained from each experiment, the average accuracy of segmentation is relatively high, reaching 0.96.

Figure 14: Brief diagram of U-Net structure
Based on U-Net, the author introduces a combined loss function which uses weighted binary cross entropy and dice loss function, and proposes a compression encoder which uses 4 × 4 maximum pooling operation which replaces the widely used 2 × 2 pooling layer and three-layer convolution layer [137]. A condensed network (CU-Net) consisting of contraction path composed of convolution, correction linear unit (ReLU), maximum pooling layer and extended path made up of convolution, convolution and ReLU is constructed. The advantage of this method ensures high amplitude activation. And a simplified network structure is necessary for feature extraction. The goal of the pre-processing stage is to provide the least noisy image for the network by normalizing the input image. The structure of the concentrated network is shown in Fig. 15. Different from U-Net, the author used the 5 × 5 convolution kernel instead of the 3 × 3 kernel of U-Net. Although only three levels of feature space were processed, the maximum pooling operation of 4 × 4 made up for the possible degradation of image feature quality. Experimental results show that standardizing intensity values during training, verification and testing is beneficial for better image detection and segmentation. The loss function in the proposed CU-Net architecture achieves high-quality segmentation results on cell images.

Figure 15: Condensed U-Net architecture
Qinghua Zhou’s team considered the deep stacked sparse autoencoder as a classifier for the model [23]. Influenced by the multi-classification model of support vector machine, a deep residual network based on U-Net structure is proposed based on U-Net convolutional neural network (CNN) and residual learning model. The depth of the network model structure is positively correlated with the samples required for training, but if the depth of the network model structure reaches a certain extent, the problem of gradient disappearance will be serious. And use U-Net structure and learn the depth of the residual as much as possible can solve the problem of the gradient disappeared which leads to the deep web cannot convergence. On the other hand, composite blocks are used as basic blocks to be integrated into the encoding and decoding structure of the original network, so the accuracy of image segmentation can be improved without deepening the network depth. The authors’ team is committed to multi-class classification and greater flexibility and accuracy, giving a three-branch architecture as shown in Fig. 16. The WT, ET, and TC in the figure represent the entire tumor, the enhanced tumor, and the tumor core, respectively. The purpose of the architecture is to simplify the complex three-class partitioning method into three binary partitions. On the other hand, Fig. 17 shows the structure of the combined convolution block in the proposed deep residual network. The data set used in the experiment was BRATS 2018, and the performance was evaluated using a quintuple cross-validation method. This network structure uses three two-class segmentation channels to identify multiple tumor regions and achieve efficient segmentation. Although the experimental results are satisfactory, there are still shortcomings, such as whether it can consider mixing convolution kernels of different sizes in the same convolution layer to improve the segmentation accuracy.

Figure 16: Overall composition of proposed three-branch structure model

Figure 17: Combination blocks of proposed network
In the paper [138], the author combined the idea of multi-scale convolution method and attention mechanism, optimized and integrated U-Net encoder, decoder, skipped connection as well as context transformation structure. The team proposed a U-Net which has multi-scale attention mechanism for liver organ segmentation in CT image, which is called MSA U-Net. MSA U-Net is mainly divided into four modules: encoder, decoder, context conversion and skip connection, as shown in Fig. 18. A structure called attention-shrinking space Pyramid (AASPP) is added to the end of the encoder. The Residual Attention Jump Module (RASM) is designed to reduce the semantic gap between encoder and decoder. On the other hand, embedding attention modules can further improve ASPP’s ability to capture context information. Experiments are carried out on public data sets and the data results are compared with some advanced semantic segmentation methods. The experimental results clearly show that the proposed method has a good segmentation effect. The data show that the dice correlation coefficient reaches 98.00% and the joint intersection reaches 96.08%.

Figure 18: Overview of MSA U-Net architecture
RU-Net is also a kind of medical image segmentation method based on the deep learning, the use of deep learning for medical image segmentation is not a new method, but to raise convolution model accuracy and to locate the boundary of the object in further digging deep learning network structure of medical image matching degree is still the important research aspects of scientific research personnel [139].
As listed in the paper [24], the authors proposed an RU-Net segmentation model which is made up of a U-Net-based segmentation network and a multi-level boundary detection network. Additionally, an image segmentation algorithm based on a deep learning model with a multi-layer boundary perception-self-attention mechanism is proposed in detail. In order to obtain global information from feature images and realize effective extraction of feature information from medical images, a new interactive self-attention module is specially introduced into the deep learning model. The author makes some modifications based on U-Net, adding feature layer elements to up-sampling symmetry layer, and improving the computational efficiency of network back propagation by using residual idea on certain level blocks. The formula for calculating characteristic residuals is as follows:
In this formula, l represents the layer feature input, l represents the feature input of the layer, and, li, j (⋅) represents the mapping model of the feature channel at the th position J.
The convolution kernel was set to the size of 3 × 3, and batch normalization and regularization were performed after each layer of convolution. Boundary-aware RU-Net network learned and realized the function of detecting organizational boundaries containing diseases. Ru-net can be regarded as a combination of two U-Nets, which respectively play the role of segmenting images and predicting the final segmentation [136,140]. An optimization algorithm is used to update model parameter values in the loss function. According to encoder-decoder, the overall structure of the whole interactive self-attention mechanism network model is divided, where the encoder part is VGG16 network structure, and the decoder takes the feature mapping of the convolution kernel as the input, which consists of the interactive self-attention module and the multi-scale horizontal output fusion module. Finally, the improved deep model is verified on IBSR dataset, JSRT dataset and lung cancer PET/CT dataset. It can avoid the occurrence of wrong segmentation to the maximum extent, and mainly solves the problem of boundary feature information extraction and adaptive extraction of medical images.
VGG 16 network is a deep convolutional neural network including convolutional layer, pooling layer, and fully connected layer. The combination of VGG 16 network structure and U-Net structure greatly improves the expressive power of the network. Ghosh et al. [141] proposed a new method which is an encoder-decoder neural network structure based on U-Net and VGG16. The decoder is a VGG16 network with three fully connected layers removed. On the other hand, the decoder is a similar architecture to the U-Net structure. The VGG16 network which consists of five sub-samples improves the capability of capturing features by deepening the depth of convolutional neural network. Dense convolution blocks can also be used in down sampling to balance the gradient vanishing problem [142] in convolution blocks. At the same time, there is a problem, which may lead to the loss of low-level features. To solve this problem, U-Net skip connection gives a satisfactory answer. The number of MN ultrasound image datasets used for testing and training was 682 and 209, respectively. The NVIDIA TITAN X graphics processing unit was used for testing and experimentation, and the Adam optimizer was used to train the model. By the way, in the papers [143–145], it is mentioned that the use of stacked sparse autoencoders as classifiers. Multiple data enhancement methods are used in handling overfitting problems.
Finally, Dice coefficient, Jaccard coefficient and Precision, which are commonly used in medical image segmentation task, were used to analyze the results. The values of the three indexes were 0.895 ± 0.036, 0.812 ± 0.057 and 0.899 ± 0.062, respectively. In contrast, U-Net results were 0.818 ± 0.081, 0.699 ± 0.112 and 0.728 ± 0.118, respectively. The segmentation effect of VGG16 U-Net is much better than the performance of pure U-Net.
This paper classifies and describes two types of medical image segmentation techniques: one is an improved clustering algorithm, and the other is the deep learning algorithm based on U-Net network. In the cluster analysis, the paper focused on effective and concise fuzzy clustering, which reflects the fuzzy theory on the membership degree of each pixel of the image to each cluster center. The difference in data depends on membership degrees. In order to achieve better performance of manual annotation and the target image, the U-Net algorithm serves as the catalyst for the development. One of the biggest advantages of U-Net is that it uses fewer data sets to achieve more accurate segmentation results. However, problems such as how to reduce U-Net’s dependence on high-quality label datasets without reducing its accuracy and how to compress network models without destroying their stability still need to be solved. Medical segmentation has a broad application prospect in biomedical engineering. Through the research and discussion of the team, thinking space is provided for the development of medical image segmentation technology. Hoping to reap a harvest for further improvement of the existing segmentation methods. It will help doctors to obtain more accurate pathological information and assist in disease diagnosis.
Funding Statement: This work was supported partly by the Open Project of State Key Laboratory of Millimeter Wave under Grant K202218, and partly by Innovation and Entrepreneurship Training Program of College Students under Grants 202210700006Y and 202210700005Z.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Gu, X. W., Chen, S. W., Zhu, H. S., Brown, M. (2022). COVID-19 imaging detection in the context of artificial intelligence and the internet of things. Computer Modeling in Engineering & Sciences, 132(2), 507–530. https://doi.org/10.32604/cmes.2022.018948 [Google Scholar] [CrossRef]
2. Despotovic, I., Goossens, B., Philips, W. (2015). MRI segmentation of the human brain: Challenges, methods, and applications. Computational and Mathematical Methods in Medicine, 2015, 450341. https://doi.org/10.1155/2015/450341 [Google Scholar] [PubMed] [CrossRef]
3. Chouhan, S., Kaul, A., Singh, U. (2018). Image segmentation using computational intelligence techniques: Review. Archives of Computational Methods in Engineering, 26(3), 533–596. https://doi.org/10.1007/s11831-018-9257-4 [Google Scholar] [CrossRef]
4. Ali, N. A., El Abbassi, A., Bouattane, O. (2022). Performance evaluation of spatial fuzzy C-means clustering algorithm on GPU for image segmentation. Multimedia Tools and Applications. https://doi.org/10.1007/s11042-022-13635-z [Google Scholar] [PubMed] [CrossRef]
5. Li, Y., Liu, X., Gong, X., Wang, M. (2021). A multi-view features hinged siamese U-net for image Co-segmentation. Multimedia Tools and Applications, 80(15), 22965–22985. https://doi.org/10.1007/s11042-020-08794-w [Google Scholar] [CrossRef]
6. Agrawal, R., Sharma, M., Singh, B. (2018). Segmentation of brain lesions in MRI and CT scan images: A hybrid approach using k-means clustering and image morphology. Journal of the Institution of Engineers (IndiaSeries B, 99, 173–180. https://doi.org/10.1007/s40031-018-0314-z [Google Scholar] [CrossRef]
7. Sumithra, M., Malathi, S. (2020). Integration of wavelet and ROI based fuzzy K-means for Mri-ct segmentation. 2020 Third International Conference on Smart Systems and Inventive Technology (ICSSIT), pp. 1031–1037. Tirunelveli, India, IEEE. https://doi.org/10.1109/ICSSIT48917.2020.9214145 [Google Scholar] [CrossRef]
8. Song, Q., Wu, C., Tian, X., Song, Y., Guo, X. (2022). A novel self-learning weighted fuzzy local information clustering algorithm integrating local and non-local spatial information for noise image segmentation. Applied Intelligence, 52(6), 6376–6397. https://doi.org/10.1007/s10489-021-02722-7 [Google Scholar] [CrossRef]
9. Kumar, P., Kumar, D., Agrawal, R. (2022). Fuzzy k-plane clustering method with local spatial information for segmentation of human brain MRI image. Neural Computing and Applications, 34, 4855–4874. https://doi.org/10.1007/s00521-021-06677-1 [Google Scholar] [CrossRef]
10. Almahfud, M. S., Sari, C., Setiadi, D., Rachmawanto, E. (2018). An effective MRI brain image segmentation using joint clustering (K-means and fuzzy C-means). 2018 International Seminar on Research of Information Technology and Intelligent Systems (ISRITI), pp. 11–16. Yogyakarta, Indonesia, IEEE. https://doi.org/10.1109/ISRITI.2018.8864326 [Google Scholar] [CrossRef]
11. Shelhamer, E., Long, J., Darrel, T. (2017). Fully convolutional networks for semantic segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 39(4), 640–651. https://doi.org/10.1109/TPAMI.2016.2572683 [Google Scholar] [PubMed] [CrossRef]
12. Chen, L. C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A. L. (2018). DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Transactions on Pattern Analysis and Machine Intelligence, 40(4), 834–848. https://doi.org/10.1109/TPAMI.2017.2699184 [Google Scholar] [PubMed] [CrossRef]
13. Badrinarayanan, V., Kendall, A., Cipolla, R. (2017). SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 39(12), 2481–2495. https://doi.org/10.1109/TPAMI.34 [Google Scholar] [CrossRef]
14. Chen, S., Hu, G., Sun, J. (2020). Medical image segmentation based on 3D U-net. 2020 19th International Symposium on Distributed Computing and Applications for Business Engineering and Science (DCABES), Xuzhou, China, 130–133. IEEE. https://doi.org/10.1109/DCABES50732.2020.00042 [Google Scholar] [CrossRef]
15. Song, Y., Yan, H. (2017). Image segmentation techniques overview. 2017 Asia Modelling Symposium (AMSpp. 103–107. Kota Kinabalu, Malaysia, IEEE. https://doi.org/10.1109/AMS.2017.24 [Google Scholar] [CrossRef]
16. Yu, L., Qin, Z., Ding, Y., Qin, Z. (2021). MIA-UNet: Multi-scale iterative aggregation U-network for retinal vessel segmentation. Computer Modeling in Engineering & Sciences, 129(2), 805–28. https://doi.org/10.32604/cmes.2021.017332 [Google Scholar] [CrossRef]
17. Hu, K., Liu, C., Yu, X., Zhang, J., He, Y. et al. (2018). A 2.5D cancer segmentation for MRI images based on U-Net. 2018 5th International Conference on Information Science and Control Engineering (ICISCE), Zhengzhou, China, pp. 6–10. https://doi.org/10.1109/ICISCE.2018.00011 [Google Scholar] [CrossRef]
18. Kittipongdaja, P., Siriborvornratanakul, T. (2022). Automatic kidney segmentation using 2.5D ResUNet and 2.5D DenseUNet for malignant potential analysis in complex renal cyst based on CT images. EURASIP Journal on Image and Video Processing, 2022(1), 5. https://doi.org/10.1186/s13640-022-00581-x [Google Scholar] [PubMed] [CrossRef]
19. Ravichandran, S. R., Nataraj, B., Huang, S., Qin, Z., Lu, Z. et al. (2019). 3D inception U-Net for aorta segmentation using computed tomography cardiac angiography. 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), pp. 1–4. Chicago, IL, USA. https://doi.org/10.1109/BHI.2019.8834582 [Google Scholar] [CrossRef]
20. Tie, J., Peng, H., Zhou, J. (2021). MRI brain tumor segmentation using 3D U-net with dense encoder blocks and residual decoder blocks. Computer Modeling in Engineering & Sciences, 128(2), 427–445. https://doi.org/10.32604/cmes.2021.014107 [Google Scholar] [CrossRef]
21. Chen, J., He, Z., Zhu, D., Hui, B., Yi, R. et al. (2022). MU-Net: Multi-path upsampling convolution network for medical image segmentation. Computer Modeling in Engineering & Sciences, 131(1), 73–95. https://doi.org/10.32604/cmes.2022.018565 [Google Scholar] [CrossRef]
22. Wang, B., Wang, S., Qiu, S., Wei, W., Wang, H. et al. (2021). CSU-Net: A context spatial U-Net for accurate blood vessel segmentation in fundus images. IEEE Journal of Biomedical and Health Informatics, 25(4), 1128–1138. https://doi.org/10.1109/JBHI.2020.3011178 [Google Scholar] [PubMed] [CrossRef]
23. Wang, S. H., Satapathy, S. C. (2022). Secondary pulmonary tuberculosis identification via pseudozernike moment and deep stacked sparse autoencoder. Journal of Grid Computing, 20(1). https://doi.org/10.1007/s10723-021-09596-6 [Google Scholar] [PubMed] [CrossRef]
24. Leclerc, S., Jodoin, P., Lovstakken, L., Bernard, O., Smistad, E. et al. (2019). RU-Net: A refining segmentation network for 2D echocardiography. 2019 IEEE International Ultrasonics Symposium (IUS), pp. 1160–1163. Glasgow, UK, IEEE. https://doi.org/10.1109/ULTSYM.2019.8926158 [Google Scholar] [CrossRef]
25. Ramkumar, M. O., Jayakumar, D., Yogesh, R. (2020). Multi Res U-Net based image segmentation of pulmonary tuberculosis using CT images. 2020 7th International Conference on Smart Structures and Systems (ICSSS), pp. 1–4. Chennai, India. https://doi.org/10.1109/ICSSS49621.2020.9202371 [Google Scholar] [CrossRef]
26. Seo, H., Huang, C., Bassenne, M., Xiao, R., Xing, L. (2020). Modified U-Net (mU-Net) with incorporation of object-dependent high level features for improved liver and liver-tumor segmentation in CT images. IEEE Transactions on Medical Imaging, 39(5), 1316–1325. https://doi.org/10.1109/TMI.42 [Google Scholar] [CrossRef]
27. Liang, K., Jiang, Y., Long, M., Liang, G. (2018). Adaptive dual threshold based moving target detection algorithm. 2018 IEEE 4th Information Technology and Mechatronics Engineering Conference (ITOEC), pp. 1111–1115. Chongqing, China. https://doi.org/10.1109/ITOEC.2018.8740402 [Google Scholar] [CrossRef]
28. Song, F., Tian, Y., Gao, X., Yang, S., Zheng, M. (2021). Research on medical image segmentation method. Big Data and Business Intelligence (MLBDBI), 577–580. https://doi.org/10.1109/MLBDBI54094.2021.00115 [Google Scholar] [CrossRef]
29. Zhang, Y., Xia, Z. (2018). Research on the image segmentation based on improved threshold extractions. 2018 IEEE 3rd International Conference on Cloud Computing and Internet of Things (CCIOT), pp. 386–389. Dalian, China. https://doi.org/10.1109/CCIOT45285.2018.9032505 [Google Scholar] [CrossRef]
30. Mohanarathinam, A. (2020). Enhanced image filtrationusing threshold based anisotropic filter for brain tumor image segmentation. 2020 3rd International Conference on Intelligent Sustainable Systems (ICISS), pp. 308–316. Thoothukudi, India. https://doi.org/10.1109/ICISS49785.2020.9315924 [Google Scholar] [CrossRef]
31. Xu, A., Wang, L., Feng, S., Qu, Y. (2010). Threshold-based level set method of image segmentation. 2010 Third International Conference on Intelligent Networks and Intelligent Systems, pp. 703–706. Shenyang, China. https://doi.org/10.1109/ICINIS.2010.181 [Google Scholar] [CrossRef]
32. Chithra, A., Renjen, R. R. U. (2018). Otsu's adaptive thresholding based segmentation for detection of lung nodules in CT image. 2018 2nd International Conference on Trends in Electronics and Informatics (ICOEI), pp. 1303–1307. Tirunelveli, India. https://doi.org/10.1109/ICOEI.2018.8553694 [Google Scholar] [CrossRef]
33. Wang, Y., Pan, Y., Yu, X. (2018). Image edge detection of medical cell based on morphology. 2018 Eighth International Conference on Instrumentation & Measurement, Computer, Communication and Control (IMCCC), pp. 1368–1372. Harbin, China. https://doi.org/10.1109/IMCCC.2018.00283 [Google Scholar] [CrossRef]
34. Schober, K., Prestin, J., Stasyuk, S. A. (2021). Edge detection with trigonometric polynomial shearlets. Advances in Computational Mathematics, 47(1). https://doi.org/10.1007/s10444-020-09838-3 [Google Scholar] [CrossRef]
35. Wan, L., Pan, D., Feng, T., Liu, W., Potapov, A. (2021). A review of dielectric optical metasurfaces for spatial differentiation and edge detection. Frontiers of Optoelectronic, 14(2), 187–200. https://doi.org/10.1007/s12200-021-1124-5 [Google Scholar] [PubMed] [CrossRef]
36. Kazi, I. M., Chowhan, S., Kulkarni, U. (2017). MRI brain image segmentation using adaptive thresholding and K-means algorithm. International Journal of Computer Applications, 167, 11–15. https://doi.org/10.5120/ijca2017914330 [Google Scholar] [CrossRef]
37. Ismail, Y. M., Yahya, A. A., Shayem, S. A., Mahmut, I., Tong, L. et al. (2018). Fuzzy C-means thresholding for a brain MRI image based on edge detection. 2018 IEEE 4th International Conference on Computer and Communications (ICCC), pp. 1562–1566. Chengdu, China. https://doi.org/10.1109/CompComm.2018.8780890 [Google Scholar] [CrossRef]
38. Hou, S., Jia, C., Li, K., Fan, L., Guo, J. et al. (2022). Edge detection of COVID-19 CT image based on GF_SSR, improved multiscale morphology, and adaptive threshold. Computer Modeling in Engineering & Sciences, 132(1), 81–94. https://doi.org/10.32604/cmes.2022.019006 [Google Scholar] [CrossRef]
39. Archa, S. P., Kumar, C. S. (2018). Segmentation of brain tumor in MRI images using CNN with edge detection. 2018 International Conference on Emerging Trends and Innovations in Engineering and Technological Research (ICETIETR), pp. 1–4. Ernakulam, India. https://doi.org/10.1109/ICETIETR.2018.8529081 [Google Scholar] [CrossRef]
40. Hua, X., Qian, J., Zhao, H., Liu, L., Wu, Y. (2018). Automatic intestinal canal segmentation based region growing with multi-scale entropy. 2018 IEEE 3rd International Conference on Image, Vision and Computing (ICIVC), pp. 273–277. Chongqing, China. https://doi.org/10.1109/ICIVC.2018.8492854 [Google Scholar] [CrossRef]
41. Vyavahare, A. J., Thool, R. C. (2012). Segmentation using region growing algorithm based on CLAHE for medical images. Fourth International Conference on Advances in Recent Technologies in Communication and Computing (ARTCom2012), pp. 182–185. Bangalore, India. https://doi.org/10.1049/cp.2012.2522 [Google Scholar] [CrossRef]
42. Angelina, S., Suresh, L. P., Veni, S. H. K. (2012). Image segmentation based on genetic algorithm for region growth and region merging. 2012 International Conference on Computing, Electronics and Electrical Technologies (ICCEET), pp. 970–974. Nagercoil, India. https://doi.org/10.1109/ICCEET.2012.6203833 [Google Scholar] [CrossRef]
43. Bresson, X., Vandergheynst, P., Thiran, J. (2003). A priori information in image segmentation: energy functional based on shape statistical model and image information. Proceedings 2003 International Conference on Image Processing (Cat. No. 03CH37429), pp. 425–428. Barcelona, Spain, IEEE. https://doi.org/10.1109/ICIP.2003.1247272 [Google Scholar] [CrossRef]
44. Cootes, T. F., Taylor, C. J. (1992). Active shape models–‘smart snakes’, pp. 266–275. London: Springer. https://doi.org/10.1007/978-1-4471-3201-1_28 [Google Scholar] [CrossRef]
45. Cootes, T. F., Edwards, G. J., Taylor, C., (2001). Active appearance models. pattern analysis and machine intelligence. IEEE Transactions, 23. 681–685. https://doi.org/10.1109/34.927467 [Google Scholar] [CrossRef]
46. Stanley, O., James, A. S. (1988). Fronts propagating with curvature-dependent speed: Algorithms based on hamilton-jacobi formulations. Journal of Computational Physics, 79(1), 12–49. https://doi.org/10.1016/0021-9991(88)90002-2 [Google Scholar] [CrossRef]
47. Chen, X., Pan, L. (2018). A survey of graph cuts/graph search based medical image segmentation. IEEE Reviews in Biomedical Engineering, 11, 112–124. https://doi.org/10.1109/RBME.2018.2798701 [Google Scholar] [PubMed] [CrossRef]
48. Li, Y., Zhang, J., Gao, P., Jiang, L., Chen, M. (2018). Grab cut image segmentation based on image region. 2018 IEEE 3rd International Conference on Image, Vision and Computing (ICIVC), pp. 311–315. Chongqing, China. https://doi.org/10.1109/ICIVC.2018.8492818 [Google Scholar] [CrossRef]
49. Rother, C., Kolmogorov, V., Blake, A. (2004). GrabCut: Interactive foreground extraction using iterated graph cuts. ACM Transactions on Graphics, 23(3), 309–314. https://doi.org/10.1145/1015706.1015720 [Google Scholar] [CrossRef]
50. Cao, Z., Zhang, X., Mei, X., (2008). Unsupervised segmentation for color image based on graph theory. 2008 Second International Symposium on Intelligent Information Technology Application, pp. 99–103. Shanghai, China, IEEE. https://doi.org/10.1109/IITA.2008.143 [Google Scholar] [CrossRef]
51. Deepa, K., Suganya, S. (2020). A novel approach of segmentation of cytomegalovirus image using K-means clustering and discrete wavelet transform. 2020 International Conference on Emerging Trends in Information Technology and Engineering (ic-ETITE). https://doi.org/10.1109/ic-ETITE47903.2020 [Google Scholar] [CrossRef]
52. Bogach, I. V., Lupiak, D. D., Ivanov, Y. Y., Stukach, O. V. (2019). Analysis and experimental research of modifications of the image segmentation method using graph theory. 2019 International Siberian Conference on Control and Communications (SIBCON), pp. 1–4. Tomsk, Russia. https://doi.org/10.1109/SIBCON.2019.8729659 [Google Scholar] [CrossRef]
53. Mamatha, S. K., Krishnappa, H. K., Shalini, N. (2022). Graph theory based segmentation of magnetic resonance images for brain tumor detection. Pattern Recognition and Image Analysis, 32(1), 153–161. https://doi.org/10.1134/S1054661821040167 [Google Scholar] [CrossRef]
54. Jiang, Y., Gu, X., Wu, D., Hang, W., Xue, J. et al. (2021). A novel negative-transfer-resistant fuzzy clustering model with a shared cross/domain transfer latent space and its application to brain CT image segmentation. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 18(1), 40–52. https://doi.org/10.1109/TCBB.2019.2963873 [Google Scholar] [PubMed] [CrossRef]
55. Abhiraj, T. K., Srilakshmi, K., Jayaraman, K., Jayaraman, S. (2021). Enhanced football game optimization-based K-means clustering for multi-level segmentation of medical images. Progress in Artificial Intelligence, 10(4), 517–28. https://doi.org/10.1007/s13748-021-00251-5 [Google Scholar] [CrossRef]
56. Mehidi, I., Chouaib Belkhiat, D. E. C., Jabri, D., (2019). An improved clustering method based on K-means algorithm for MRI brain tumor segmentation. 2019 6th International Conference on Image and Signal Processing and Their Applications (ISPA), pp. 1–6. Mostaganem, Algeria, IEEE. https://doi.org/10.1109/ISPA48434.2019.8966891 [Google Scholar] [CrossRef]
57. Arjmand, A., Meshgini, S., Afrouzian, R., Farzamnia, A., (2019). Breast tumor segmentation using K-means clustering and cuckoo search optimization. 2019 9th International Conference on Computer and Knowledge Engineering (ICCKE), pp. 305–308. Mashhad, Iran, IEEE. https://doi.org/10.1109/ICCKE48569.2019.8964794 [Google Scholar] [CrossRef]
58. Islam, M. Z., Nahar, S., Islam, S. M. S., Islam, S., Mukherjee, A. et al. (2021). Customized K-means clustering based color image segmentation measuring PRI. 2021 International Conference on Electronics, Communications and Information Technology (ICECIT), pp. 1–4. Khulna, Bangladesh, IEEE. https://doi.org/10.1109/ICECIT54077.2021.9641094 [Google Scholar] [CrossRef]
59. Bharathi, B. S., Swamy, K. V., (2020). Effective image segmentation using modified K-means technique. 2020 4th International Conference on Trends in Electronics and Informatics (ICOEI)(48184), pp. 757–762. Tirunelveli, India, IEEE. https://doi.org/10.1109/ICOEI48184.2020.9142910 [Google Scholar] [CrossRef]
60. Faris, M., Javid, T., Fatima, K., Azhar, M., Kamran, R. (2019). Detection of tumor region in MR image through fusion of Dam construction and K-mean clustering algorithms. 2019 2nd International Conference on Computing, Mathematics and Engineering Technologie, pp. 1–16. Sukkur, Pakistan, IEEE. https://doi.org/10.1109/ICOMET.2019.8673511 [Google Scholar] [CrossRef]
61. Khilkhal, R., Ismael, M. (2022). Brain tumor segmentation utilizing thresholding and K-means clustering. 2022 Muthanna International Conference on Engineering Science and Technology (MICEST), pp. 43–48. Samawah, Iraq, IEEE. https://doi.org/10.1109/MICEST54286.2022.9790103 [Google Scholar] [CrossRef]
62. Asyrofi, R., Winata, Y. A., Sarno, R., Fajar, A. (2020). Cerebellum and frontal lobe segmentation based on K-means clustering and morphological transformation. 2020 International Seminar on Application for Technology of Information and Communication (iSemantic), pp. 149–154. Semarang, Indonesia, IEEE. https://doi.org/10.1109/iSemantic50169.2020.9234262 [Google Scholar] [CrossRef]
63. Kumar, D. M., Satyanarayana, D., Prasad, M. N. G. (2021). An improved gabor wavelet transform and rough K-means clustering algorithm for MRI brain tumor image segmentation. Multimedia Tools and Applications, 80(5), 6939–6957. https://doi.org/10.1007/s11042-020-09635-6 [Google Scholar] [CrossRef]
64. Huang, C., Wan, M. (2022). Automated segmentation of brain tumor based on improved U-Net with residual units. Multimedia Tools and Applications, 81(9), 12543–12566. https://doi.org/10.1007/s11042-022-12335-y [Google Scholar] [CrossRef]
65. Yang, L., Jiang, W., Ji, H., Zhao, Z., Zhu, X. et al. (2019). Automatic brain tumor segmentation using cascaded FCN with DenseCRF and K-means. 2019 IEEE/CIC International Conference on Communications in China (ICCC), pp. 545–549. Changchun, China, IEEE. https://doi.org/10.1109/ICCChina.2019.8855974 [Google Scholar] [CrossRef]
66. Kaur, P. (2017). Intuitionistic fuzzy sets based credibilistic fuzzy C-means clustering for medical image segmentation. International Journal of Information Technology, 9, 345–351. https://doi.org/10.1007/s41870-017-0039-2 [Google Scholar] [CrossRef]
67. Bai, X., Zhang, Y., Liu, H., Wang, Y. (2019). Intuitionistic center-free FCM clustering for MR brain image segmentation. IEEE Journal of Biomedical and Health Informatics, 23(5), 2039–2051. https://doi.org/10.1109/JBHI.6221020 [Google Scholar] [CrossRef]
68. Kumar, D., Agrawal, R. K., Singh Kirar, J. (2019). Intuitionistic fuzzy clustering method with spatial information for MRI image segmentation. 2019 IEEE International Conference on Fuzzy Systems (FUZZ-IEEE), pp. 1–7. Orleans, LA, USA. https://doi.org/10.1109/FUZZ-IEEE.2019.8858865 [Google Scholar] [CrossRef]
69. Kumar, D., Agrawal, R. K., Kumar, P. (2022). Bias-corrected intuitionistic fuzzy C-means with spatial neighborhood information approach for human brain MRI image segmentation. IEEE Transactions on Fuzzy Systems, 30(3), 687–700. https://doi.org/10.1109/TFUZZ.2020.3044253 [Google Scholar] [CrossRef]
70. Kala, R., Deepa, P. (2021). Spatial rough intuitionistic fuzzy C-means clustering for MRI segmentation. Neural Processing Letters, 53(2), 1305–53. https://doi.org/10.1007/s11063-021-10441-w [Google Scholar] [CrossRef]
71. Kumar, D., Verma, H., Mehra, A., Agrawal, R. K. (2019). A modified intuitionistic fuzzy c-means clustering approach to segment human brain MRI image. Multimedia Tools and Applications, 78(10), 12663–87. https://doi.org/10.1007/s11042-018-5954-0 [Google Scholar] [CrossRef]
72. Verma, O. P., Hooda, H. (2020). A novel intuitionistic fuzzy co-clustering algorithm for brain images. Multimedia Tools and Applications, 79(41–42), 31517–31540. https://doi.org/10.1007/s11042-020-09320-8 [Google Scholar] [CrossRef]
73. Huang, C. W., Lin, K. P., Wu, M. C., Hung, K. C., Liu, G. S. et al. (2015). Intuitionistic fuzzy cc-means clustering algorithm with neighborhood attraction in segmenting medical image. Soft Computing Volume, 19, 459–470. https://doi.org/10.1007/s00500-014-1264-2 [Google Scholar] [CrossRef]
74. Xiangxiao, L., Honglin, O., Lijuan, X. (2019). Kernel-distance-based intuitionistic fuzzy c-means clustering algorithm and its application. Pattern Recognition and Image Analysis, 29(4), 592–7. https://doi.org/10.1134/S1054661819040199 [Google Scholar] [CrossRef]
75. Jiang, Y., Zhao, K., Xia, K., Qian, P. (2019). A novel distributed multitask fuzzy clustering algorithm for automatic MR brain image segmentation. Journal of Medical Systems, 43(118), 1–9. https://doi.org/10.1007/s10916-019-1245-1 [Google Scholar] [PubMed] [CrossRef]
76. Yugander, P., Reddy, G. R. (2017). Liver tumor segmentation in noisy CT images using distance regularized level set evolution based on fuzzy C-means clustering. 2017 2nd IEEE International Conference on Recent Trends in Electronics, Information & Communication Technology (RTEICT), pp. 1530–1534. Bangalore, India. https://doi.org/10.1109/RTEICT.2017.8256854 [Google Scholar] [CrossRef]
77. Wan, C., Ye, M., Yao, C., Wu, C. (2017). Brain MR image segmentation based on Gaussian filtering and improved FCM clustering algorithm. BioMedical Engineering and Informatics (CISP-BMEI), 1–5. https://doi.org/10.1109/CISP-BMEI.2017.8301978 [Google Scholar] [CrossRef]
78. Xin, G., Yan, Z., Hua, W., Yujuan, S., Feng, Z. et al. (2022). A modified fuzzy clustering algorithm based on dynamic relatedness model for image segmentation. The Visual Computer. https://doi.org/10.1007/s00371-022-02430-4 [Google Scholar] [CrossRef]
79. Kahali, S., Sing, J. K., Saha, P. K. (2019). A new entropy-based approach for fuzzy c-means clustering and its application to brain MR image segmentation. Soft Computing, 23, 10407–10414. https://doi.org/10.1007/s00500-018-3594-y [Google Scholar] [CrossRef]
80. Kollem, S., Reddy, K. R., Rao, D. S. (2021). An optimized SVM based possibilistic fuzzy cmeans clustering algorithm for tumor segmentation. Multimed Tools Application, 80, 409–437. https://doi.org/10.1007/s11042-020-09675-y [Google Scholar] [CrossRef]
81. Zhao, C., Zhang, Z., Hu, J., Fan, B., Wu, S. (2018). Image segmentation based on improved fuzzy clustering algorithm. 2018 Chinese Control and Decision Conference (CCDC), pp. 495–500. Shenyang, China. https://doi.org/10.1109/CCDC.2018.8407183 [Google Scholar] [CrossRef]
82. Devanathan, K., Samhithan, E. (2020). Lesion segmentation in dermoscopic images using superpixel based fast fuzzy C-means clustering. 2020 IEEE Congreso Bienal de Argentina (ARGENCON), pp. 1–6. Resistencia, Argentina. https://doi.org/10.1109/ARGENCON49523.2020.9505562 [Google Scholar] [CrossRef]
83. Halder, A., Guha, S. (2017). Medical image segmentation using rough-spatial kernelized FCM algorithm. 2017 International Conference on Advances in Computing, Communications and Informatics (ICACCI), pp. 818–823. Udupi, India. https://doi.org/10.1109/ICACCI.2017.8125943 [Google Scholar] [CrossRef]
84. Santos, L., Veras, R., Machado, V. (2018). Medical image segmentation using seeded fuzzy C-means: A semi-supervised clustering algorithm. 2018 International Joint Conference on Neural Networks (IJCNN), pp. 1–7. Rio de Janeiro, Brazil. https://doi.org/10.1109/IJCNN.2018.8489401 [Google Scholar] [CrossRef]
85. Li, M., Xiang, Z., Zhang, L., Lian, Z., Xiao, L. (2017). Robust segmentation of brain MRI images using a novel fuzzy c-means clustering method. Fuzzy Systems and Knowledge Discovery (ICNC-FSKD), 1158–1162. https://doi.org/10.1109/FSKD.2017.8392927 [Google Scholar] [CrossRef]
86. Srinivasan, A., Sadagopan, S. (2021). RETRACTED ARTICLE: Rough fuzzy region based bounded support fuzzy C-means clustering for brain MR image segmentation. Journal of Ambient Intelligence and Humanized Computing, 12, 3775–3788. https://doi.org/10.1007/s12652-019-01672-w [Google Scholar] [CrossRef]
87. Zhang, X., Sun, Y., Wang, G., Guo, Q., Zhang, C. et al. (2016). Improved fuzzy clustering algorithm with non-local information for image segmentation. Multimedia Tools and Applications, 76, 7869–7895. https://doi.org/10.1007/s11042-016-3399-x [Google Scholar] [CrossRef]
88. Mithra, K., Emmanuel, W. R. S. (2017). An efficient approach to sputum image segmentation using improved fuzzy local information C means clustering algorithm for tuberculosis diagnosis. 2017 International Conference on Inventive Computing and Informatics (ICICI), pp. 126–130. Coimbatore, India. https://doi.org/10.1109/ICICI.2017.8365321 [Google Scholar] [CrossRef]
89. Gharieb, R. R., Gendy, G., Abdelfattah, A. (2016). A robust local data and membership information based FCM algorithm for noisy image segmentation. 2016 12th International Computer Engineering Conference (ICENCO), pp. 93–98. Cairo, Egypt: IEEE. https://doi.org/10.1109/ICENCO.2016.7856451 [Google Scholar] [CrossRef]
90. Mahata, N., Sing, J. K. (2019). Global and spatially constrained local entropy-based fuzzy clustering algorithm for segmentation of 3D brain MR image volume with high noise and intensity inhomogeneity. TENCON 2019—2019 IEEE Region 10 Conference (TENCON), pp. 1963–1968. Kochi, India. https://doi.org/10.1109/TENCON.2019.8929520 [Google Scholar] [CrossRef]
91. Khosravanian, A., Rahmanimanesh, M., Keshavarzi, P., Mozaffari, S. (2020). Fuzzy local intensity clustering (FLIC) model for automatic medical image segmentation. The Visual Computer, 37, 1185–1206. https://doi.org/10.1007/s00371-020-01861-1 [Google Scholar] [CrossRef]
92. Zhang, H., Liu, J. (2022). Fuzzy c-means clustering algorithm with deformable spatial information for image segmentation. Multimed Tools Application, 81, 11239–11258. https://doi.org/10.1007/s11042-022-11904-5 [Google Scholar] [CrossRef]
93. Kumar, S. N., Fred, A. L., Varghese, P. S. (2019). Suspicious lesion segmentation on brain, mammograms and breast MR images using new optimized spatial feature based super-pixel fuzzy C-means clustering. Journal of Digital Imaging, 32, 322–335. https://doi.org/10.1007/s10278-018-0149-9 [Google Scholar] [PubMed] [CrossRef]
94. Xu, J., Zhao, T., Feng, G., Ni, M., Ou, S. (2021). A fuzzy C-means clustering algorithm based on spatial context model for image segmentation. International Journal of Fuzzy Systems, 23, 816–832. https://doi.org/10.1007/s40815-020-01015-4 [Google Scholar] [CrossRef]
95. Sajith, A. G., Hariharan, S. (2015). Spatial fuzzy C-means clustering based segmentation on CT images. 2015 2nd International Conference on Electronics and Communication Systems (ICECS), pp. 414–417. Coimbatore, India, IEEE. https://doi.org/10.1109/ECS.2015.7124937 [Google Scholar] [CrossRef]
96. Zhang, L., Gui, Z., Wang, J., Zhang, P., Qin, Z. et al. (2022). Spatial information-based intuitionistic fuzzy kernel clustering algorithm for cerebral hemorrhage image segmentation. Signal, Image and Video Processing, 16, 983–991. https://doi.org/10.1007/s11760-021-02043-9 [Google Scholar] [CrossRef]
97. Liu, Z., Song, Y. (2014). An adapted spatial information kernel-based fuzzy C-means clustering method. 2014 7th International Congress on Image and Signal Processing, pp. 370–374. Dalian, China, IEEE. https://doi.org/10.1109/CISP.2014.7003808 [Google Scholar] [CrossRef]
98. Gendy, G. (2018). Adaptive fuzzy C-means algorithm using the hybrid spatial information for medical image segmentation. 2018 9th Cairo International Biomedical Engineering Conference (CIBEC), pp. 25–28. Cairo, Egypt, IEEE. https://doi.org/10.1109/CIBEC.2018.8641801 [Google Scholar] [CrossRef]
99. Sun, J. Y., Wu, T. H., Liu, H., Liu, Y. H. (2018). A novel three-dimensional clustering method incorporated with ellipsoid model fitting for personalizing myocardial perfusion positron emission tomography image segmentation. 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC), pp. 1–4. Sydney, NSW, Australia. https://doi.org/10.1109/NSSMIC.2018.8824346 [Google Scholar] [CrossRef]
100. Ghogge, R., Ambedkar Institue (2014). Brain tumour detection using K-means and fuzzy C-means clustering algorithm. computing science. International Journal of Computer Science and Information Technology Research Excellence, 3(2). https://doi.org/10.13140/RG.2.1.3961.5841 [Google Scholar] [CrossRef]
101. Wu, C., Huang, C., Zhang, J. (2021). Intuitionistic fuzzy information-driven total bregman divergence fuzzy clustering with multiple local information constraints for image segmentation. The Visual Computer, https://doi.org/10.1007/s00371-021-02319-8 [Google Scholar] [CrossRef]
102. Yin, S., Li, H., Liu, D., Karim, S. (2020). Active contour modal based on density-oriented BIRCH clustering method for medical image segmentation. Multimedia Tools and Applications, 79(41–42), 31049–31068. https://doi.org/10.1007/s11042-020-09640-9 [Google Scholar] [CrossRef]
103. Zhang, X., Wang, H., Zhang, Y., Gao, X., Wang, G. et al. (2021). Improved fuzzy clustering for image segmentation based on a low-rank prior. Computational Visual Media, 7, 513–528. https://doi.org/10.1007/s41095-021-0239-3 [Google Scholar] [CrossRef]
104. Lian, C., Ruan, S., Denoeux, T., Li, H., Vera, P. (2019). Joint tumor segmentation in PET-CT images using Co-clustering and fusion based on belief functions. IEEE Transactions on Image Processing, 28(2), 755–766. https://doi.org/10.1109/TIP.2018.2872908 [Google Scholar] [PubMed] [CrossRef]
105. Mariena, A. A., Sathiaseelan, J. G. R., Abraham, J. T. (2018). Hybrid approach for image segmentation usingregion splitting and clustering techniques. 2018 International Conference on Circuits and Systems in Digital Enterprise Technology (ICCSDET), pp. 1–4. Kottayam, India. https://doi.org/10.1109/ICCSDET.2018.8821144 [Google Scholar] [CrossRef]
106. Ronneberger, O. (2017). Invited talk: U-Net convolutional networks for biomedical image segmentation, vol. 3. Berlin Heidelberg: Springer. https://doi.org/10.1007/978-3-662-54345-0_3 [Google Scholar] [CrossRef]
107. Nazi, Z. A., Abir, T. A. (2020). Automatic skin lesion segmentation and melanoma detection: transfer learning approach with U-Net and DCNN-SVM, pp. 371–81. Singapore: Springer. https://doi.org/10.1007/978-981-13-7564-4 [Google Scholar] [CrossRef]
108. Zhao, C., Zhao, Z., Feng, Y. (2020). MVP U-Net: Multi-view pointwise U-Net for brain tumor segmentation. In: Lecture notes in computer science, pp. 99–103. https://doi.org/10.1007/978-3-030-72087-2_9 [Google Scholar] [CrossRef]
109. Albishri, A. A., Shah, S. J., Kang, S. S., Lee, Y. (2022). AM-UNet: Automated mini 3D end-to-end U-netbased network for brain claustrum segmentation. Multimedia Tools and Applications, 81, 36171–36194. https://doi.org/10.1007/s11042-021-11568-7 [Google Scholar] [PubMed] [CrossRef]
110. Chen, X., Jiang, S., Guo, L., Chen, Z., Zhang, C. (2021). Whole brain segmentation method from 2.5D brain MRI slice image based on triple U-Net. The Visual Computer. https://doi.org/10.1007/s00371-021-02326-9 [Google Scholar] [CrossRef]
111. Li, K., Kong, L., Zhang, Y. (2020). 3D U-Net brain tumor segmentation using VAE skip connection. 2020 IEEE 5th International Conference on Image, Vision and Computing (ICIVC), pp. 97–101. Beijing, China. https://doi.org/10.1109/ICIVC50857.2020.9177441 [Google Scholar] [CrossRef]
112. Zhang, Y. D., Satapathy, S. C. (2022). Fruit category classification by fractional Fourier entropy with rotation angle vector grid and stacked sparse autoencoder. Expert Systems, 39(3). https://doi.org/10.1111/exsy.12701 [Google Scholar] [CrossRef]
113. Zhao, L., Feng, X., Meyer, C. H., Alsop, D. C. (2020). Choroid plexus segmentation using optimized 3D U-Net. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pp. 381–384. Iowa City, IA, USA. https://doi.org/10.1109/ISBI45749.2020.9098443 [Google Scholar] [CrossRef]
114. Liu, X., Sun, Z., Han, C., Cui, Y., Huang, J. et al. (2021). Development and validation of the 3D U-netalgorithm for segmentation of pelvic lymph nodes on diffusion-weighted images. BMC Medical Imaging, 21(1), 170. https://doi.org/10.1186/s12880-021-00703-3 [Google Scholar] [PubMed] [CrossRef]
115. Wu, Y., Shen, H., Tan, Y., Shi, Y. (2022). Automatic liver tumor segmentation used the cascade multi-scaleattention architecture method based on 3D U-Net. International Journal of Computer Assisted Radiology and Surgery, 17(10), 1915–1922. https://doi.org/10.1007/s11548-022-02653-9 [Google Scholar] [PubMed] [CrossRef]
116. Habijan, M., Leventic, H., Galic, I., Babin, D. (2019). Whole heart segmentation from CT images using 3D U-Net architecture. 2019 International Conference on Systems, Signals and Image Processing (IWSSIP), pp. 121–126. Osijek, Croatia. https://doi.org/10.1109/IWSSIP.2019.8787253 [Google Scholar] [CrossRef]
117. Zhang, K., Li, J., Ma, R., Li, G. (2020). An end-to-end segmentation network for the temporomandibular joints CBCT image based on 3D U-Net. 2020 13th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), pp. 664–668. Chengdu, China. https://doi.org/10.1109/CISP-BMEI51763.2020.9263566 [Google Scholar] [CrossRef]
118. Bazgir, O., Barck, K. H., Carano, R. A., Weimer, R. M., Xie, L. (2020). Kidney segmentation using 3D U-Net localized with expectation maximization. 2020 IEEE Southwest Symposium on Image Analysis and Interpretation (SSIAI), pp. 22–25. Albuquerque, NM, USA. https://doi.org/10.1109/SSIAI49293.2020.9094601 [Google Scholar] [CrossRef]
119. Pandey, M., Gupta, A. (2022). Tumorous kidney segmentation in abdominal CT images using activecontour and 3D-UNet. Irish Journal of Medical Science. https://doi.org/10.1007/s11845-022-03113-8 [Google Scholar] [PubMed] [CrossRef]
120. Gamal, A., Bedda, K., Ashraf, N., Ayman, S., AbdAllah, M. et al. (2021). Brain tumor segmentation using 3D U-Net with hyperparameter optimization. 2021 3rd Novel Intelligent and Leading Emerging Sciences Conference (NILES), pp. 269–272. Giza, Egypt. https://doi.org/10.1109/NILES53778.2021.9600556 [Google Scholar] [CrossRef]
121. Popa, D. L., Popa, R. T., Barbulescu, L. F., Ivanescu, R. C., Mocanu, M. L. (2022). Segmentation of different human organs on 3D computer tomography and magnetic resonance imaging using an open source 3D U-Net framework. 2022 23rd International Carpathian Control Conference (ICCC), pp. 54–57. Sinaia, Romania. https://doi.org/10.1109/ICCC54292.2022.9805911 [Google Scholar] [CrossRef]
122. Francis, S., Jayaraj, P. B., Pournami, P. N., Thomas, M., Jose, A. T. et al. (2022). ThoraxNet: A 3D Unet based two-stage framework for OAR segmentation on thoracic CT images. Physical and Engineering Sciences in Medicine, 45(1), 189–203. https://doi.org/10.1007/s13246-022-01101-x [Google Scholar] [PubMed] [CrossRef]
123. Habijan, M., Galic, I., Leventic, H., Romic, K., Babin, D. (2020). Abdominal aortic aneurysm segmentation from CT images using modified 3D U-Net with deep supervision. 2020 International Symposium ELMAR, pp. 123–128. Zadar, Croatia. https://doi.org/10.1109/ELMAR49956.2020.9219015 [Google Scholar] [CrossRef]
124. Ahmed, S. F., Rahman, F. S., Tabassum, T., Bhuiyan, M. T. I. (2019). 3D U-Net: fully convolutional neural network for automatic brain tumor segmentation. 2019 22nd International Conference on Computer and Information Technology (ICCIT), pp. 1–6. Dhaka, Bangladesh. https://doi.org/10.1109/ICCIT48885.2019.9038237 [Google Scholar] [CrossRef]
125. Manoj, K. C., Dhas, D. A. S. (2022). Automated brain tumor malignancy detection via 3D MRI using adaptive-3-D U-Net and heuristic-based deep neural network. Multimedia Systems, 28(6), 2247–2273. https://doi.org/10.1007/s00530-022-00952-4 [Google Scholar] [CrossRef]
126. Kong, L., Zhang, Y. (2021). Multi-modal brain tumor segmentation using cascaded 3D U-Net. 2021 6th International Conference on Image, Vision and Computing (ICIVC), pp. 129–133. Qingdao, China. https://doi.org/10.1109/ICIVC52351.2021.9526932 [Google Scholar] [CrossRef]
127. Lin, R., Wang, S., Chen, Q., Cai, Z., Zhu, Y. et al. (2021). Combining K-means attention and hierarchical mimicking strategy for 3D U-Net based brain tumor segmentation. 2021 IEEE International Conference on Information Communication and Software Engineering (ICICSE), pp. 92–96. Chengdu, China. https://doi.org/10.1109/ICICSE52190.2021.9404138 [Google Scholar] [CrossRef]
128. Liang, J., Yang, C., Zhong, J., Ye, X. (2022). BTSwin-Unet: 3D U-shaped symmetrical swin transformer-based network for brain tumor segmentation with self-supervised Pre-training. Neural Processing Letters, https://doi.org/10.1007/s11063-022-10919-1 [Google Scholar] [CrossRef]
129. Wang, Y., Zhao, L., Wang, M., Song, Z. (2019). Organ at risk segmentation in head and neck CT images using a Two-stage segmentation framework based on 3D U-Net. IEEE Access, 7, 144591–144602. https://doi.org/10.1109/Access.6287639 [Google Scholar] [CrossRef]
130. Hu, C., Kang, G., Hou, B., Ma, Y., Labeau, F. et al. (2020). Acu-Net: A 3D attention context U-Net for multiple sclerosis lesion segmentation. ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 1384–1388. Barcelona, Spain. https://doi.org/10.1109/ICASSP40776.2020.9054616 [Google Scholar] [CrossRef]
131. Leng, J., Liu, Y., Zhang, T., Quan, P., Cui, Z. (2018). Context-aware U-Net for biomedical image segmentation. 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 2535–2538. Madrid, Spain. https://doi.org/10.1109/BIBM.2018.8621512 [Google Scholar] [CrossRef]
132. Fu, X., Zhao, N. (2022). AGC-UNet: A global context feature fusion method based on U-Net for retinal vessel segmentation. 2022 IEEE 2nd International Conference on Information Communication and Software Engineering (ICICSE), pp. 94–99. Chongqing, China. https://doi.org/10.1109/ICICSE55337.2022.9828894 [Google Scholar] [CrossRef]
133. Jha, D., Riegler, M., Johansen, D., Halvorsen, P., Johansen, H. (2020). DoubleU-Net: A deep convolutional neural network for medical image segmentation. Computer Vision and Pattern Recognition. https://doi.org/10.1109/CBMS49503.2020 [Google Scholar] [CrossRef]
134. Zuo, B., Lee, F., Chen, Q. (2022). An efficient U-shaped network combined with edge attention module and context pyramid fusion for skin lesion segmentation. Medical & Biological Engineering & Computing, 60(7), 1987–2000. https://doi.org/10.1007/s11517-022-02581-5 [Google Scholar] [PubMed] [CrossRef]
135. Fan, T., Wang, G., Wang, X., Li, Y., Wang, H. (2021). MSN-Net: A multi-scale context nested U-Net for liver segmentation. Signal, Image and Video Processing, 1–9. https://doi.org/10.1007/s11760-020-01835-9 [Google Scholar] [CrossRef]
136. Yagi, N., Nii, M., Kobashi, S. (2019). Abdominal organ area segmentation using U-Net for cancer radiotherapy support. 2019 IEEE International Conference on Systems, Man and Cybernetics (SMC), pp. 1210–1214. Bari, Italy. https://doi.org/10.1109/SMC.2019.8913943 [Google Scholar] [CrossRef]
137. Akbas, C. E., Kozubek, M. (2020). Condensed U-Net (Cu-Netan improved U-Net architecture for cell segmentation powered by 4 × 4 max-pooling layers. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pp. 446–450. Iowa City, IA, USA. https://doi.org/10.1109/ISBI45749.2020.9098351 [Google Scholar] [CrossRef]
138. Wu, J., Zhou, S., Zuo, S., Chen, Y., Sun, W. et al. (2021). U-Net combined with multi-scale attention mechanism for liver segmentation in CT images. BMC Medical Informatics and Decision Making, 21(1), 283. https://doi.org/10.1186/s12911-021-01649-w [Google Scholar] [PubMed] [CrossRef]
139. An, F., Liu, J. (2021). Medical image segmentation algorithm based on multilayer boundary perception-self attention deep learning model. Multimedia Tools and Applications, 80, 15017–15039. https://doi.org/10.1007/s11042-021-10515-w [Google Scholar] [CrossRef]
140. Huang, A., Wang, Q., Jiang, L., Zhang, J. (2021). Automatic segmentation of median nerve in ultrasound image by a combined Use of U-net and VGG16. 2021 IEEE International Ultrasonics Symposium (IUS), pp. 1–4. Xi'an, China. https://doi.org/10.1109/IUS52206.2021.9593861 [Google Scholar] [CrossRef]
141. Ghosh, S., Chaki, A., Santosh, K. (2021). Improved U-Net architecture with VGG-16 for brain tumor segmentation. Physical and Engineering Sciences in Medicine, 44, 703–712. https://doi.org/10.1007/s13246-021-01019-w [Google Scholar] [PubMed] [CrossRef]
142. Zhang, Z. D., Khan, M. A. (2021). Pseudo zernike moment and deep stacked sparse autoencoder for COVID-19 diagnosis. Computers, Materials & Continua, 69(3), 3145–3162. https://doi.org/10.32604/cmc.2021.018040 [Google Scholar] [CrossRef]
143. Farahani, A., Mohseni, H. (2021). Medical image segmentation using customized U-net with adaptive activation functions. Neural Computing and Applications, 33, 6307–6323. https://doi.org/10.1007/s00521-020-05396-3 [Google Scholar] [CrossRef]
144. Yang, J., Qiu, K. (2021). An improved segmentation algorithm of CT image based on U-Net network and attention mechanism. Multimedia Tools and Applications, 81, 35983–36006. https://doi.org/10.1007/s11042-021-10841-z [Google Scholar] [CrossRef]
145. Mishra, A., Gupta, P., Tewari, P. (2022). Global U-Net with amalgamation of inception model and improved kernel variation for MRI brain image segmentation. Multimedia Tools and Applications, 81, 23339–23354. https://doi.org/10.1007/s11042-022-12094-w [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools