 Open Access
Open Access
ARTICLE
Inner Cascaded U2-Net: An Improvement to Plain Cascaded U-Net
1 Tongji University, Shanghai, 201804, China
2 Jiading District Central Hospital Affiliated to Shanghai University of Medicine and Health Sciences, Shanghai, 201800, China
* Corresponding Author: Guanjun Liu. Email:
Computer Modeling in Engineering & Sciences 2023, 134(2), 1323-1335. https://doi.org/10.32604/cmes.2022.020428
Received 23 November 2021; Accepted 14 March 2022; Issue published 31 August 2022
Abstract
Deep neural networks are now widely used in the medical image segmentation field for their performance superiority and no need of manual feature extraction. U-Net has been the baseline model since the very beginning due to a symmetrical U-structure for better feature extraction and fusing and suitable for small datasets. To enhance the segmentation performance of U-Net, cascaded U-Net proposes to put two U-Nets successively to segment targets from coarse to fine. However, the plain cascaded U-Net faces the problem of too less between connections so the contextual information learned by the former U-Net cannot be fully used by the latter one. In this article, we devise novel Inner Cascaded U-Net and Inner Cascaded U2-Net as improvements to plain cascaded U-Net for medical image segmentation. The proposed Inner Cascaded U-Net adds inner nested connections between two U-Nets to share more contextual information. To further boost segmentation performance, we propose Inner Cascaded U2-Net, which applies residual U-block to capture more global contextual information from different scales. The proposed models can be trained from scratch in an end-to-end fashion and have been evaluated on Multimodal Brain Tumor Segmentation Challenge (BraTS) 2013 and ISBI Liver Tumor Segmentation Challenge (LiTS) dataset in comparison to related U-Net, cascaded U-Net, U-Net++, U2-Net and state-of-the-art methods. Our experiments demonstrate that our proposed Inner Cascaded U-Net and Inner Cascaded U2-Net achieve better segmentation performance in terms of dice similarity coefficient and hausdorff distance as well as get finer outline segmentation.Keywords
The first deep neural network applied in image segmentation field is Fully Convolutional Network (FCN) proposed by Long et al. in 2015 [1]. FCN tackles the challenge of semantic image segmentation by classifying each pixel as target or background and can accept input images of any size, whose variants get excellent segmentation performance on natural image segmentation field. However, the semantics of medical images are simpler compared to natural images and the structures are relatively fixed. The greatest difficulty in medical image segmentation field is the acquisition and labels of medical images, leading to a relatively small dataset. Thus, overfitting will be caused if the model is too large and contains overmuch parameters. The most well-known deep learning baseline model in medical image segmentation field is U-Net proposed by Ronneberger et al. in 2015 [2]. The main differences between FCN and U-Net can be summarized as two aspects, one is a symmetrical encoder-decoder structure of U-Net which combines the low-resolution features in the encoder and the high-resolution features in the decoder, the other is skip connections in U-Net to provide supplementary information in the up-sampling operations. And the model size of U-Net is relatively smaller. These features make U-Net been the first choice for medical image segmentation.
Since then, a mass of variants of U-Net have been proposed to boost its performance in various kinds of medical scenarios, including but not limited to improvements to skip pathways [3,4], more powerful and sophisticated U-blocks [5–9], cascaded U-structures [10–16], combining with attention mechanism [17–20] and so on.
Zhou et al. (UNet++) [3] proposed to redesign the skip connections using a sequence of conventional blocks to fill the gaps as opposed to straight connections in U-Net, which aims to decrease the semantic gap between the encoder and decoder feature maps. Huang et al. [4] further improved with full-scale skip connections to get performance gain with fewer parameters compared to UNet++, where each convolution block in the encoder is connected to the convolution block in the decoder of the same or lower level.
Zhang et al. [5] added residual connections in each convolution block, aiming to solve the problem of gradient vanishing and thus allowing to train deeper network. Ibtehaz et al. [6] propose MultiRes block to replace the two continuous convolution blocks. Inspired by Inception block [8], a sequence of three layers with 3 × 3 filters is applied to approximate 5 × 5 and 7 × 7 convolution operation, which is introduced to predict targets at different scales. Qin et al. [7] (U2-Net) developed a two-level nested U-structure for salient object detection by replacing conventional two convolutions in each level of U-Net with a residual U-block to extract intra-stage multi-scale features while remaining comparable memory and computation cost.
The most intuitive and simplest method to improve the segmentation performance is cascading several same or different U-Nets. The representation of cascaded U-Net architecture can be grossly divided into two groups, one is segmenting regions of interest firstly followed by target segmentation [10,11] and the other is rough pre-segmentation followed by explicit segmentation [12–16]. Christ et al. [10] first applied cascaded U-Net for automatic liver and lesion segmentation, in which the predicted liver ROIs segmented by the former U-Net are fed into the latter one as inputs to segment lesions only. Liu et al. [11] proposed Cascaded Atrous Dual-Attention UNet to leverage the inter-slice context information and emphasize the salient features of tumors. 3D AU-Net is first employed to get the coarse localization of the liver, whose prediction results are then concatenated with the input 3D feature map to preserve the z-axis information, followed by 2D ADAU-Net for precise tumor segmentation. Jiang et al. [12] proposed a two-stage cascaded 3D U-Net concentrating on the segmentation of substructures of brain tumors from rough to fine. Besides the coarse segmentation map from the first-stage U-Net, the raw images are fed together into the second U-Net too. Qin et al. [13] devised BASNet consisting of two U-Net-like networks adopted for salient object detection, with the prior U-Net learning to predict saliency map followed by residual refinement module which is also a U-Net refining the boundary. Li et al. [14] proposed a hybrid two-stage densely connected U-Net named H-DenseUNet for liver and tumor segmentation. The network includes a 2D DenseUNet for extracting intra-slice features due to memory limitation of 3D volume training and a 3D DenseUNet for extracting inter-slice features, which are fused eventually through a hybrid feature fusion layer. Liu et al. [15] devised a cascaded U-Net with residual mapping named CR-U-Net for liver segmentation, in which morphological techniques, including opening and closing operation are used between the two U-Nets as an intermediate-processing module to refine the boundary. Hu et al. [16] proposed a multi-cascaded CNN to capture multi-scale features on brain MRI images with multi-modality. Specifically, three sub-networks are trained separately to segment the brain tumor from the axial, coronal, and sagittal views, respectively. The segmentation results are then fused to get the final segmentation masks.
UNet++ [3] proposed in 2018 replaces the initial straight skip connections with a series of nested convolution blocks. Inspired by UNet++, we present Inner Cascaded U-Net, an improved cascaded network that places the encoder of the latter U-Net between the encoder and decoder rather than the end of the former U-Net. The inputs of each except the first and the last convolution block of the latter U-Net come from three aspects, which are the output from the previous layer of the same U-Net, the output from the former U-Net of the same level, and the corresponding up-sampled output of the lower convolution block of the former U-Net, respectively. By involving more between connections, the two U-Nets could share their learned features and the contextual information learned by the former U-Net can be incorporated into the latter one to enhance feature selecting and combination for better segmentation performance.
To further boost performance of Inner Cascaded U-Net, we propose Inner Cascaded U2-Net, which adopts residual U-block (RSU) proposed by Qin et al. [7] to replace the sequence of two plain convolution layers. A RSU block is a small U-Net like structure with residual connections, which can capture intra-stage multi-scale features while the residual connections allow to train deeper networks.
There are many improvements to plain U-Net from various aspects, among which cascaded U-Net adopts the most intuitive method to cascade two successive U-Nets for the consideration of extracting features from coarse to fine. The main purpose of this article is to analyze the deficiency of the plain cascaded way and propose an improved inner cascaded method, which outperforms cascaded U-Net in segmentation accuracy and outline refinement. The contributions of the paper can be summarized as follows:
• We analyze different ways to improve the network architecture of the baseline deep learning model U-Net and the drawback of plain cascaded way.
• We propose a novel U-Net like architecture named Inner Cascaded U-Net, which organizes two U-Nets in an inner cascaded way and adds more between connections.
• We further propose Inner Cascaded U2-Net, which distinguishes from Inner Cascaded U-Net by residual U-block as an improvement to plain convolution block.
In this section, we first analyze the problem faced by plain cascaded U-Net. Then, we introduce Inner Cascaded U-Net, an improved combination method of two successive U-Nets. The inputs of the encoder and decoder convolution block of the latter U-Net are defined, respectively. Finally, we refer residual U-block and incorporate it into the network to get Inner Cascaded U2-Net.
2.1 The Problem of Plain Cascaded U-Net
The conventional cascaded U-Net combines a sequence of two U-Nets in an end-to-end way to predict feature maps from coarse to fine. The latter U-Net plays a role in improving outline segmentation of the former U-Net by inputting roughly segmented image masks. As shown in Fig. 1, the red arrow between the two U-Nets denotes the between connection where the input feature maps of the latter U-Net are generated by the former one. However, beyond that there are no other connections of the two U-Nets, which means the features that the former U-Net extracted cannot be properly shared with the latter one.
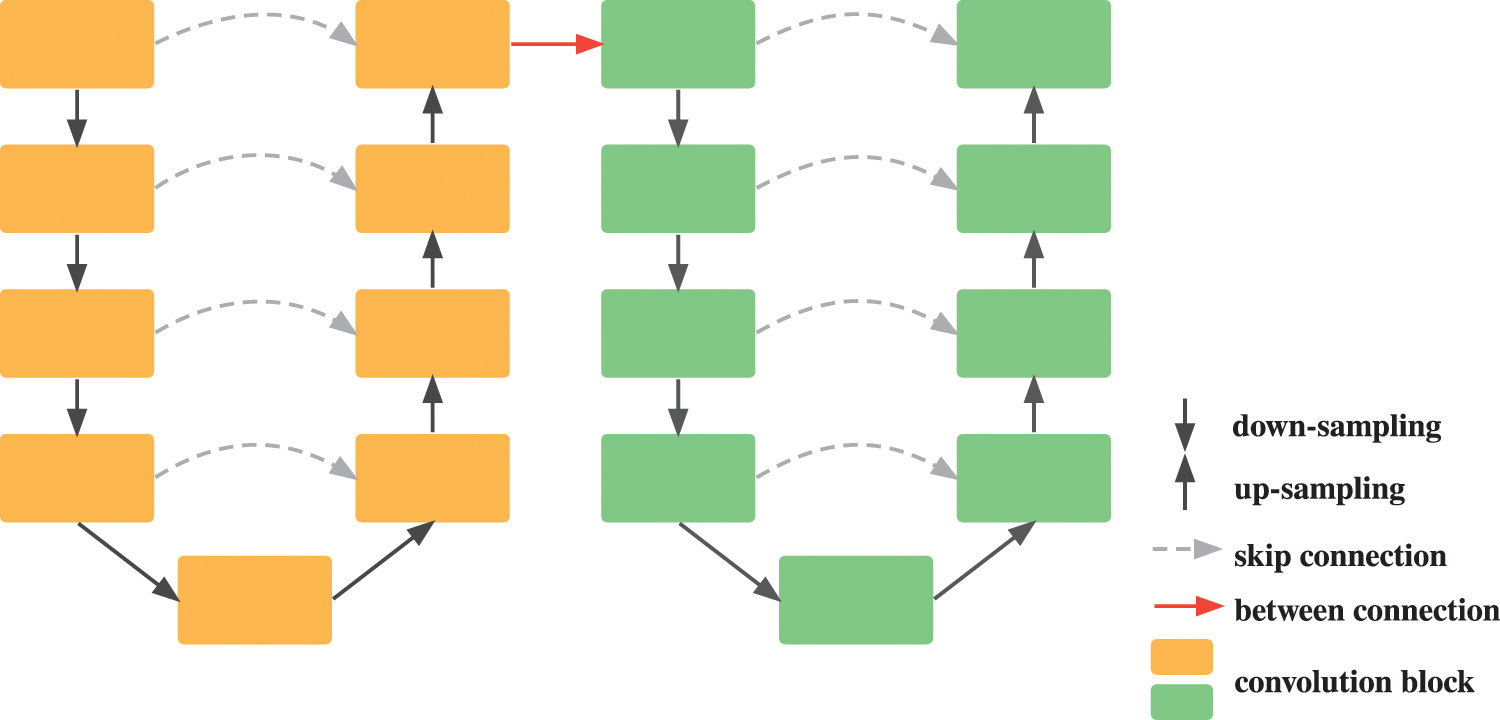
Figure 1: Illustration of cascaded U-Net architecture
Enlightened by the nested dense skip pathways of UNet++, we design our Inner Cascaded U-Net to fuse the two successive U-Nets into a single U-Net like network, as shown in Fig. 2. Besides the down-sampled feature maps of the higher layer, the inputs of the encoder convolution block in the latter U-Net also involve the output of the convolution block of the same layer and the corresponding up-sampled output of the lower layer in the former U-Net. Similarly, the inputs of the decoder convolution block comprise the output of the same layer in the former U-Net as well. UNet++ designs nested dense skip pathways in consideration of decreasing the semantic gap between the feature maps of the encoder and decoder. Here, we declare our purpose is to improve the cascade ways of two U-Nets by fusing more contextual information learned by the former U-Net into the latter one, instead of only taking the output of the former U-Net into consideration.
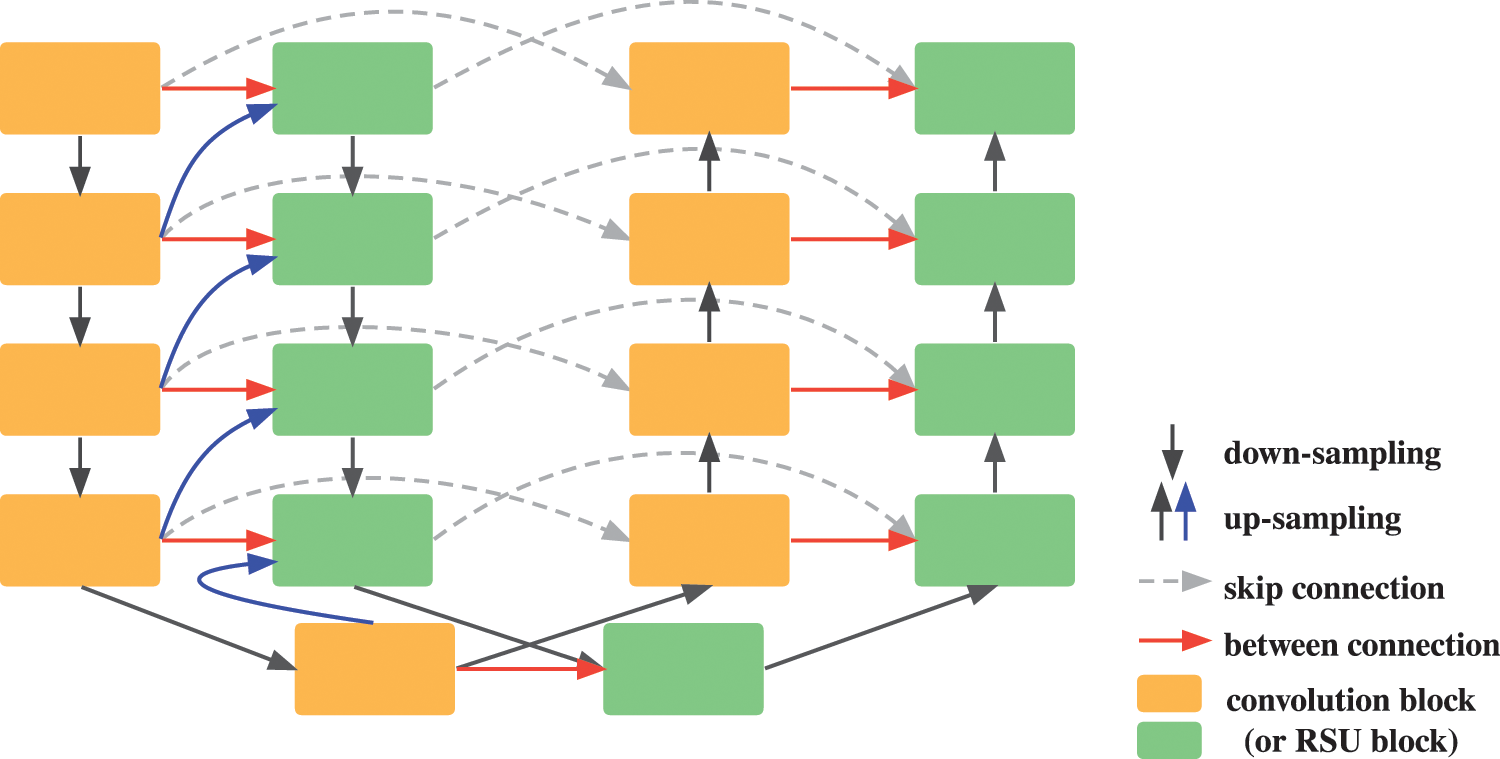
Figure 2: Illustration of our proposed inner cascaded U-Net architecture. The red and blue arrows denote added between connections from the former U-Net to the latter one
Formally, the inputs of the encoder convolution block which are represented as e in the latter U-Net can be formulated in Eq. (1):
where
While the inputs of the decoder convolution block which are represented as d in the latter U-Net are described in Eq. (2):
where all definitions are the same as encoder convolution block. Each encoder convolution block receives input feature maps from the corresponding encoder, the decoder of the former U-Net and the up-sampling feature maps from the lower decoder.
To further capture more global contextual information for better segmentation of small lesions in medical images, we propose Inner Cascaded U2-Net, which replaces the initial two successive convolution layers with Residual U-blocks (RSU) proposed in U2-Net [7]. The structure of RSU is shown in Fig. 3b, where H and W are the height and width of the input images,
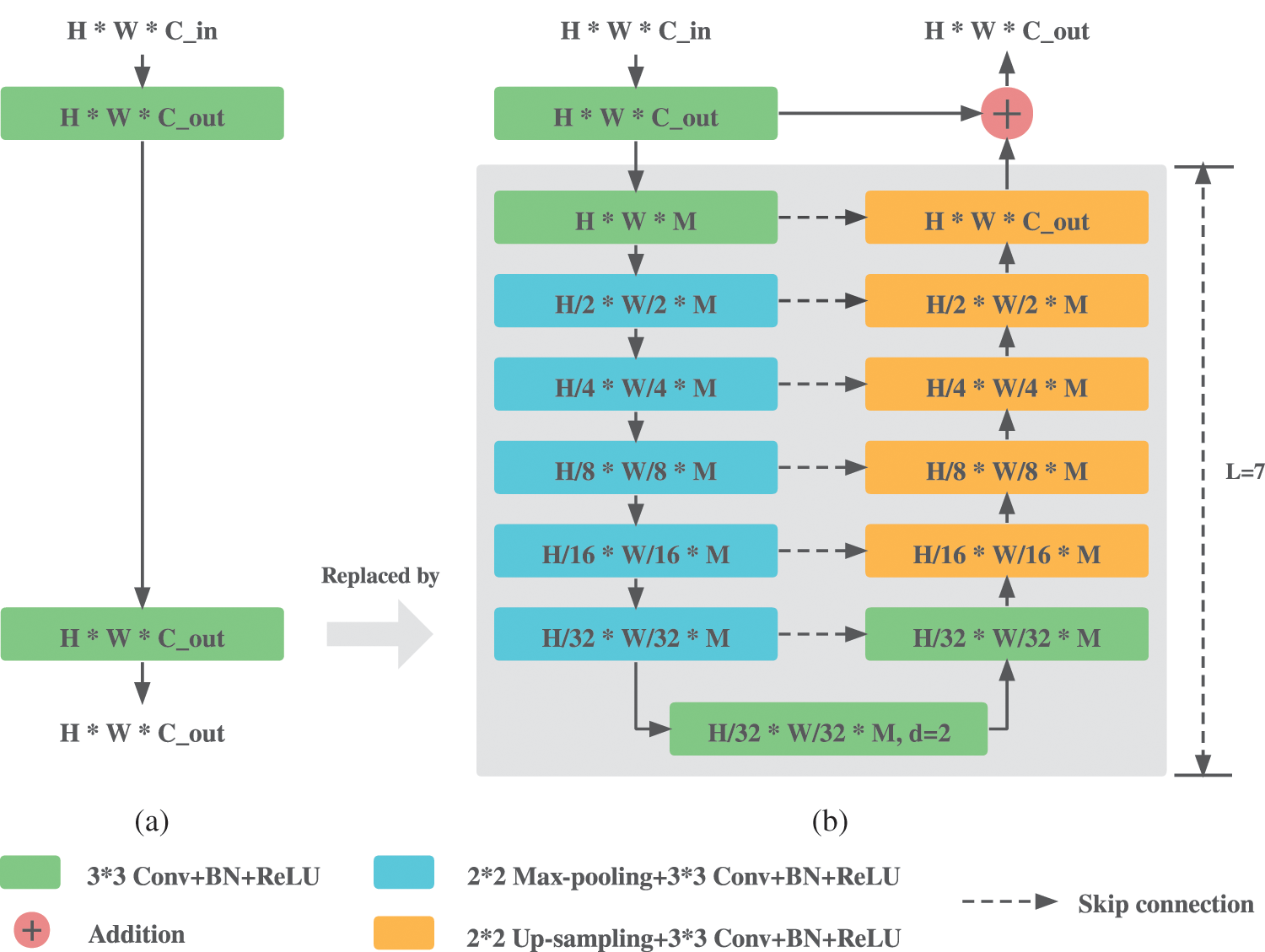
Figure 3: Comparisons between the plain convolution block and RSU. (a) Origin plain convolution block in U-Net. (b) Residual U-block RSU
Compared to the plain convolution block shown in Fig. 3a, RSU replaces the second convolution layer with a U-Net like network whose output is then added to the output of the first convolution block, aiming to extract multi-stage features at small memory and computation cost while keeping a consistent output dimension with plain convolution block. Deeper levels with larger L lead to larger receptive field and thus more multi-stage contextual information can be extracted during progressive down-sampling and up-sampling process. However, larger L also means more encoder and decoder layers, leading to more memory and computation cost. L is set to 7 as recommend in [7].
3 Experimental Results and Analysis
We conduct experiments and validate our methods on two public medical imaging datasets including brain MRI and liver CT. In this section, we first introduce the two datasets and preprocessing operations to each dataset, respectively. Then we present two evaluation metrics which are widely used in medical image segmentation field and the experiments environment in which we conduct our experiments. Finally, we give the performance analysis based on the quantitative and qualitative results.
3.1 Datasets and Preprocessing
The dataset for brain segmentation in MRI images is the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2013 dataset. The dataset contains four modal images for each patient, which are T1, T1c, T2, Flair, respectively. It is composed of real and synthetic images and we only use the real images, including 20 high-grade gliomas (HG) patients and 10 low-grade gliomas (LG) patients. There are 4 different types of brain tumor in the dataset, which are all regarded as tumor positive for simplicity. The four modal images of the same size 240 × 240 are first resized to 256 × 256 using bilinear interpolation and then concatenated to one single image with four channels to enhance the segmentation performance. Considering the dataset is relatively small, it is randomly divided into training, validation and testing set at 60%, 20% and 20%, resulting in 3361, 1110, 1014 images, respectively.
The dataset for liver segmentation in CT images is the ISBI Liver Tumor Segmentation Challenge (LiTS) 2017 dataset. The dataset contains 131 patients so we use more data for training in view of the abundant images in the dataset, of which the first 100, the next 15 and the last 15 except the last patients are used for training, validation and testing set, adding up to a total number of 13144, 3308 and 2518 images, respectively. The tumor areas are labelled as the liver for binary segmentation. And the CT values of the images in the dataset are preprocessed to liver window settings [−200, 250] to reduce distractions from other organs.
All images are normalized finally to set CT values between [0, 1] before inputting them to the network to accelerate the training process. The overall experiment pipeline on the BraTS 2013 and LiTS dataset is shown in Fig. 4, from which we can see the proposed architecture is trained in an end-to-end fashion with preprocessing steps as few as possible.
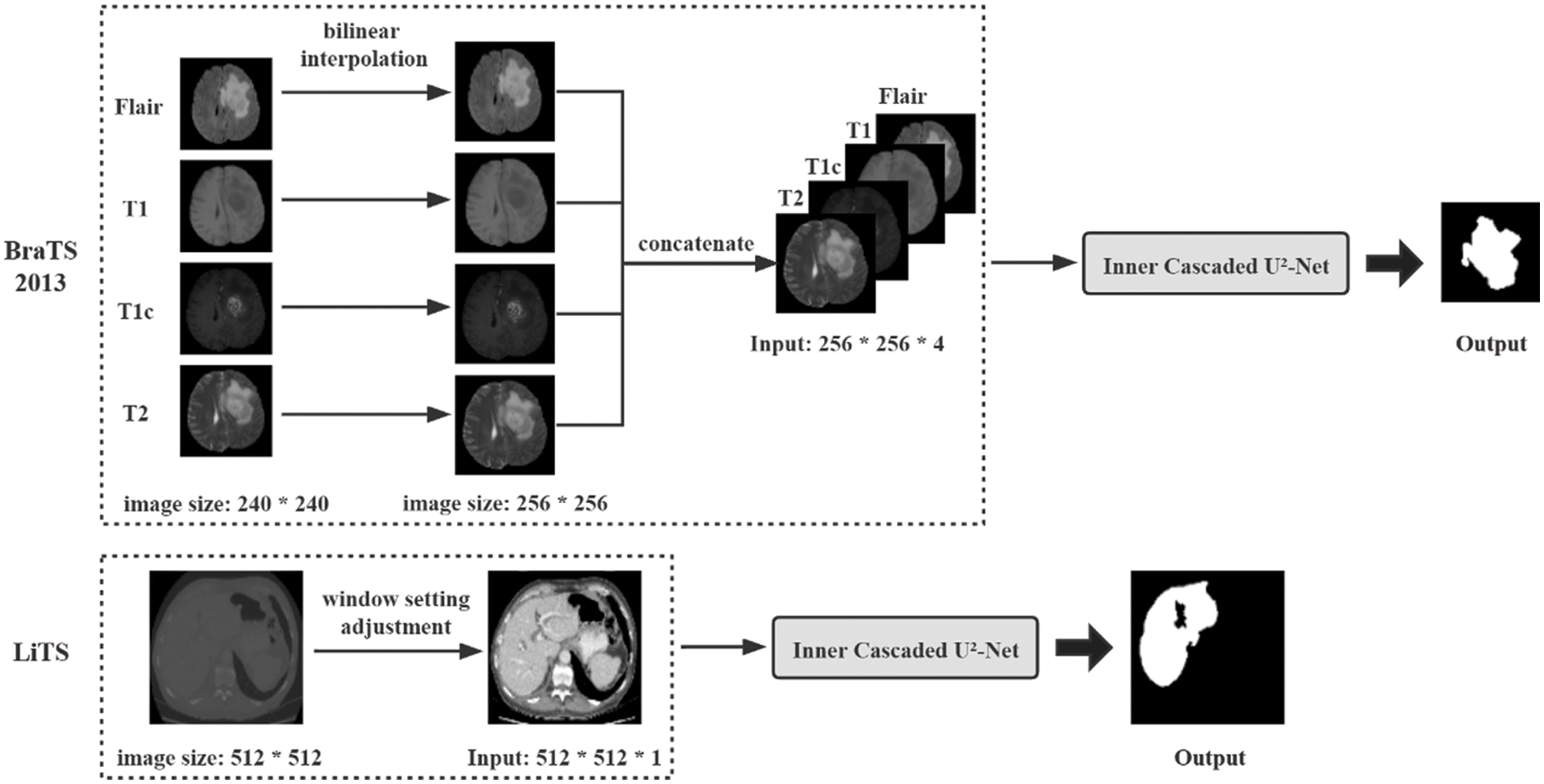
Figure 4: Overview of the proposed pipeline of brain tumor segmentation on the BraTS 2013 dataset and liver segmentation on the LiTS dataset. The operations within the dotted line denote the preprocessing steps
Dice Similarity Coefficient (DSC) and Hausdorff Distance (HD) [22] are used to evaluate the segmentation performance of our proposed method. DSC is widely used in medical image segmentation to measure the overlap degree between the predicted feature maps and ground truth, which is defined in Eq. (3):
where P represents the predicted feature maps, G is the ground truth labels,
HD is a measurement to the similarity degree of two groups of point sets. And here are the predicted feature maps and ground truth labels. HD is conventionally multiplied by 95% (HD95) to eliminate the influence of outliers. HD95 is defined in Eq. (4):
where P and G are the predicted feature maps and the ground truth labels, respectively,
The implementation of our proposed model is based on Python 3.6 and Keras frame with Tensorflow backend. All experiments are conducted on a single machine with an Intel Core i9-10900X 3.70 GHz (64 GB RAM) and an NVIDIA RTX 2080Ti GPU (11 GB memory). In particular, the initial learning rate is set to 1e-4 and Adam optimizer is adopted to update the weights of the network, with learning rate decay of 0.1 and patience of 10 in LiTS or 4 in BraTS 2013 monitored on the loss of validation set. In the training process of LiTS dataset, the maximum training epochs is set to 20 and a mini batch size of 2 is applied due to limit of GPU memory, while the models are trained for 50 epochs using batch size of 8 for the training of BraTS 2013 dataset. All models in our experiments are trained from scratch with no data augmentation before training or post-processing after training.
Focal Tversky Loss (FTL) [23] rather than Dice loss is implemented in our experiments for the consideration of achieving a better tradeoff between precision and recall. FTL is defined in Eq. (5):
where larger
3.4 Quantitative and Qualitative Results
Table 1 compares the segmentation performance on BraTS 2013 and LiTS of our proposed Inner Cascaded U-Net and Inner Cascaded U2-Net with related 4 existing methods whose main ideas are referenced. As seen, cascaded U-Net outperforms U-Net on both datasets, which is the advantage of adding another U-Net with the same structure to refine the segmentation mask predicted by the single U-Net. While our proposed Inner Cascaded U-Net further improves the segmentation performance to cascaded U-Net by 0.5% and 0.2% respectively in DSC. Inner cascaded method distinguishes from plain cascaded U-Net by replacing the coarse-to-fine improved way with a single-stage method in which the latter U-Net plays a more important role in predicting where more contextual information learned by the former U-Net is incorporated into the latter U-Net to boost the overall segmentation performance. It is worth mentioning that Inner Cascaded U-Net gets the smallest value of HD95 on both datasets, which means the predicted results of Inner Cascaded U-Net is best-matched to the ground truth and shows the effectiveness of inner cascaded method. Our proposed Inner Cascaded U2-Net achieves the best performance in case of DSC on both datasets, which outperforms cascaded U-Net, U2-Net, Inner Cascaded U-Net by 0.6%, 0.5%, 0.1% respectively on BraTS 2013 and 0.9%, 1.1%, 0.7% respectively on LiTS. Standard convolution block consists of two successive convolution layers with 3 * 3 kernel, which is equal to a single convolution layer with 5 * 5 kernel [9] and thus is inadequate to extract multi-scale features especially the features of small lesions. While RSU in Inner Cascaded U2-Net is able to capture multi-scale features and incorporate more global contextual information through a small U-Net like network for better segmentation of small lesions, which further boosts the performance on medical image segmentation.

The proposed Inner Cascaded U-Net and Inner Cascaded U2-Net are benchmarked against existing state-of-the-art methods on the BraTS 2013 and LiTS dataset in terms of DSC, as shown in Table 2. The results of BraTS 2013 dataset show that two cascaded methods including Cascaded DCNN and MCCNN achieve DSC of 0.8 and 0.89, respectively, while our proposed Inner Cascaded U2-Net outperforms the two methods a lot with DSC of 0.9474. The comparison results obtained from the LiTS dataset demonstrate our proposed Inner Cascaded U2-Net achieve the second highest DSC within the two-dimensional methods, among which CR-U-Net and Cascaded modified U-Net are designed in a cascaded method, which shows the effectiveness of Inner Cascaded U2-Net. Furthermore, two cascaded methods fusing 2D and 3D networks namely 2.5D methods are also presented in the table, which are H-DenseUNet and 3D AUNet + 2D ADAU-Net, respectively. The results demonstrate the 2.5D and 3D methods can lead to an enormous performance gain, which is due to the inherent defects in 2D models that they are incompetent to capture the spatial context information along z-axis, leading to segmentation performance degradation. Incorporating the inner cascaded method into three-dimensional models has not yet been implemented due to limitation by computing resources, which can be further researched.

Fig. 5 shows a qualitative comparison between our proposed models and other related 4 models. As we can see, our proposed Inner Cascaded U-Net and Inner Cascaded U2-Net can get more precise segmentation masks and finer outlines compared to other related models. Inner Cascaded U2-Net has a better handle on the segmentation of different types of objects at different scales, which is attributed to the ability of RSU to capture multi-scale features. The second case from BraTS 2013 demonstrates promising segmentation performance of Inner Cascaded U2-Net, with other models all omitting the outline details of small objects. But, our Inner Cascaded U2-Net can still capture the main features and have a finer outline segmentation. Here the global contextual information extracted by RSU is utilized for boundary refinement. The last case from LiTS dataset shows less false positive predictions in UNet++ and our proposed methods compared to other three models, indicating the significance of shared contextual information. The single between connection in cascaded U-Net is insufficient to share the learned contextual information from the former U-Net to the latter one, leading to insensitivity to false positive predictions, which is improved by inner cascaded method where more contextual information is incorporated in each corresponding layer.

Figure 5: Qualitative comparison of proposed Inner Cascaded U-Net and Inner cascaded U2-Net with U-Net, cascaded U-Net, U-Net++ and U2-Net. The first two cases are visualization of segmentation masks from BraTS 2013 dataset, while the last two cases are from the LiTS dataset
In this paper, we propose Inner Cascaded U-Net and Inner Cascaded U2-Net. Inner Cascaded U-Net refines the plain cascaded U-Net which connects in an end-to-end way by adding inner nested connections between the corresponding encoders and decoders of the two U-Nets, thus fusing more contextual information learned by the former U-Net into the latter one. Inner Cascaded U2-Net further enhances the segmentation performance by replacing the plain convolution block with residual U-block to capture more global contextual information for the sake of better segmentation of small regions. Experiments on the BraTS 2013 and LiTS datasets demonstrate that both models achieve excellent segmentation performance in terms of DSC, HD95 as well as refined segmentation masks.
Compared to the plain cascaded U-Net and U2-Net, however, the number of increased parameters of our proposed models are relatively large, leading to more computation and memory cost. Our future work will aim at exploring optimization strategy to decrease the model size and thus save the training as well as prediction time.
Availability of Data and Materials: The two datasets used to support our research are both publicly available. The BraTS 2013 dataset is available at https://www.smir.ch/BRATS/Start2013#!#download, accessed on 13 November 2021. And the LiTS dataset is available at https://competitions.codalab.org/competitions/17094#participate, accessed on 13 November 2021. All models mentioned in the experiments and code generated during the study by the corresponding author are available at https://github.com/FreedomXL/Inner-Cascaded-U-2-Net.
Funding Statement: This paper was supported in part by the National Nature Science Foundation of China (No. 62172299), in part by the Shanghai Municipal Science and Technology Major Project (No. 2021SHZDZX0100), and in part by the Fundamental Research Funds for the Central Universities of China.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Long, J., Shelhamer, E., Darrell, T. (2015). Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440. Boston, MA, USA. [Google Scholar]
2. Ronneberger, O., Fischer, P., Brox, T. (2015). U-Net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 234–241. Munich, Germany. [Google Scholar]
3. Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., Liang, J. (2018). UNet++: A nested U-Net architecture for medical image segmentation. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp. 3–11. Springer: Cham. [Google Scholar]
4. Huang, H., Lin, L., Tong, R., Hu, H., Zhang, Q. et al. (2020). UNet 3+: A full-scale connected unet for medical image segmentation. 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 1055–1059. Barcelona, Spain. [Google Scholar]
5. Zhang, Z., Liu, Q., Wang, Y. (2018). Road extraction by deep residual U-Net. IEEE Geoscience and Remote Sensing Letters, 15(5), 749–753. DOI 10.1109/LGRS.2018.2802944. [Google Scholar] [CrossRef]
6. Ibtehaz, N., Rahman, M. S. (2020). MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Networks, 121, 74–87. DOI 10.1016/j.neunet.2019.08.025. [Google Scholar] [CrossRef]
7. Qin, X., Zhang, Z., Huang, C., Dehghan, M., Zaiane, O. R. et al. (2020). U2-Net: Going deeper with nested U-structure for salient object detection. Pattern Recognition, 106, 107404. DOI 10.1016/j.patcog.2020.107404. [Google Scholar] [CrossRef]
8. Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S. et al. (2015). Going deeper with convolutions. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9. Boston, MA, USA. [Google Scholar]
9. Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z. (2016). Rethinking the inception architecture for computer vision. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826. Las Vegas, Nevada, USA. [Google Scholar]
10. Christ, P. F., Elshaer, M. E. A., Ettlinger, F., Tatavarty, S., Bickel, M. et al. (2016). Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 415–423. Athens, Greece. [Google Scholar]
11. Liu, Y. C., Shahid, M., Sarapugdi, W., Lin, Y. X., Chen, J. C. et al. (2021). Cascaded atrous dual attention U-Net for tumor segmentation. Multimedia Tools and Applications, 80(20), 30007–30031. DOI 10.1007/s11042-020-10078-2. [Google Scholar] [CrossRef]
12. Jiang, Z., Ding, C., Liu, M., Tao, D. (2019). Two-stage cascaded U-Net: 1st place solution to brats challenge 2019 segmentation task. International MICCAI Brainlesion Workshop, pp. 231–241. Shenzhen, China. [Google Scholar]
13. Qin, X., Zhang, Z., Huang, C., Gao, C., Dehghan, M. et al. (2019). BASNet: Boundary-aware salient object detection. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 7479–7489. Long Beach, CA, USA. [Google Scholar]
14. Li, X., Chen, H., Qi, X., Dou, Q., Fu, C. W. et al. (2018). H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Transactions on Medical Imaging, 37(12), 2663–2674. DOI 10.1109/TMI.2018.2845918. [Google Scholar] [CrossRef]
15. Liu, Y., Qi, N., Zhu, Q., Li, W. (2019). CR-U-Net: Cascaded U-Net with residual mapping for liver segmentation in CT images. 2019 IEEE Visual Communications and Image Processing (VCIP), pp. 1–4. Sydney, Australia. [Google Scholar]
16. Hu, K., Gan, Q., Zhang, Y., Deng, S., Xiao, F. et al. (2019). Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access, 7, 92615–92629. DOI 10.1109/ACCESS.2019.2927433. [Google Scholar] [CrossRef]
17. Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich, M. et al. (2018). Attention U-Net: Learning where to look for the pancreas. arXiv preprint arXiv: 1804.03999. [Google Scholar]
18. Ni, Z. L., Bian, G. B., Zhou, X. H., Hou, Z. G., Xie, X. L. et al. (2019). RAUNet: Residual attention U-Net for semantic segmentation of cataract surgical instruments. International Conference on Neural Information Processing, pp. 139–149. Sydney, NSW, Australia. [Google Scholar]
19. Roy, A. G., Navab, N., Wachinger, C. (2018). Concurrent spatial and channel ‘squeeze & excitation’ in fully convolutional networks. International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 421–429. Granada, Spain. [Google Scholar]
20. Wang, Q., Wu, B., Zhu, P., Li, P., Zuo, W. et al. (2020). ECA-Net: Efficient channel attention for deep convolutional neural networks. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 11534–11542. Seattle, WA, USA. [Google Scholar]
21. Yu, F., Koltun, V. (2015). Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv: 1511.07122. [Google Scholar]
22. Moradi, S., Ghelich-Oghli, M., Alizadehasl, A., Shiri, I., Oveisi, N. et al. (2019). A novel deep learning based approach for left ventricle segmentation in echocardiography. MFP-Unet. arXiv preprint arXiv: 1906.10486. [Google Scholar]
23. Abraham, N., Khan, N. M. (2019). A novel focal tversky loss function with improved attention U-Net for lesion segmentation. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), pp. 683–687. Venice, Italy. [Google Scholar]
24. Hussain, S., Anwar, S. M., Majid, M. (2017). Brain tumor segmentation using cascaded deep convolutional neural network. 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 1998–2001. IEEE, Jeju Island, Korea. [Google Scholar]
25. Zhou, Z., He, Z., Shi, M., Du, J., Chen, D. (2020). 3D dense connectivity network with atrous convolutional feature pyramid for brain tumor segmentation in magnetic resonance imaging of human heads. Computers in Biology and Medicine, 121, 103766. DOI 10.1016/j.compbiomed.2020.103766. [Google Scholar] [CrossRef]
26. Chithra, P. L., Dheepa, G. (2020). Di-phase midway convolution and deconvolution network for brain tumor segmentation in MRI images. International Journal of Imaging Systems and Technology, 30(3), 674–686. DOI 10.1002/ima.22407. [Google Scholar] [CrossRef]
27. Amin, J., Sharif, M., Raza, M., Saba, T., Sial, R. (2020). Brain tumor detection: A long short-term memory (LSTM)-based learning model. Neural Computing and Applications, 32(20), 15965–15973. DOI 10.1007/s00521-019-04650-7. [Google Scholar] [CrossRef]
28. Debnath, S., Talukdar, F. A., Islam, M. (2022). Complete 3D brain tumour detection using a two-phase method along with confidence function evaluation. Multimedia Tools and Applications, 81, 437–458. DOI 10.1007/s11042-021-11443-5. [Google Scholar] [CrossRef]
29. Guo, X., Schwartz, L. H., Zhao, B. (2019). Automatic liver segmentation by integrating fully convolutional networks into active contour models. Medical Physics, 46(10), 4455–4469. DOI 10.1002/mp.13735. [Google Scholar] [CrossRef]
30. Wang, Z., Zou, Y., Liu, P. X. (2021). Hybrid dilation and attention residual U-Net for medical image segmentation. Computers in Biology and Medicine, 134, 104449. DOI 10.1016/j.compbiomed.2021.104449. [Google Scholar] [CrossRef]
31. Mourya, G. K., Bhatia, D., Gogoi, M., Handique, A. (2021). CT guided diagnosis: Cascaded U-Net for 3D segmentation of liver and tumor. IOP Conference Series: Materials Science and Engineering, 1128(1), 012049. Chennai, India, IOP Publishing. [Google Scholar]
32. Qayyum, A., Lalande, A., Meriaudeau, F. (2020). Automatic segmentation of tumors and affected organs in the abdomen using a 3D hybrid model for computed tomography imaging. Computers in Biology and Medicine, 127, 104097. DOI 10.1016/j.compbiomed.2020.104097. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools