 Open Access
Open Access
ARTICLE
Differentiate Xp11.2 Translocation Renal Cell Carcinoma from Computed Tomography Images and Clinical Data with ResNet-18 CNN and XGBoost
1 Department of Urology, Nanjing Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, 210008, China
2 School of Data Science, Perdana University, Serdang, 43400, Malaysia
3 State Key Laboratory for Novel Software Technology, Nanjing University, Nanjing, 210008, China
* Corresponding Authors: Tong Lu. Email: ; Weidong Gan. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Computer Modeling of Artificial Intelligence and Medical Imaging)
Computer Modeling in Engineering & Sciences 2023, 136(1), 347-362. https://doi.org/10.32604/cmes.2023.024909
Received 13 June 2022; Accepted 23 August 2022; Issue published 05 January 2023
Abstract
This study aims to apply ResNet-18 convolutional neural network (CNN) and XGBoost to preoperative computed tomography (CT) images and clinical data for distinguishing Xp11.2 translocation renal cell carcinoma (Xp11.2 tRCC) from common subtypes of renal cell carcinoma (RCC) in order to provide patients with individualized treatment plans. Data from 45 patients with Xp11.2 tRCC from January 2007 to December 2021 are collected. Clear cell RCC (ccRCC), papillary RCC (pRCC), or chromophobe RCC (chRCC) can be detected from each patient. CT images are acquired in the following three phases: unenhanced, corticomedullary, and nephrographic. A unified framework is proposed for the classification of renal masses. In this framework, ResNet-18 CNN is employed to classify renal cancers with CT images, while XGBoost is adopted with clinical data. Experiments demonstrate that, if applying ResNet-18 CNN or XGBoost singly, the latter outperforms the former, while the framework integrating both technologies performs similarly or better than urologists. Especially, the possibility of misclassifying Xp11.2 tRCC, pRCC, and chRCC as ccRCC by the proposed framework is much lower than urologists.Keywords
Renal cell carcinoma (RCC) is the most common subtype of primary renal cancer, accounting for 80%–90% of renal malignancies [1]. Clear cell renal cell carcinoma (ccRCC), papillary renal cell carcinoma (pRCC), and chromophobe renal cell carcinoma (chRCC) are common subtypes of renal cancer, accounting for 70%, 15%–20%, and 6%–11% of all renal cancers, respectively [2–4]. Xp11.2 translocation renal cell carcinoma (Xp11.2 tRCC) is an independent and rare subtype of RCCs, formed by the balanced translocation of the TFE3 gene on the short arm of the X chromosome and other genes, accompanied by the overexpression of TFE3 protein [5]. Xp11.2 tRCC and RCC associated with t (6; 11)/TFEB gene fusions were classified as MiT family translocation RCC in the 2016 World Health Organization kidney tumor classification scheme [6]. Recent studies have reported that Xp11.2 tRCC was more aggressive than ccRCC in adult patients [7,8]. Due to the aggressive nature of Xp11.2 tRCC, adult patients with strong positive expression of TFE3 protein tend to have a worse prognosis than patients with TFE3-negative RCC, namely ccRCC, pRCC, and chRCC. In addition, the 2019 European Urological Association guidelines strongly recommended nephron-sparing surgery for clinical stage T1 renal cancer. However, multi-center clinical studies have shown that Xp11.2 tRCC patients who undergo nephron-sparing surgery at clinical stage T1b are more likely to suffer disease progression than radical nephrectomy [9,10]. The treatment for Xp11.2 tRCC should be different from common subtypes of RCCs when the surgical plan is selected for the first time. However, there is still a lack of effective preoperative diagnosis in differentiating Xp11.2 tRCC from three main subtypes of RCCs. Therefore, accurate preoperative prediction of Xp11.2 tRCC is essential to realize the individualized surgical plan for patients.
Artificial intelligence (AI) for image classification through deep learning (DL) models and machine learning (ML) has been established in health and medicine and achieved rapid development. The diagnostic performance of AI technology in several diseases has exceeded those of manual methods with advantages of automated quantitative evaluation, low error probability, and consistent and stable diagnosis results [11–14]. In recent years, the applications of ML and DL in urology increased and succeeded in image recognition diagnostics, personalized medicine, and clinical decision-making, and the development of AI technology will further improve disease prediction accuracy and promote precision medicine [15–18]. The deep neural network model trained and tested by histopathological images can automatically differentiate Xp11.2 tRCC and ccRCC with high performance [19], simplifying the traditional pathological diagnosis model of Xp11.2 tRCC. However, the pathological diagnosis was performed by pathologists or AI technology after surgery, which cannot meet the urgent clinical needs of preoperative diagnosis. Therefore, the development of a DL neural network model based on preoperative abdominal computed tomography (CT) scan to assist in the differential diagnosis of Xp11.2 tRCC has greater clinical significance and practical value.
In this study, we proposed an AI model based on preoperative CT images and clinical data to distinguish between Xp11.2 tRCC and common subtypes of renal cancer and then provided patients with individualized treatment plans.
The complete CT imaging data and postoperative pathological data of patients with Xp11.2 tRCC diagnosed in the Nanjing Drum Tower Hospital were retrospectively analyzed from January 2007 to December 2021. 45 cases of Xp11.2 tRCC with complete clinicopathological data were enrolled in the study, including complete medical histories, clinical information, and pathological data. According to both tumor size and pathological stage, the matched patients included 45 with ccRCC, pRCC, or chRCC, respectively. The study was approved by the Institutional Review Board of Nanjing Drum Tower Hospital.
Inclusion criteria:
1. Patients underwent radical or partial nephrectomy.
2. Preoperative CT examination of renal tumors and complete examination records.
3. The reviewed pathological diagnosis was consistent with the initial postoperative diagnosis and the pathological diagnosis of Xp11.2 tRCC was validated by the next generation sequence.
4. These patients were without cardiovascular and cerebrovascular diseases and chronic kidney disease that affected the blood supply of the kidney.
5. These patients had a unilateral single renal tumor.
Exclusion criteria:
1. Incomplete clinical and CT imaging data.
2. After reviewing the pathological diagnosis, it was inconsistent with the initial pathological diagnosis.
3. These patients had cardiovascular and cerebrovascular diseases, chronic kidney disease, and other diseases which affected the blood supply of the kidney.
4. These patients had multiple renal tumors.
2.2 CT Technical Parameters and Image Preprocessing
All patients in our hospital used 64-slice CT (Light-Speed; GE Healthcare, Princeton, NJ, USA) for CT examination. In each case, images inputted into the model were acquired at three phases, including unenhanced, corticomedullary, and nephrographic phases. Both corticomedullary and nephrographic phases were included in the enhanced phase. Most of the image layer thicknesses in the unenhanced phase were 5 and 10 mm, and the image layer thicknesses in the corticomedullary and nephrographic phases were 1 and 6 mm. We exported anonymous images in digital imaging and communication in medicine (DICOM) format from Picture Archiving and Communication System (PACS) for each patient. We manually segmented the kidney cancer area in each CT image and removed the segmented non-tumor area using MATLAB software (MATLAB R2016a). The CT images in the folder were automatically read, manually scribed by two experienced urologists, executed the segmentation algorithm, removed the background, and automatically saved the segmented tumor image according to the original path. The procedure of segmentation is shown in Figs. 1 and 2. In order to ensure that all kidney cancers were in the segmentation area, we set the mouse click position 2 mm within the edge of the tumor to ensure the accuracy of the segmentation image. Since the boundaries of the renal tumors in the unenhanced CT images were difficult to distinguish from the normal kidney tissue, we decided to use the enhanced image to assist the segmentation. For ambiguous cases, determining the boundaries of renal cancers were jointly determined by two urologists with more than 10 years of work experience.
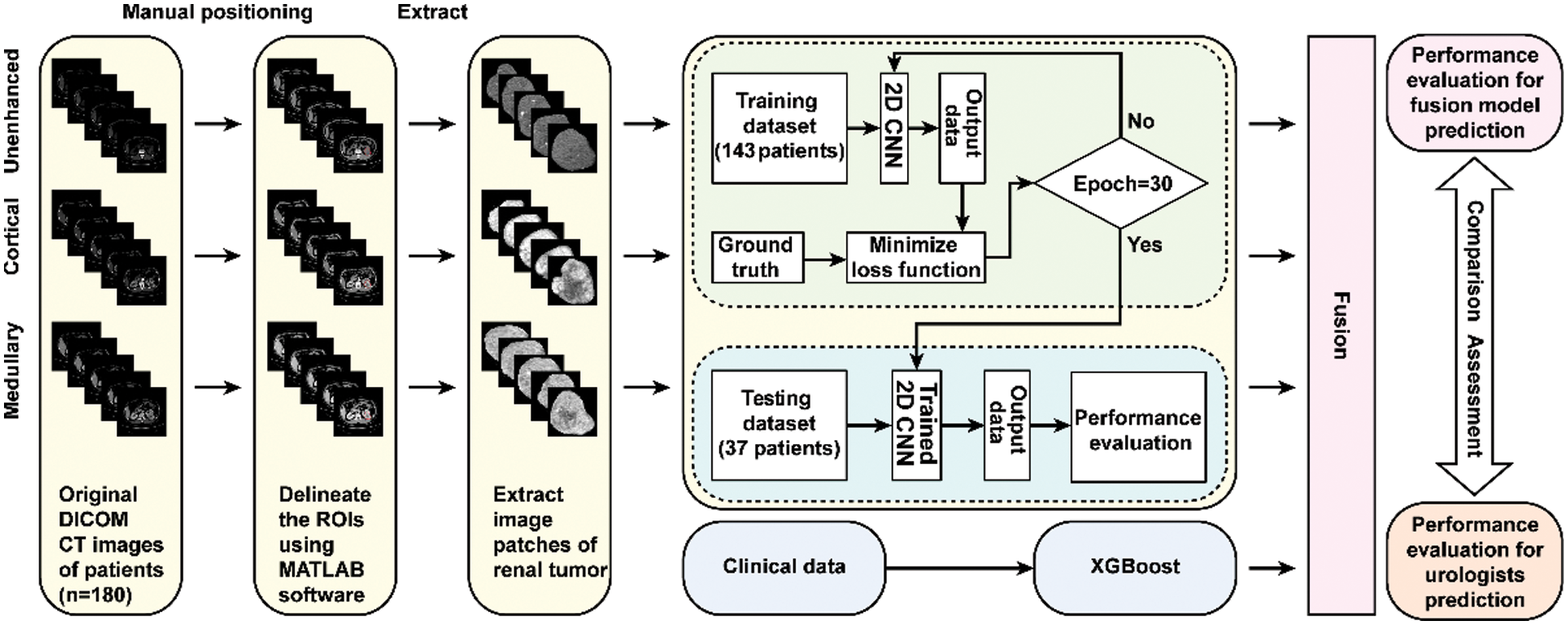
Figure 1: Flowchart illustrating the design of the research. The ResNet-18 CNN model took image patches of renal tumor extracted from a multi-phase CT scan as an input. The Clinical data was applied to train and test the XGBoost model. The fusion model was constructed from the ResNet-18 CNN combined with the XGBoost model and compared with urologists in predictive performance
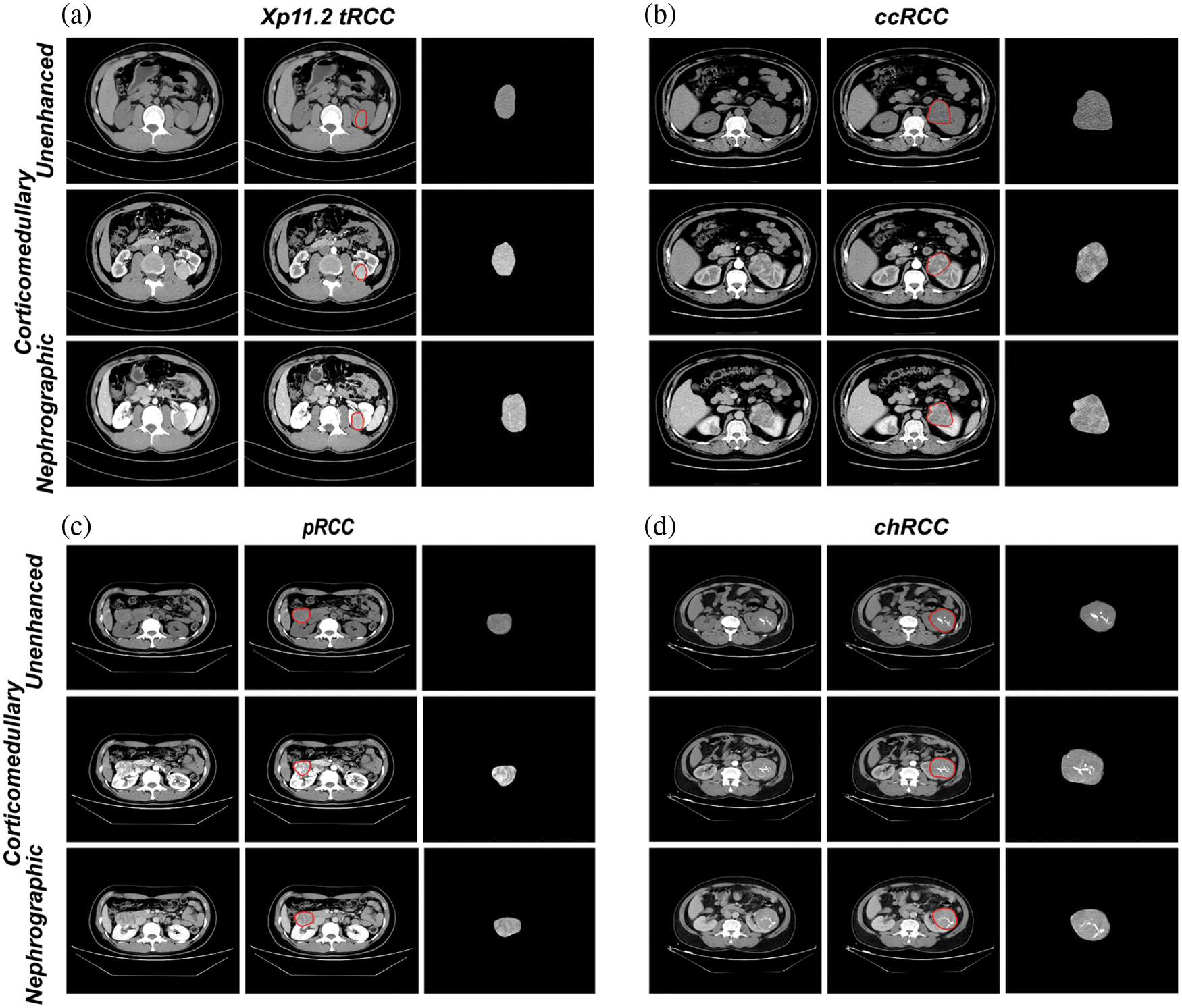
Figure 2: Representative axial CT images of delineating and extracting renal tumor in Xp11.2 tRCC (a), ccRCC (b), pRCC (c), chRCC (d): unenhanced phase, corticomedullary phase, and nephrographic phase were used as the input data of the model, respectively. For every single renal cancer subtype, the left was the original CT axial image, the middle was the segmented image of the kidney tumor, and the right was the kidney tumor after removing the non-tumor area
2.3 Clinical Data Preprocessing
Since the clinical data of patients was an essential basis for differentiating Xp11.2 tRCC from the common subtypes of RCCs, we inputted the clinical data of patients into the model, including age, gender, tumor size, hypertension, hematuria, hyperuricemia, abdominal pain, and abdominal mass. Among them, age data was converted into integer type, and the rest of the clinical data was represented by 1 for yes and 0 for no.
Our model consisted of two modules: a CT image classifier and a clinical data classifier. Both modules operated in parallel. First, each patient’s CT images and clinical data were input into a CT image classifier and a clinical data classifier, respectively. Then, each of the two classifiers classified the data and outputted the probability of the type of renal cancer. Finally, the results of the two classifiers were fused to obtain the final predicted probability of kidney tumor.
2.4.1 ResNet-18 Convolutional Neural Network
We used deep convolutional neural networks (CNN) to classify CT image data, of which we chose ResNet-18 CNN to extract image features and used Softmax for four-classification. ResNet-18 CNN was formed by stacking a series of convolutional blocks with shortcuts, with different suffixes depending on the number of convolutional layers. ResNet-18 CNN indicated that this network consisted of 18 convolutional layers. At the end of the network, the activation layer Softmax directly gave the probability of each renal cell carcinoma. For each patient, we had the CT image sequence
Since slices at different positions of the renal cell carcinoma had different contributions to distinguishing tumor types, the closer to the center, the stronger the degree of discrimination, so we used the normal distribution function to calculate the fusion weight of each image. For the i-th slice, we defined its weight as:
Therefore, the classification result of each phase can be formulated as follows:
Clinical experience showed that the reliability of CT at different phases varies widely. The reliability of unenhanced, corticomedullary, and nephrographic renal CT increased sequentially. Therefore, we empirically set the phases of unenhanced, corticomedullary, and nephrographic weights to be 0.2, 0.3, and 0.5 in the three phases fusion step.
We used XGBoost to classify clinical data. XGBoost is a variant of random forest, which is widely used for the mining of structured data. Different from the random forest, XGBoost first builds all the subtrees that can be established from top to bottom and then reversely prunes from the bottom to the top to avoid the model from falling into the optimal local solution. For clinical data, it only contained 7 fields, and each decision tree of XGBoost randomly discarded some fields, so XGBoost with a maximum depth of 3 was enough to cover all fields. For each case, we got the corresponding probabilities of the four tumors. The process was represented as follows:
Considering the importance of CT images and clinical data for the judgment of renal cancer types, we added the results of CNN and the results of XGBoost to obtain the final probability.
For CT images, we train each image separately. We use the ResNet-18 pre-trained on ImageNet [20] as the backbone. All cropped CT images are resized to 224 × 224 with the randomized horizontal flip data augmentation. We use SGD as the optimizer with a weight decay of 0.0001 and a momentum of 0.9 and set the batch size to 256. The model is trained for 30 epochs. The learning rate is set as 0.01 and divided by 10 at epochs 15 and 25. For clinical data, we set the learning rate of XGBoost to 0.1, the number of estimators to 100, and the maximum depth to 3.
In the inference, we use the fusion method mentioned in Section 2.4.3 to fuse the CT images probability and fuse with the probability of XGBoost to get the final result.
We used a permutation test to compare the performance of the ensemble model with that of the urologists. Specifically, we randomly sampled 35 times from 45 patients with replacement to obtain 35 bootstrap samples and then calculated sensitivity, specificity, accuracy, and AUCs for these 35 samples, respectively. This process was repeated 100 times to generate the distribution of sensitivity, specificity, accuracy, and AUCs, and then the mean and 95 confidence intervals (CI) were calculated.
3.1 Patients and Lesion Features
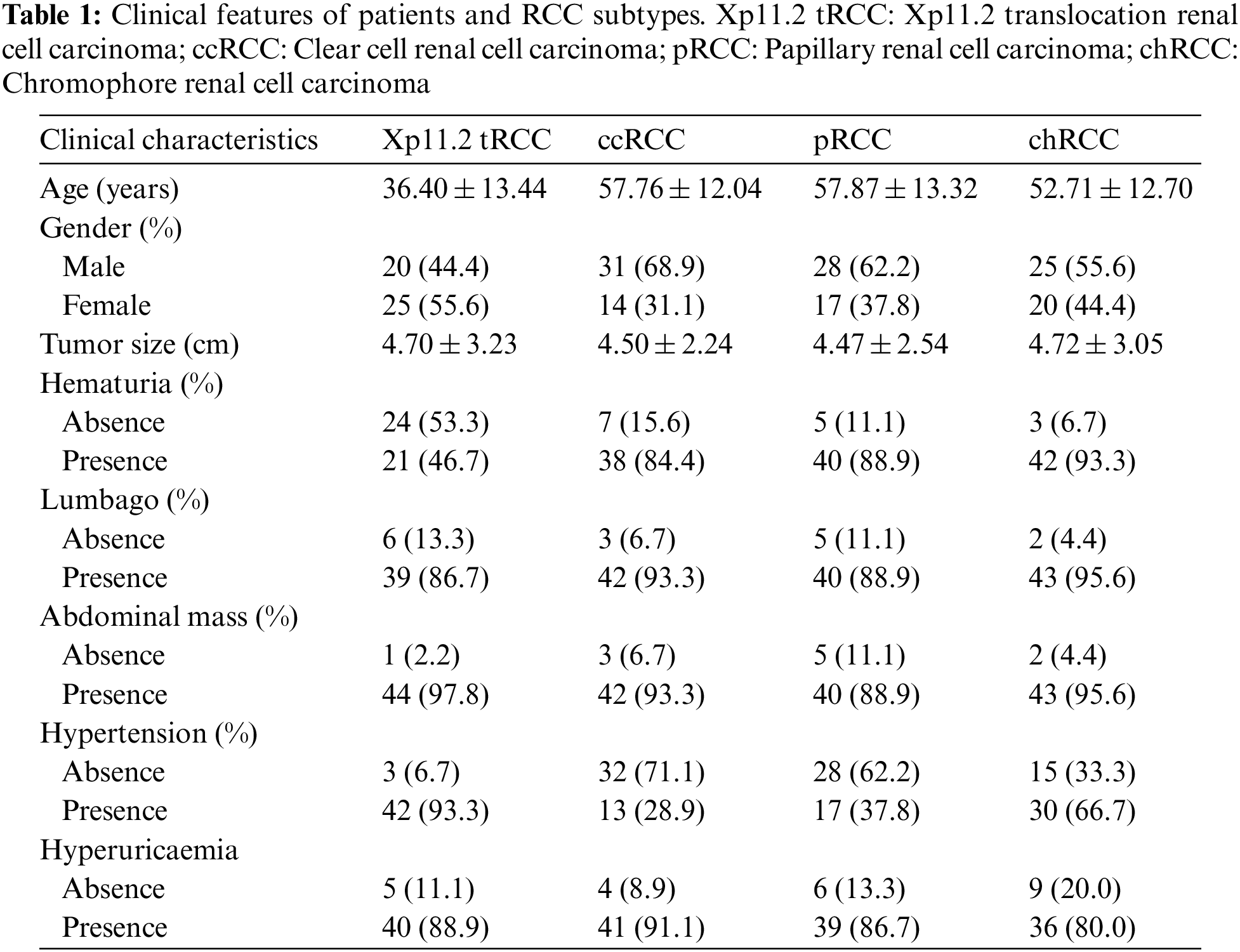
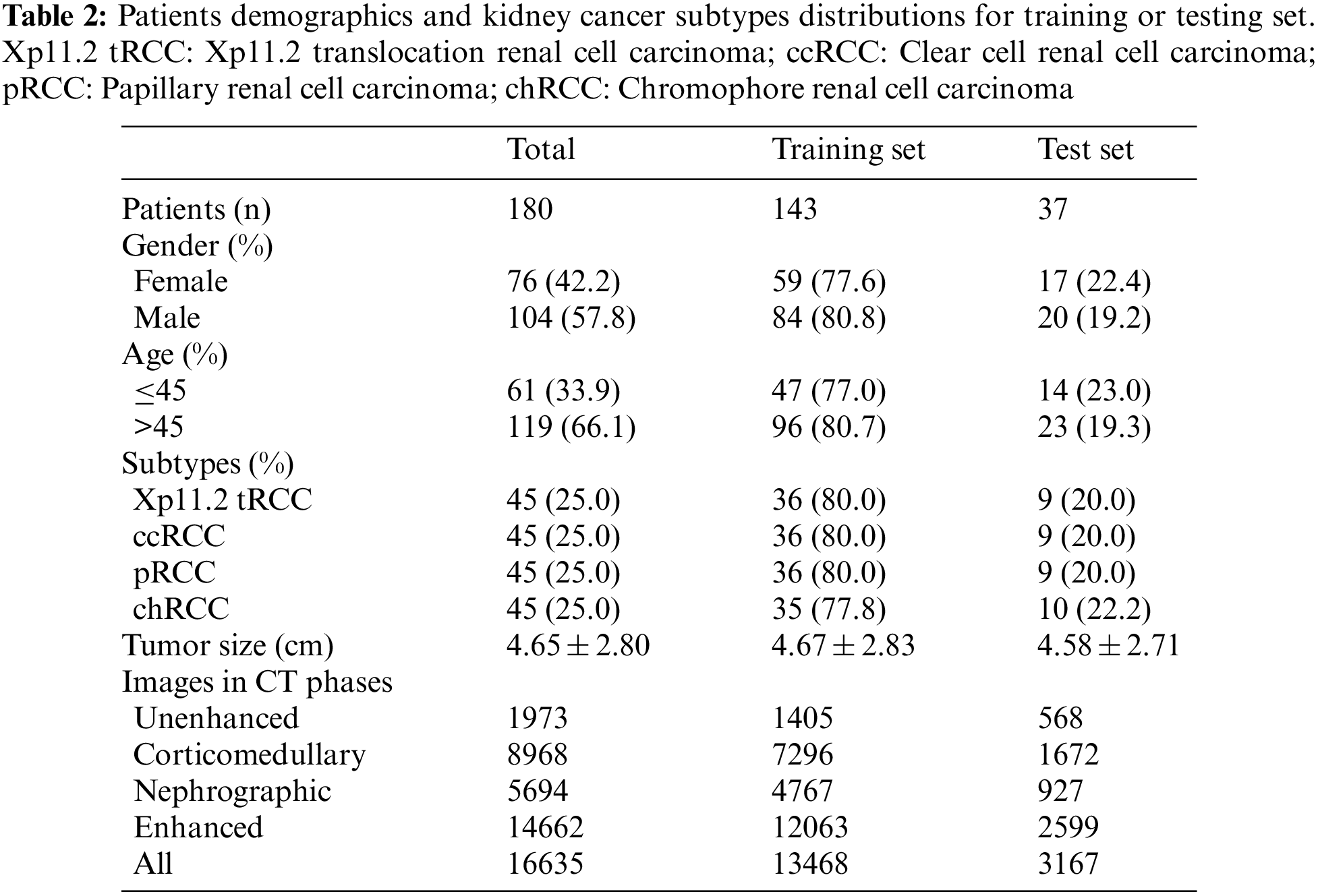
We screened all RCC patients treated in Nanjing Drum Tower Hospital from January 2007 to December 2021 and collected 45 cases of Xp11.2 tRCC with complete clinicopathological data, which included complete medical histories, clinical information, and pathological data. The matched patients included 45 with ccRCC, pRCC, or chRCC, respectively. The clinicopathologic characteristics of the entire cohort were displayed in Table 1. The mean age of Xp11.2 tRCC patients was 36.40 ± 13.44 years (range 21–71 years), and young female patients were more common in Xp11.2 tRCC. Conversely, ccRCC, pRCC, and chRCC affected older men more frequently. The mean diameter of the lesion for Xp11.2 tRCC patients was 4.70 ± 3.23 cm, and there was no significant difference in tumor size between Xp11.2 tRCC and the three main RCCs. The clinicopathologic records, such as hematuria, lumbago, abdominal mass, hypertension, and hyperuricemia, were retrospectively investigated and listed as categorical variables in the models. Patient demographics and kidney cancer subtypes distributions for training or testing set were summarized in Table 2. Thirty-seven cases with three CT phases were randomly selected to test these models, and the remaining cases were performed on the training test. The mean tumor size and kidney cancer subtypes distributions were not significantly different between the training and testing sets. The age and gender distribution in the training or testing set were consistent with the entire cohort.
3.2 Predictive Performance Metrics
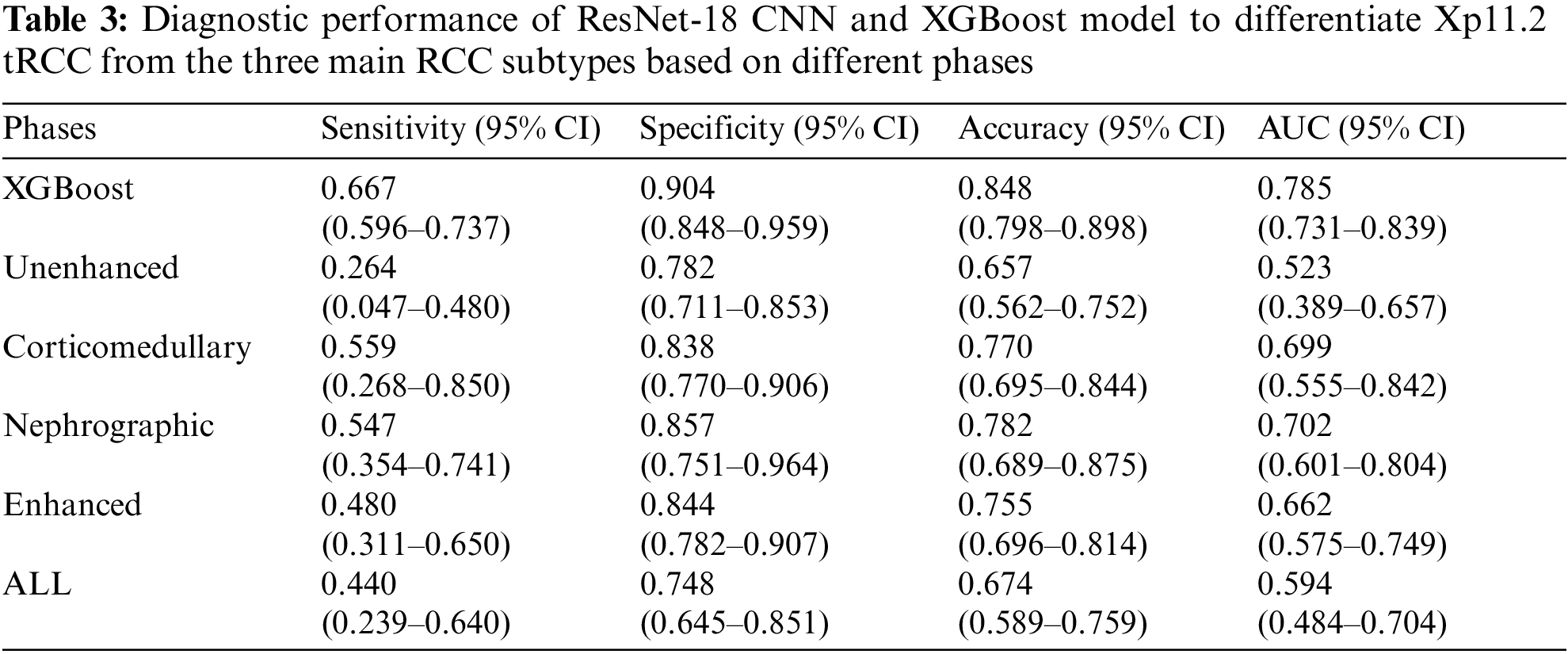
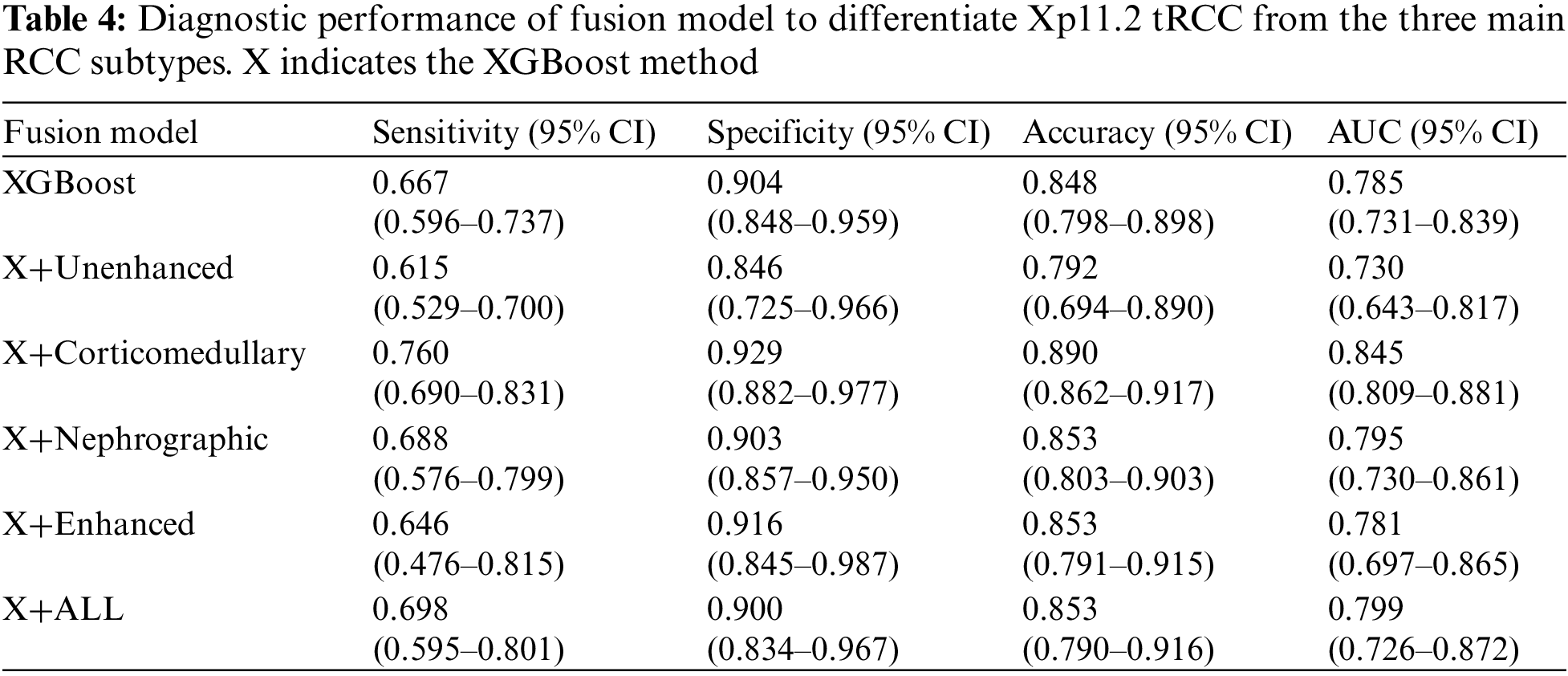
During the training stage, 30 epochs were performed to converge the ResNet-18 CNN. For prediction using a ResNet-18 CNN model, the subtypes of RCCs were differentiated by the average probability scores of their image patches. The predictive performance metrics of the ResNet-18 CNN model in classifying subtypes of RCCs during the testing stage were displayed in Table 3. Among the predictive performance metrics of the ResNet-18 CNN model based on different CT phases, the average sensitivity, specificity, accuracy, and AUC were 0.547 (95% CI, 0.354–0.741), 0.857 (95% CI, 0.751–0.964), 0.782 (95% CI, 0.689–0.875) and 0.702 (95% CI, 0.601–0.804) respectively in the nephrographic phase, which exhibited the high performance in comparison with single phase. However, when we ensembled the classification results of CT images in different phases (enhanced phases or all phases), we found that the model’s performance trained by enhanced phases or all phases data did not improve further. Surprisingly, compared with ResNet-18 CNN model, XGBoost model trained by clinical data achieved the top performance with the average sensitivity, specificity, accuracy and AUC: 0.667 (95% CI, 0.596–0.737), 0.904 (95% CI, 0.848–0.959), 0.848 (95% CI, 0.798–0.898) and 0.785 (95% CI, 0.731–0.839), respectively. In addition, the diagnostic performance of the ResNet-18 CNN fused XGBoost model to differentiate Xp11.2 tRCC from three main RCC subtypes is shown in Table 4. The ensemble model trained by unenhanced phase and clinical data achieved a test sensitivity of 0.615 (95% CI, 0.529–0.700), specificity of 0.846 (95% CI, 0.725–0.966), accuracy of 0.792 (95% CI, 0.694–0.890), precision-recall AUC of 0.730 (95% CI, 0.643–0.817), which showed better performance than the ResNet-18 CNN model trained by unenhanced phase. Similarly, other ensemble models outperform the compared ResNet-18 CNN model. The ensemble model trained by corticomedullary phase and clinical data achieved optimal performance with a test sensitivity of 0.760 (95% CI, 0.690–0.831), specificity of 0.929 (95% CI, 0.882–0.977), accuracy of 0.890 (95% CI, 0.862–0.917), precision-recall AUC of 0.845 (95% CI, 0.809–0.881).
3.3 Performance Compared between the Ensemble Model and Urologists
The confusion matrices for the ensemble model and three individual urologists were depicted in Figs. 3a–3d. Urologist 1 achieved a test sensitivity of 0.495 (95% CI, 0.128–0.861), specificity of 0.830 (95% CI, 0.655–1.004) and accuracy of 0.744 (95% CI, 0.661–0.826). Urologist 2 had a test sensitivity of 0.492 (95% CI, 0.225–0.758), specificity of 0.829 (95% CI, 0.680–0.979) and accuracy of 0.744 (95% CI, 0.681–0.806). Urologist 3 had a test sensitivity of 0.414 (95% CI, 0.001–0.827), specificity of 0.802 (95% CI, 0.640–0.964) and accuracy of 0.703 (95% CI, 0.614–0.791). Compared with all urologists averaged, the ensemble deep learning model had higher test sensitivity (0.667 vs. 0.467, P = 0.0036, specificity (0.893 vs. 0.820, P = 0.2173, and accuracy (0.838 vs. 0.730, P = 0.0272). Moreover, the results displayed that Xp11.2 tRCC, pRCC, and chRCC were frequently misclassified as ccRCC by the urologists, whereas they were more correctly predicted by the ensemble deep learning model. As was depicted in Figs. 3e–3g, compared to the average urologists, the ensemble model demonstrated statistically significant improvements in sensitivity for pRCC and chRCC (P = 0.0167 and P < 0.05, respectively) and even in specificity for ccRCC (P = 0.0056). In addition, there were statistically significant improvements in accuracy for ccRCC and chRCC (P = 0.0038 and P = 0.0027). The receiver operating characteristic (ROC) curves of the ensemble model and the performance of the urologists are depicted in Fig. 4. The AUCs with 95% CI for each curve were computed. The ensemble model achieved an average AUC of 0.799 (95% CI, 0.726–0.872) and outperformed the urologists in most cases. Especially most of the points indicating the performance of the urologists fell on or below the ROC curves of the ensemble model for four subtypes of RCCs.
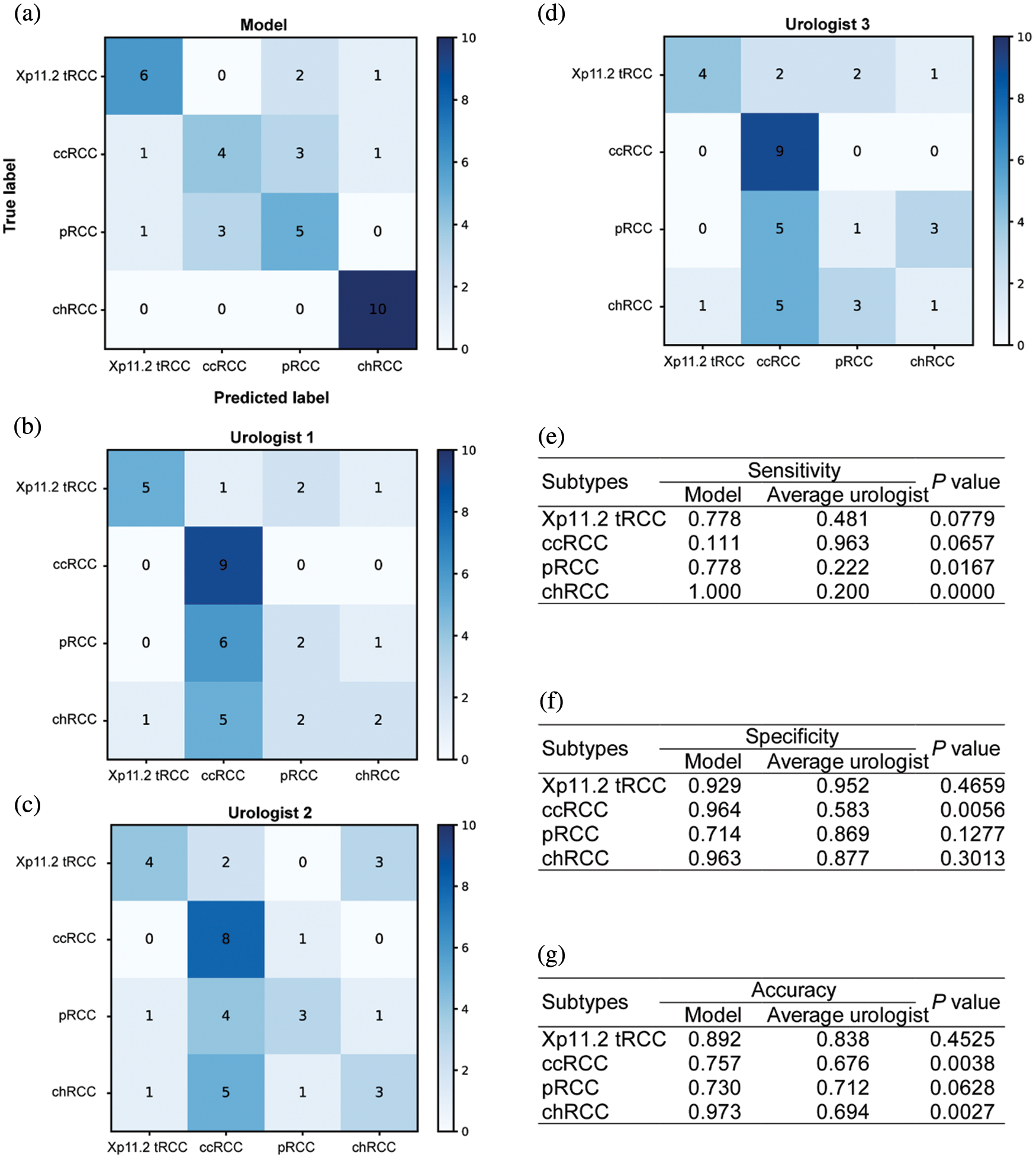
Figure 3: Diagnostic performance of the ensemble model and individual urologists for kidney cancer. Confusion matrices for the ensemble model (a) and individual urologists (b–d). The ensemble model compares with urologists, including the sensitivity (e), specificity (f), and accuracy (g)
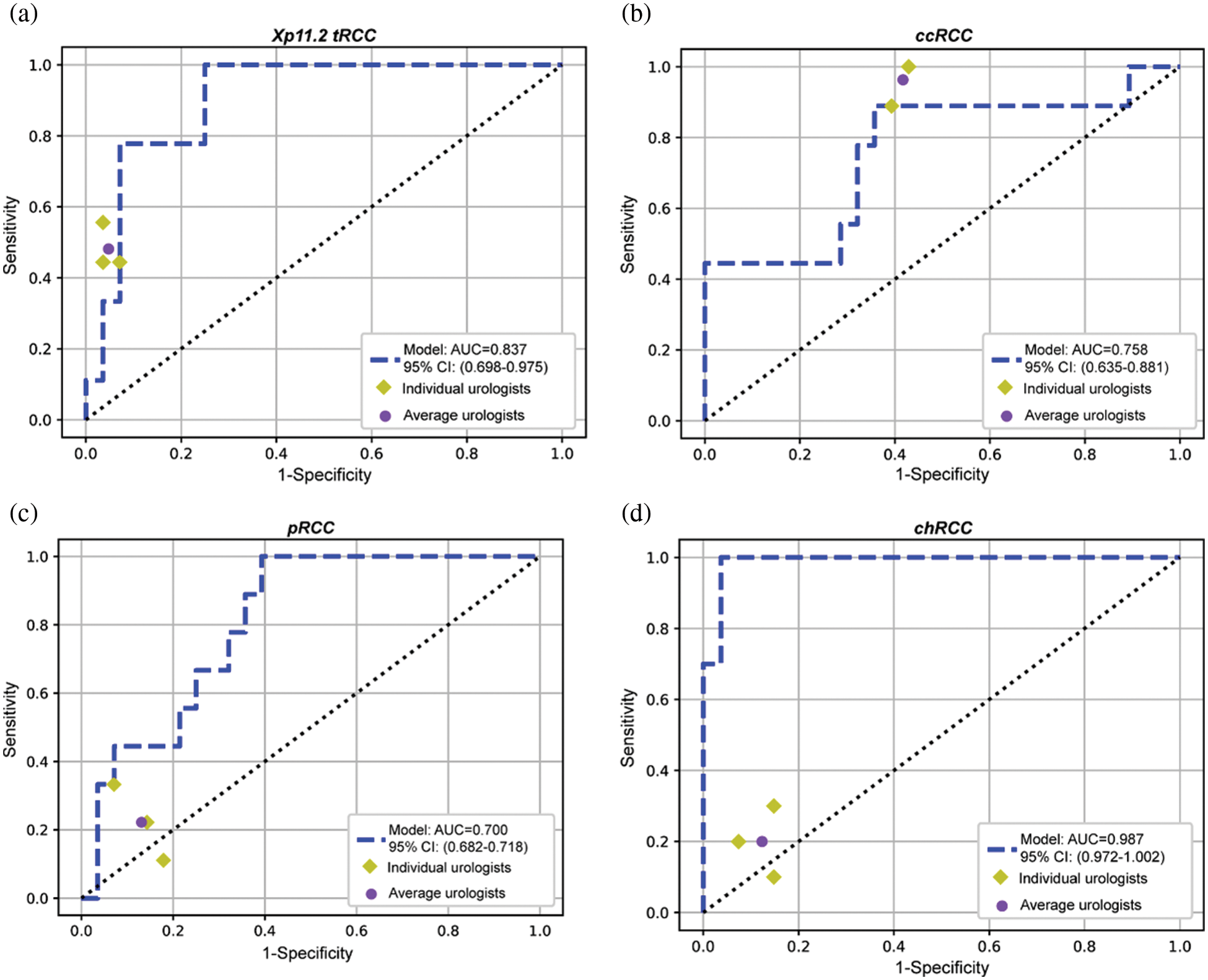
Figure 4: The ROC curves of the ensemble model and the performance of the three urologists are plotted for four tumor subtypes, including Xp11.2 tRCC (a), ccRCC (b), pRCC (c), and chRCC (d)
Compared with other common subtypes of renal tumor, Xp11.2 tRCC is highly aggressive and has a poor clinical prognosis. Therefore, the treatment for Xp11.2 tRCC should vary. However, the current diagnosis of Xp11.2 tRCC depends on postoperative pathology, and there are still no effective preoperative diagnosis methods. As the understanding and acceptance of AI grows in health and medicine, so does our imagination in ways to improve diagnostic accuracy, expedite clinical processes, and decrease human resource costs by assisting medical professionals in what once were time-consuming problems [21]. Therefore, we proposed a preoperative diagnosis prediction model based on an artificial intelligence algorithm. In the present study, we described automated methods based on decision fusion of a slice-based ResNet-18 CNN or clinical data-based XGBoost model to differentiate Xp11.2 tRCC from the three main RCC subtypes on multi-phase abdominal computed tomography. Also, we evaluated the performance of the ensemble model from both the ResNet-18 CNN model and the XGBoost model for the classification of renal masses. Although the ensemble model slightly outperformed the urologists based on small samples, our proposed deep learning methods assisted them in swiftly and noninvasively distinguishing Xp11.2 tRCC. For experienced urologists, the proposed method may be useful for reducing workload and increasing consistency of diagnostic results, and it also really does help those who are not fully trained in urology and shorten their learning curves. To our knowledge, a definitive diagnosis is vital for Xp11.2 tRCC, which has different surgical plans. The study is the first to investigate the use of artificial neural networks to differentiate Xp11.2 tRCC from common RCC subtypes on multiphasic CT and thus provides individualized treatment plans. In addition, our data consisting of CT images and detailed medical history is extremely intact, which is beneficial to the accuracy and reliability of our model.
Artificial neural networks have been increasingly and successfully applied to image recognition diagnostics, personalized medicine, and clinical decision-making. In recent years, powerful CNN has been used to explore complex interactions in clinical and imaging data to provide diagnosis, treatment planning, and prognosis for RCCs. For example, Uhm et al. proposed a DL model for differentiating five major histologic subtypes of renal tumors that outperformed radiologists for most subtypes, achieving an AUC of 0.889 [11], and ML also showed the same performance according to the literature [22,23]. In our study, the ensemble deep learning model showed good performance for discriminating Xp11.2 tRCC from common RCC subtypes with a test AUC of 0.845, and it also achieved similar or better diagnostic performance than urologists. Additionally, the previous study has shown that deep learning models trained by CT images can accurately distinguish between high and low nuclear grades for ccRCC with the accuracy of 0.82 [24], but more studies displayed that ML models constructed from CT imaging texture features can accurately predict the nuclear grades for ccRCC or pRCC with good performance [25–28]. We speculated that ML has much better performance than DL for nuclear-grade prediction, but ML based on artificial neural networks showed the greatest accuracy for differentiating low- and high-grade for ccRCCs [29]. Moreover, DL was also applied to doing treatment planning. Chen et al. evaluated epithelial-mesenchymal transition molecular classification of ccRCC tissue to predict prognosis and the effect of immunotherapy using DL convolutional neural networks [30], and a commercial AI-based contouring model was trained to provide prostate segmentation and demonstrated good performance in the implementation of an automated prostate treatment planning process [31]. Another important application of AI is the prediction of prognosis for RCCs. The AI prediction model based on multiple gene expression signatures has been developed to predict overall survival in ccRCC [32], and most previous studies also proposed AI model-based histopathological images or clinical data to evaluate prognosis for RCC patients [33–35]. Although the application of AI has been wildly studied in ccRCC, it deserves further exploration in other more aggressive renal cancer subtypes, such as pRCC, renal collecting duct carcinoma, and Xp11.2 tRCC. These are promising and broad prospects for developing AI models of histopathologic subtypes, nuclear grade, therapy response, and prognosis, which benefit patients.
XGBoost is a highly effective ML method used widely by data scientists to achieve state-of-the-art results on many ML challenges. As a novel type of ML algorithm, XGBoost can achieve better performance and accuracy than ML algorithms, and it comes from an improved gradient lifting decision tree and has great advantages in preventing over-fitting, parallel processing, cross-validation, and processing missing values [36]. XGBoost is growingly gaining popularity in health and medicine and is applied to predict cancer early diagnosis, subtypes, and prognosis. Schieda et al. proposed the XGBoost ML model trained by texture analysis features from CT images to distinguish ccRCC from other RCC subtypes or benign tumors with high performance [37]. The results from our study displayed that the XGBoost model constructed from clinical data also achieved the top performance with an average AUC of 0.785, which had similar or better diagnostic performance than previous studies. Furthermore, the XGBoost model was applied to predict the overall survival of genitourinary cancer patients in recent studies [38,39]. Wei et al. designed the XGBoost model based on clinical data to predict the prognosis of renal collecting duct carcinoma, and the results implied that the models had the highest predictive accuracy and helped clinicians to make clinical decisions for patients [40]. Consistent with our study, clinical data should be considered a very important basis for diagnosing diseases when building models. Although some clinical data may be subjective, they still have reference value. Our results showed that the performance of the ResNet-18 CNN model was not very outstanding, but the clinical characteristics between Xp11.2 tRCC and the common subtypes of RCCs were significantly different. Therefore, clinical data were used to train the XGBoost model, and finally, the ensemble model was established and achieved great performance than urologists, especially in Xp11.2 tRCC, pRCC, and chRCC.
Despite several strengths, our research has certain limitations. Firstly, the tumor location cannot be automatically detected by our model, and manually segmenting the renal lesions was time-consuming and labor-intensive work. Further study is required to combine automatic renal mass recognition strategies with our ensemble model. Secondly, the ensemble model was validated with our dataset; therefore, it lacks external validation. The applicability of the model in other datasets requires further validation. Thirdly, the study was retrospective, and the patients in the entire cohort were small and not consecutive. Therefore, it may introduce potential population bias, and validation of our model is needed to validate in other populations, especially in prospective and consecutive large clinical samples.
In this research, we proposed a ResNet-18 CNN combined with an XGBoost model that can achieve urologist-level performance for distinguishing Xp11.2 tRCC from three main RCC subtypes using CT images and clinical data. Moreover, the ensemble model successfully performed fine-grained classification of renal tumors into four pathological subtypes, including rare and common kidney cancer. These results highlight the potential for the ensemble model to assist urologists in diagnosing Xp11.2 tRCC patients. Further studies with a larger number of cases will be needed to validate the applicability of the model in clinical practice and required to validate in further research with large prospective clinical data. Additionally, we believe the presented DL model could also be applied to analyze other cancer types and other multimodality imaging, such as magnetic resonance imaging and contrast-enhanced ultrasound.
Acknowledgement: Thank you for the technical support from Dr. Lei Yang, Dr. Yindong Zheng, and Dr. Guo Chen.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding Statement: This study was supported by Beijing Ronghe Medical Development Foundation.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Moch, H., Cubilla, A. L., Humphrey, P. A., Reuter, V. E., Ulbright, T. M. (2016). The 2016 who classification of tumours of the urinary system and male genital organs–Part A: Renal, penile, and testicular tumours. European Urology, 70(1), 93–105. DOI 10.1016/j.eururo.2016.02.029. [Google Scholar] [CrossRef]
2. Ledezma, R. A., Negron, E., Paner, G. P., Rjepaj, C., Lascano, D. et al. (2016). Clinically localized type 1 and 2 papillary renal cell carcinomas have similar survival outcomes following surgery. World Journal of Urology, 34(5), 687–693. DOI 10.1007/s00345-015-1692-3. [Google Scholar] [CrossRef]
3. Tsui, K. H., Shvarts, O., Smith, R. B., Figlin, R. A., deKernion, J. B. et al. (2000). Prognostic indicators for renal cell carcinoma: A multivariate analysis of 643 patients using the revised 1997 TNM staging criteria. The Journal of Urology, 163(4), 1090–1095. DOI 10.1016/S0022-5347(05)67699-9. [Google Scholar] [CrossRef]
4. Volpe, A., Novara, G., Antonelli, A., Bertini, R., Billia, M. et al. (2012). Chromophobe renal cell carcinoma (RCCOncological outcomes and prognostic factors in a large multicentre series. BJU International, 110(1), 76–83. DOI 10.1111/j.1464-410X.2011.10690.x. [Google Scholar] [CrossRef]
5. Wang, X. T., Xia, Q. Y., Zhou, X. J., Rao, Q. (2017). Incidence, clinicopathological features and fusion transcript landscape of translocation renal cell carcinomas. Histopathology, 71(5), 835–836. DOI 10.1111/his.13256. [Google Scholar] [CrossRef]
6. Argani, P. (2015). MiT family translocation renal cell carcinoma. Seminars in Diagnostic Pathology, 32(2), 103–113. DOI 10.1053/j.semdp.2015.02.003. [Google Scholar] [CrossRef]
7. Argani, P., Olgac, S., Tickoo, S. K., Goldfischer, M., Moch, H. et al. (2007). Xp11 translocation renal cell carcinoma in adults: Expanded clinical, pathologic, and genetic spectrum. The American Journal of Surgical Pathology, 31(8), 1149–1160. DOI 10.1097/PAS.0b013e318031ffff. [Google Scholar] [CrossRef]
8. Xu, L., Yang, R., Gan, W., Chen, X., Qiu, X. et al. (2015). Xp11.2 translocation renal cell carcinomas in young adults. BMC Urology, 15(1), 1–6. [Google Scholar]
9. Liu, N., Qu, F., Shi, Q., Zhuang, W., Ma, W. et al. (2021). Nephron-sparing surgery for adult Xp11.2 translocation renal cell carcinoma at clinical t1 stage: A multicenter study in China. Annals of Surgical Oncology, 28(2), 1238–1246. DOI 10.1245/s10434-020-08813-y. [Google Scholar] [CrossRef]
10. Ljungberg, B., Albiges, L., Abu-Ghanem, Y., Bensalah, K., Dabestani, S. et al. (2019). European association of urology guidelines on renal cell carcinoma: The 2019 update. European Urology, 75(5), 799–810. DOI 10.1016/j.eururo.2019.02.011. [Google Scholar] [CrossRef]
11. Uhm, K. H., Jung, S. W., Choi, M. H., Shin, H. K., Yoo, J. I. et al. (2021). Deep learning for end-to-end kidney cancer diagnosis on multi-phase abdominal computed tomography. npj Precision Oncology, 5(1), 1–6. DOI 10.1038/s41698-021-00195-y. [Google Scholar] [CrossRef]
12. Wang, S. H., Tang, C., Sun, J., Yang, J., Huang, C. et al. (2018). Multiple sclerosis identification by 14-layer convolutional neural network with batch normalization, dropout, and stochastic pooling. Frontiers in Neuroscience, 12, 818. DOI 10.3389/fnins.2018.00818. [Google Scholar] [CrossRef]
13. Wang, S. H., Zhang, Y. D. (2020). Densenet-201-based deep neural network with composite learning factor and precomputation for multiple sclerosis classification. ACM Transactions on Multimedia Computing, Communications, and Applications, 16(2 s), 1–19. DOI 10.1145/3341095. [Google Scholar] [CrossRef]
14. Zhang, Y. D., Pan, C., Chen, X., Wang, F. (2018). Abnormal breast identification by nine-layer convolutional neural network with parametric rectified linear unit and rank-based stochastic pooling. Journal of Computational Science, 27, 57–68. DOI 10.1016/j.jocs.2018.05.005. [Google Scholar] [CrossRef]
15. Baghdadi, A., Aldhaam, N. A., Elsayed, A. S., Hussein, A. A., Cavuoto, L. A. et al. (2020). Automated differentiation of benign renal oncocytoma and chromophobe renal cell carcinoma on computed tomography using deep learning. BJU International, 125, 553–560. DOI 10.1111/bju.14985. [Google Scholar] [CrossRef]
16. Coy, H., Hsieh, K., Wu, W., Nagarajan, M. B., Young, J. R. et al. (2019). Deep learning and radiomics: The utility of google tensorflow™ inception in classifying clear cell renal cell carcinoma and oncocytoma on multiphasic CT. Abdominal Radiology, 44(6), 2009–2020. DOI 10.1007/s00261-019-01929-0. [Google Scholar] [CrossRef]
17. Cui, E. M., Lin, F., Li, Q., Li, R. G., Chen, X. M. et al. (2019). Differentiation of renal angiomyolipoma without visible fat from renal cell carcinoma by machine learning based on whole-tumor computed tomography texture features. Acta Radiologica, 60(11), 1543–1552. DOI 10.1177/0284185119830282. [Google Scholar] [CrossRef]
18. Suarez-Ibarrola, R., Hein, S., Reis, G., Gratzke, C., Miernik, A. (2020). Current and future applications of machine and deep learning in urology: A review of the literature on urolithiasis, renal cell carcinoma, and bladder and prostate cancer. World Journal of Urology, 38(10), 2329–2347. DOI 10.1007/s00345-019-03000-5. [Google Scholar] [CrossRef]
19. Cheng, J., Han, Z., Mehra, R., Shao, W., Cheng, M. et al. (2020). Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11. 2 translocation renal cell carcinoma. Nature Communications, 11(1), 1–9. [Google Scholar]
20. Deng, J., Dong, W., Socher, R., Li, L., Li, K. et al. (2009). Imagenet: A large-scale hierarchical image database. CVPR, 248–255. DOI 10.1109/CVPR.2009.5206848. [Google Scholar] [CrossRef]
21. Tran, B. X., Vu, G. T., Ha, G. H., Vuong, Q. H., Ho, M. T. et al. (2019). Global evolution of research in artificial intelligence in health and medicine: A bibliometric study. Journal of Clinical Medicine, 8(3), 360. DOI 10.3390/jcm8030360. [Google Scholar] [CrossRef]
22. Feng, Z., Rong, P., Cao, P., Zhou, Q., Zhu, W. et al. (2018). Machine learning-based quantitative texture analysis of ct images of small renal masses: Differentiation of angiomyolipoma without visible fat from renal cell carcinoma. European Radiology, 28(4), 1625–1633. DOI 10.1007/s00330-017-5118-z. [Google Scholar] [CrossRef]
23. Yan, L., Liu, Z., Wang, G., Huang, Y., Liu, Y. et al. (2015). Angiomyolipoma with minimal fat: Differentiation from clear cell renal cell carcinoma and papillary renal cell carcinoma by texture analysis on CT images. Academic Radiology, 22(9), 1115–1121. DOI 10.1016/j.acra.2015.04.004. [Google Scholar] [CrossRef]
24. Lin, F., Ma, C., Xu, J., Lei, Y., Li, Q. et al. (2020). A CT-based deep learning model for predicting the nuclear grade of clear cell renal cell carcinoma. European Journal of Radiology, 129, 109079. DOI 10.1016/j.ejrad.2020.109079. [Google Scholar] [CrossRef]
25. Haji-Momenian, S., Ricker, R., Chen, Z., Houser, M., Adusumilli, N. et al. (2021). Prediction of histologic grade and type of small (<4 cm) papillary renal cell carcinomas using texture and neural network analysis: A feasibility study. Abdominal Radiology, 46(9), 4266–4277. DOI 10.1007/s00261-021-03044-5. [Google Scholar] [CrossRef]
26. Lai, S., Sun, L., Wu, J., Wei, R., Luo, S. et al. (2021). Multiphase contrast-enhanced ct-based machine learning models to predict the fuhrman nuclear grade of clear cell renal cell carcinoma. Cancer Management and Research, 13, 999–1008. DOI 10.2147/CMAR.S290327. [Google Scholar] [CrossRef]
27. Luo, S., Wei, R., Lu, S., Lai, S., Wu, J. et al. (2022). Fuhrman nuclear grade prediction of clear cell renal cell carcinoma: Influence of volume of interest delineation strategies on machine learning-based dynamic enhanced ct radiomics analysis. European Radiology, 32(4), 2340–2350. DOI 10.1007/s00330-021-08322-w. [Google Scholar] [CrossRef]
28. Xv, Y., Lv, F., Guo, H., Zhou, X., Tan, H. et al. (2021). Machine learning-based ct radiomics approach for predicting who/isup nuclear grade of clear cell renal cell carcinoma: An exploratory and comparative study. Insights into Imaging, 12(1), 1–14. DOI 10.1186/s13244-021-01107-1. [Google Scholar] [CrossRef]
29. He, X., Wei, Y., Zhang, H., Zhang, T., Yuan, F. et al. (2020). Grading of clear cell renal cell carcinomas by using machine learning based on artificial neural networks and radiomic signatures extracted from multidetector computed tomography images. Academic Radiology, 27(2), 157–168. DOI 10.1016/j.acra.2019.05.004. [Google Scholar] [CrossRef]
30. Chen, Q., Kuai, Y., Wang, S., Zhu, X., Wang, H. et al. (2021). Deep learning–based classification of epithelial–mesenchymal transition for predicting response to therapy in clear cell renal cell carcinoma. Frontiers in Oncology, 11, 782515. DOI 10.3389/fonc.2021.782515. [Google Scholar] [CrossRef]
31. Duan, J., Bernard, M., Downes, L., Willows, B., Feng, X. et al. (2022). Evaluating the clinical acceptability of deep learning contours of prostate and organs-at-risk in an automated prostate treatment planning process. Medical Physics, 49(4), 2570–2581. DOI 10.1002/mp.15525. [Google Scholar] [CrossRef]
32. Peng, Q., Shen, Y., Fu, K., Dai, Z., Jin, L. et al. (2021). Artificial intelligence prediction model for overall survival of clear cell renal cell carcinoma based on a 21-gene molecular prognostic score system. Aging, 13(5), 7361–7381. DOI 10.18632/aging.202594. [Google Scholar] [CrossRef]
33. Tabibu, S., Vinod, P., Jawahar, C. (2019). Pan-renal cell carcinoma classification and survival prediction from histopathology images using deep learning. Scientific Reports, 9(1), 1–9. DOI 10.1038/s41598-019-46718-3. [Google Scholar] [CrossRef]
34. Marostica, E., Barber, R., Denize, T., Kohane, I. S., Signoretti, S. et al. (2021). Development of a histopathology informatics pipeline for classification and prediction of clinical outcomes in subtypes of renal cell carcinoma. Clinical Cancer Research, 27(10), 2868–2878. DOI 10.1158/1078-0432.CCR-20-4119. [Google Scholar] [CrossRef]
35. Chen, S., Zhang, N., Jiang, L., Gao, F., Shao, J. et al. (2021). Clinical use of a machine learning histopathological image signature in diagnosis and survival prediction of clear cell renal cell carcinoma. International Journal of Cancer, 148(3), 780–790. DOI 10.1002/ijc.33288. [Google Scholar] [CrossRef]
36. Chen, T., Guestrin, C. (2016). Xgboost: A scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 785–794. San Francisco, CA, USA, ACM. DOI 10.1145/2939672.2939785. [Google Scholar] [CrossRef]
37. Schieda, N., Nguyen, K., Thornhill, R. E., McInnes, M. D., Wu, M. et al. (2020). Importance of phase enhancement for machine learning classification of solid renal masses using texture analysis features at multi-phasic CT. Abdominal Radiology, 45(9), 2786–2796. DOI 10.1007/s00261-020-02632-1. [Google Scholar] [CrossRef]
38. Nazari, M., Shiri, I., Zaidi, H. (2021). Radiomics-based machine learning model to predict risk of death within 5-years in clear cell renal cell carcinoma patients. Computers in Biology and Medicine, 129, 104135. DOI 10.1016/j.compbiomed.2020.104135. [Google Scholar] [CrossRef]
39. Nayan, M., Salari, K., Bozzo, A., Ganglberger, W., Carvalho, F. et al. (2021). Predicting survival after radical prostatectomy: Variation of machine learning performance by race. The Prostate, 81(16), 1355–1364. DOI 10.1002/pros.24233. [Google Scholar] [CrossRef]
40. Wei, L., Huang, Y., Chen, Z., Li, J., Huang, G. et al. (2021). A novel machine learning algorithm combined with multivariate analysis for the prognosis of renal collecting duct carcinoma. Frontiers in Oncology, 11, 777735. DOI 10.3389/fonc.2021.777735. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools