
 | Computer Modeling in Engineering & Sciences |  |
DOI: 10.32604/cmes.2022.018565
ARTICLE
Mu-Net: Multi-Path Upsampling Convolution Network for Medical Image Segmentation
1School of Information and Software Engineering, University of Electronic Science and Technology of China, Chengdu, 610054, China
2Department of Economics and Finance/Sustainable Real Estate Research Center, Hong Kong Shue Yan University, Hongkong, China
3Rattanakosin International College of Creative Entrepreneurship, Rajamangala University of Technology Rattanakosin, Nakhon Pathom, 73170, Thailand
4Department of Computer Science and Engineering, School of Sciences, European University Cyprus, Nicosia, 1516, Cyprus
5CIICESI, ESTG, Politécnico do Porto, 4610-156, Felgueiras, Portugal
*Corresponding Author: Bei Hui. Email: bhui@uestc.edu.cn
Received: 02 August 2021; Accepted: 21 October 2021
Abstract: Medical image segmentation plays an important role in clinical diagnosis, quantitative analysis, and treatment process. Since 2015, U-Net-based approaches have been widely used for medical image segmentation. The purpose of the U-Net expansive path is to map low-resolution encoder feature maps to full input resolution feature maps. However, the consecutive deconvolution and convolutional operations in the expansive path lead to the loss of some high-level information. More high-level information can make the segmentation more accurate. In this paper, we propose MU-Net, a novel, multi-path upsampling convolution network to retain more high-level information. The MU-Net mainly consists of three parts: contracting path, skip connection, and multi-expansive paths. The proposed MU-Net architecture is evaluated based on three different medical imaging datasets. Our experiments show that MU-Net improves the segmentation performance of U-Net-based methods on different datasets. At the same time, the computational efficiency is significantly improved by reducing the number of parameters by more than half.
Keywords: Medical image segmentation; MU-Net (multi-path upsampling convolution network); U-Net; clinical diagnosis; encoder-decoder networks
The purpose of medical image segmentation is to aid radiologists and clinicians in making the diagnostic and treatment process more efficient. However, manually segmenting large numbers of medical images is a time-consuming, error-prone task and requires a professional physician. To deal with this problem, an automated medical image segmentation method is proposed. Automated medical image segmentation can be applied to lung segmentation [1], brain segmentation in computed tomography (CT) [2], blood vessel detection in retinal images [3], etc. Previous methods for medical image segmentation are usually based on threshold segmentation, region segmentation, edge detection and segmentation of specific theories. With the development of artificial intelligence [4], deep learning is widely used in such research domains as image classification, segmentation and detection. Convolutional neural networks (CNNs) [5–7] have been introduced into the field of image segmentation and have achieved great success. At present, medical image segmentation is dominated by deep convolutional neural networks.
Based on fully convolutional networks (FCN) [8] and U-Net [9], CNNs have made significant improvements in biomedical image segmentation. These encoder-decoder networks have become a benchmark for medical image segmentation. The encoder-decoder architectures used for medical image segmentation share a similar critical structure: skip connections. High-level, deep, coarse-grained feature maps from the expansive path (decoder) can be combined with low-level, shallow, fine-grained feature maps from the contracting path (encoder) through skip connections. The skip connections can recover fine-grained details lost in the contracting path. The encoder-decoder networks have been proved to be well applied to medical image segmentation. The state-of-the-art models for image segmentation are variants of the encoder-decoder architecture like U-Net. However, this U-Net structure still has shortcomings.
As we know, both low-level features and high-level features are important in the U-Net, and only effective use of these features can achieve better segmentation results. Nevertheless, in the U-Net series network, the feature map size is reduced in the contracting path, leading to the loss of some low-level spatial information. Similarly, we argue that the consecutive deconvolution and convolutional operations in the expansive path lead to the loss of some high-level information. While the low-level, fine-grained information lost in the contracting path can be supplemented by a skip connection, the high-level semantic information lost in the single expansive path cannot be remedied. The single expansive path of U-Net-based approaches does not make efficient use of high-level information. Meanwhile, there is redundancy in the expansive path of the U-Net series network. The U-shaped symmetric structure leads to a large number of convolution operations in the expansive path, thus increasing the parameter amount to make the model more complicated.
To overcome the shortcomings in the U-Net series network for medical image segmentation, we propose the MU-Net, a new segmentation architecture based on multi-path upsampling and skip connections. The multi-path fusion of shallow and deep features in the up-sampling process can more effectively use low-level and high-level information. We argue that the skip connections can make up for the lost fine-grained information in the down-sampling process, multi-path up-sampling can make up for the lost high-level information in the upsampling process. The contracting path and multiple expansive paths are connected by skip connections, which will result in the fusion of different level features. The effective fusion of such different level features is beneficial to obtain more accurate segmentation. The experimental results show that the proposed architecture is effective and greatly reduces the number of parameters in the model meanwhile it can improve the performance of U-Net series networks. The contributions of our work can be summarized as follows:
1) Based on the U-Net series, we propose a novel convolutional network MU-Net for medical image segmentation, using multi-expansive paths to implement upsampling.
2) We have solved the defect that high-level information lost in the expansive path. The features of different layers in the contracting path are fused with the features of multiple expansion paths to improve the segmentation performance.
3) We remove the convolution operation in the expansive path, which greatly reduces the number of parameters while ensuring the performance of the model. Our proposed method outperforms the state-of-the-art methods in these different medical datasets.
The remainder of the paper is organized as follows. Section 2 introduces the related recent literature. In Section 3, we describe the MU-Net architecture and its analysis. In Section 4, we evaluate the performance of MU-Net on three different medical imaging datasets. In Section 5, we draw some conclusions.
Long et al. [8] first propose fully convolutional networks (FCN) for semantic segmentation and it shares a novel idea: skip connections. Then, Ronneberger et al. [9] introduce U-Net which uses skip connections to combine low-level features from the current layer with high-level feature maps. U-Net has achieved great success in the segmentation field due to its good performance. The variations have been proposed on U-Net for different image segmentation tasks. Vladimir et al. [10] propose the TernausNet model, which introduces the pre-trained encoder. A pre-trained encoder can be used when the data set is small. Milletari et al. [11] propose V-Net, an approach to 3D image segmentation based on a volumetric, fully convolutional, neural network. V-Net is a 3D segmentation implementation based on U-Net. Zhou et al. [12] reduce the semantic gap between the feature maps of the encoder and decoder sub-networks by a series of nested dense skip path connections, propose the UNet++.
In addition, some other structures and mechanisms have been introduced to improve the performance of U-Net. Oktay et al. [13] propose Attention U-Net and introduce attention gate (AG) which can be easily integrated into standard CNN architecture. Attention gate can suppress the learning of the part that is not related to the task while aggravating the learning of the related task. Inspired by the densely connected [14] and inception architecture [15], Dolz et al. [16] propose Dense Multi-path U-Net for Ischemic Stroke Lesion Segmentation. Guan et al. [17] propose Fully Dense UNet for 2D Sparse Photoacoustic Tomography Artifact Removal. Zhang et al. [18] propose a Multi-scale Densely Connected U-Net which directly fuses the neighboring different scale feature maps from both higher layers and lower layers to strengthen feature propagation in the current layer. Alom et al. [19] propose the R2U-Net model utilizing the power of U-Net, Residual Network, as well as RCNN. Gu et al. [20] argue that capturing higher-level features in the encoder and preserving more spatial information in the decoder improve the performance of image segmentation. They propose a Context Extractor Module that employs different receptive fields and a residual [21] multi-kernel pooling to segment targets of different sizes. Ibtehaz et al. [22] propose MultiResUNet, which uses MultiRes blocks and Res paths to solve the scale diversity in medical images and the semantic gap between the corresponding layers of the encoder-decoder.
Despite the above models have achieved excellent segmentation, there are some common flaws. 1) To capture more high-level features in the encoder, some spatial information is lost in the contracting path but can be compensated by skip connections. However, the high-level features will also be partially missed through the expansive path. The deep abstract features cannot be utilized more efficiently. 2) There is redundancy in the expansive path of U-Net-based networks, where convolution operations are not necessarily required.
In this section, we will introduce the specific structural design of the model. MU-Net has an encoder sub-network and a corresponding decoder sub-network, followed by a pixel-wise classification layer. Fig. 1 shows the overall architecture of our model, which mainly consists of three major parts: (1) Contracting Path (left): extract deep and high-level features; (2) Skip Connection (middle): reduce the semantic gap; (3) Multi-Expansive Paths (right): enable precise localization. There are four upsampling paths marked by purple arrows. The first three expansive paths are upsampled twice, and the last only needs to be upsampled once. Each blue box corresponds to a multi-channel feature map. The number of channels is denoted on top of the box.

Figure 1: The illustration of MU-Net architecture: multi-path upsampling network
In the contracting path of the U-Net network, each block contains two convolution layers and one max pooling layer. In our proposed network, we replace the two convolution layers with a MultiRes block [22], as is shown in Fig. 2. It uses three successive 3 × 3 filters and adds a residual connection (along with a 1 × 1 filter for conserving dimensions). The three consecutive convolutional layers have different numbers of filters, which are 1/6, 1/3, 1/2 of the number of input feature map channels. The gradual increase in filters prevents the memory requirement of the earlier layers from extremely spreading to the deeper parts of the network. After a MultiRes block [22], the number of filters is doubled. Specifically, after 4 times of MultiRes blocks in the encoder, the number of filters remains from 32 to 512. The max-pooling operation in the encoder sub-network is 4 times, and each downsampling will halve the size of the feature map. In the contracting path, the resolution of the feature map is continuously reduced, which will lead to the loss of some spatial information. Fortunately, the lost spatial information can be made up by skip connections. The contracting path can automatically extract the high-level features of the image.
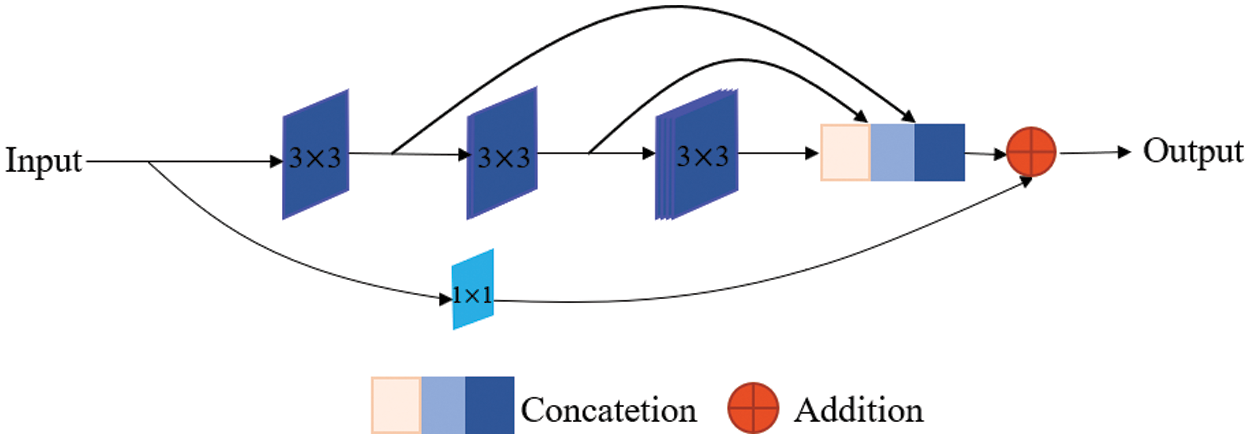
Figure 2: MultiRes block
The U-Net architecture connects the encoder to the decoder through skip connection, enabling the spatial information lost in the max pooling operation of the contracting path to be propagated to the decoder. Skip connections can combine low-level, fine-grained feature maps from the contracting path with high-level, coarse-grained feature maps from expansive paths [12]. However, features from the contracting path should be lower-level features, while features from the expansive path should be higher-level features. Maybe there is a semantic gap between features of encoder and decoder when feature merging is done through skip connection. The fusion of incompatible feature maps may adversely affect the prediction process. Hence, we replace the ordinary skip connection with the Res path [22], so that the features of the encoder can be processed more, making the corresponding layer features stitched through the skip connection more semantically similar.
The structure of the Res path is shown in Fig. 3. There are four Max-pooling between the top layer and the bottom layer, so there are 4 repeats of 3 × 3 and 1 × 1 convolution in the Res path. Connect the feature map of the encoder to the decoder via a chain of convolutional layers with residual connections. The residual connections [23] make the learning process easier.
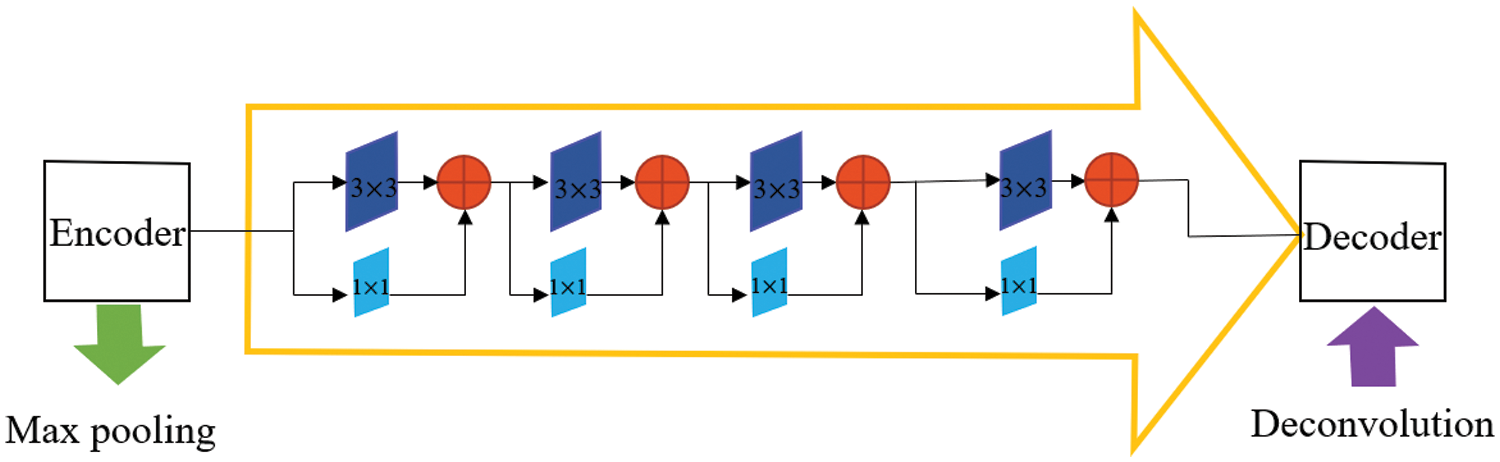
Figure 3: The Res path structure of the top-level skip connection
The expansive path is mainly used to restore the resolution of the image to its original size. The typical decoder structures in the U-Net-based networks are shown in Fig. 4, containing a single expansive path only. The deepest abstract feature reduces the number of feature channels by deconvolution, then merges with the feature map from the skip connection. Next, the feature maps are reduced by convolution. This process repeats until the original image size is restored. However, there are two main defects in the decoder structure of a typical U-Net network.
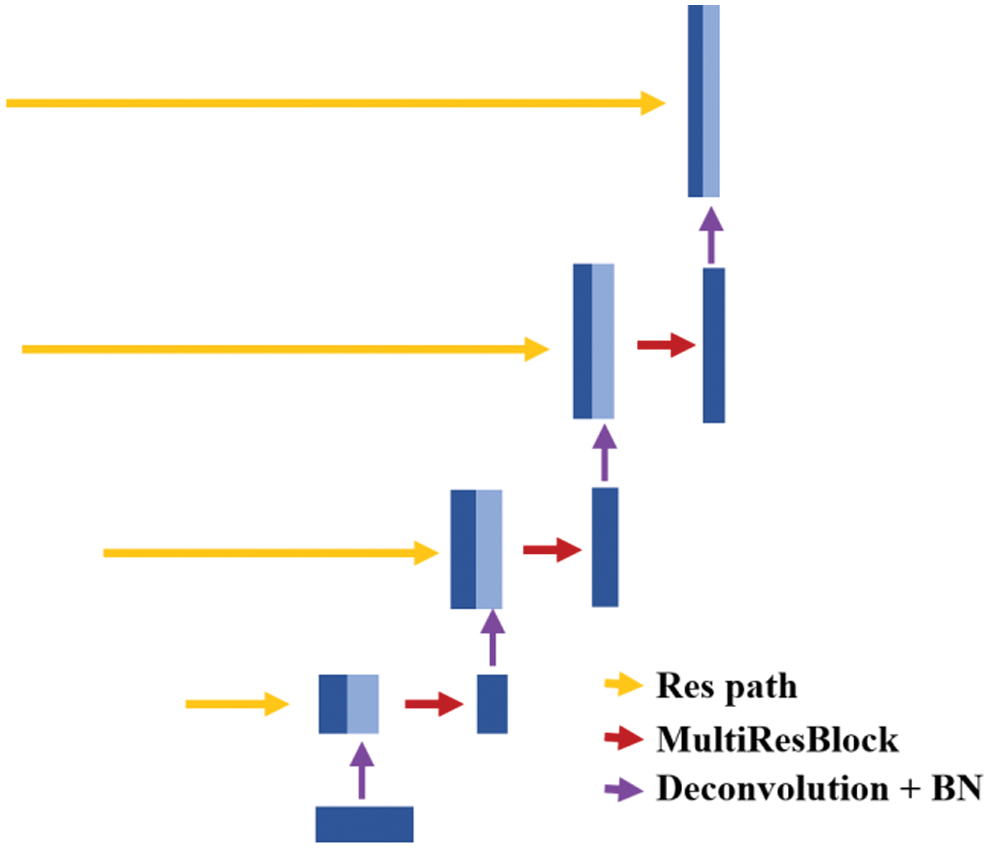
Figure 4: The decoder of U-Net based networks
Firstly, the high-level information is partially lost in the single expansive path. The high-level information lost in the expansive path mainly comes from two aspects. The one is that the number of feature channels is reduced due to the deconvolution operation which results in the loss of high-level information and the other one is that the convolution of the merged feature map will reduce feature channels. Secondly, there is a lot of redundancy due to the convolution operation in the expansive path. Plenty of convolution operations in the expansive path increase the number of model parameters, but these convolutions are not necessary and may not necessarily improve the performance of the model.
To solve these two problems, we have re-designed the expansive path. The most important modification in our network is to change the single expansive path in the decoder into multi-expansive paths while removing the convolution operation in the expansive path, as shown in Fig. 1. The four expansive paths are connected by skip connection with different layers in the contracting path respectively. The first expansive path is restored to the resolution of the original image only by one deconvolution, and the last three expansive paths are deconvolved twice. The features of different scales are extracted from different layers in the contracting path and then fused with the features obtained by deconvolution of different expansive paths. Formally, let
where F indicates the lowest level features,
Effective use of shallow and high-level features can improve image segmentation performance. The skip connections take some fine-grained information from encoder to decoder to remedy the information loss in the extracting path, which allows the shallow features to be fully utilized. Our proposed multi-expansive paths can remedy the loss of high-level feature information in the decoder. Our decoder is a novel architecture.
To demonstrate the performance of the MU-Net, we have tested our model on three different medical imaging datasets. These include cell contour segmentation, blood vessel segmentation from retina images (DRIVE), skin cancer lesion segmentation. The network structure parameter table is detailed in the Appendix.
In this section, we first introduce some implementation details of the model. The task of semantic segmentation is to predict each pixel to be foreground or background, which is a pixel-wise classification problem. Our network architecture can be trained end-to-end. The encoder and decoder weights of the network are initialized using the technique described by He et al. [25]. We train all parameters through the Keras platform, using stochastic gradient descent with a fixed learning rate of 0.1 and a momentum of 0.9. Gradient updates are computed using small batch sizes of 2 samples.
The most common loss function is the cross-entropy loss function [8]. To verify the high performance of our network model, we choose this loss function as the objective function for training the network. For quantitative analysis of the experimental results, several performance metrics are considered, including accuracy, recall (sensitivity), Jaccard Index, and Hausdorff Distance. Jaccard Index and Hausdorff Distance are two of the most commonly used indicators for segmentation. We compare our network architecture with a state-of-the-art segmented network. A comparative experiment is conducted with the MultiResUnet [22] architecture. MultiResUnet is based on the U-Net network modification and has achieved good segmentation results. Table 1 shows a comparison between our method and others. The proposed MU-Net model is benchmarked against the MulitResUNet and U-Net for different training and testing splits. To display the change in the number of model parameters, we display the original image size and the input size of the training into the table.
1) Cell Contour Segmentation
We first evaluate the proposed MU-Net on cell contour segmentation. The dataset is provided by the EM challenge, which started at ISBI 2012. The training set contains 30 images. The testing set consists of 30 images and is publicly available as well. The original image size was 512 × 512, however, we resized the images to 256 × 256 pixels. We trained this data set using the 5-fold cross-validation to ensure a balance between bias and variance. Data augmentation [9] is essential to teach the network the desired invariance and robustness properties when only a few training samples are available. During the training, the properties of elastic deformation including image horizontal, vertical and diagonal flips [26,27] were used to enhance the data.

From the comparison, the MU-Net achieves 0.9233, 0.9625, 0.9111, and 148.56 in Accuracy, Recall, Jaccard index, and Hausdorff distance respectively, better than U-Net and MultiResUNet. Meanwhile, the parameter quantity of our model is 3,445,126, which is greatly reduced compared with the parameter quantity of 7,759,585 of U-Net and 7,260,504 of MultiResUnet. From Table 1, we can conclude that the accuracy of our proposed model can be increased by up to 1% when the parameters are halved, which shows that the training cost of our model has been reduced. These experimental results show that our redesigned decoder module not only reduces the number of model parameters but also facilitates cell contour segmentation. We argue that the multi-path upsampling network can make more effective use of the high-level features, to improve the performance of the model. We also give examples for visual comparison in Figs. 5 and 6. In Fig. 5, the first column shows the original image, the second column shows the segmentation output using U-Net, the third column shows the segmentation output using MultiResUNet [22], the fourth column shows our architecture MU-Net segmentation result, and the last column is ground truth. Fig. 6 shows the outline of our segmentation result is more consistent with the ground truth. The result of U-Net segmentation has more noise, MultiResUNet is easy to mislabel part of the nucleus, and MU-Net is relatively better. From the comparison, we can conclude that the segmentation contour of our proposed model fits better with the real label. Also as shown in Fig. 7, the MU-Net model has the best segmentation effects among the three models when segmenting the cell neuron structure.
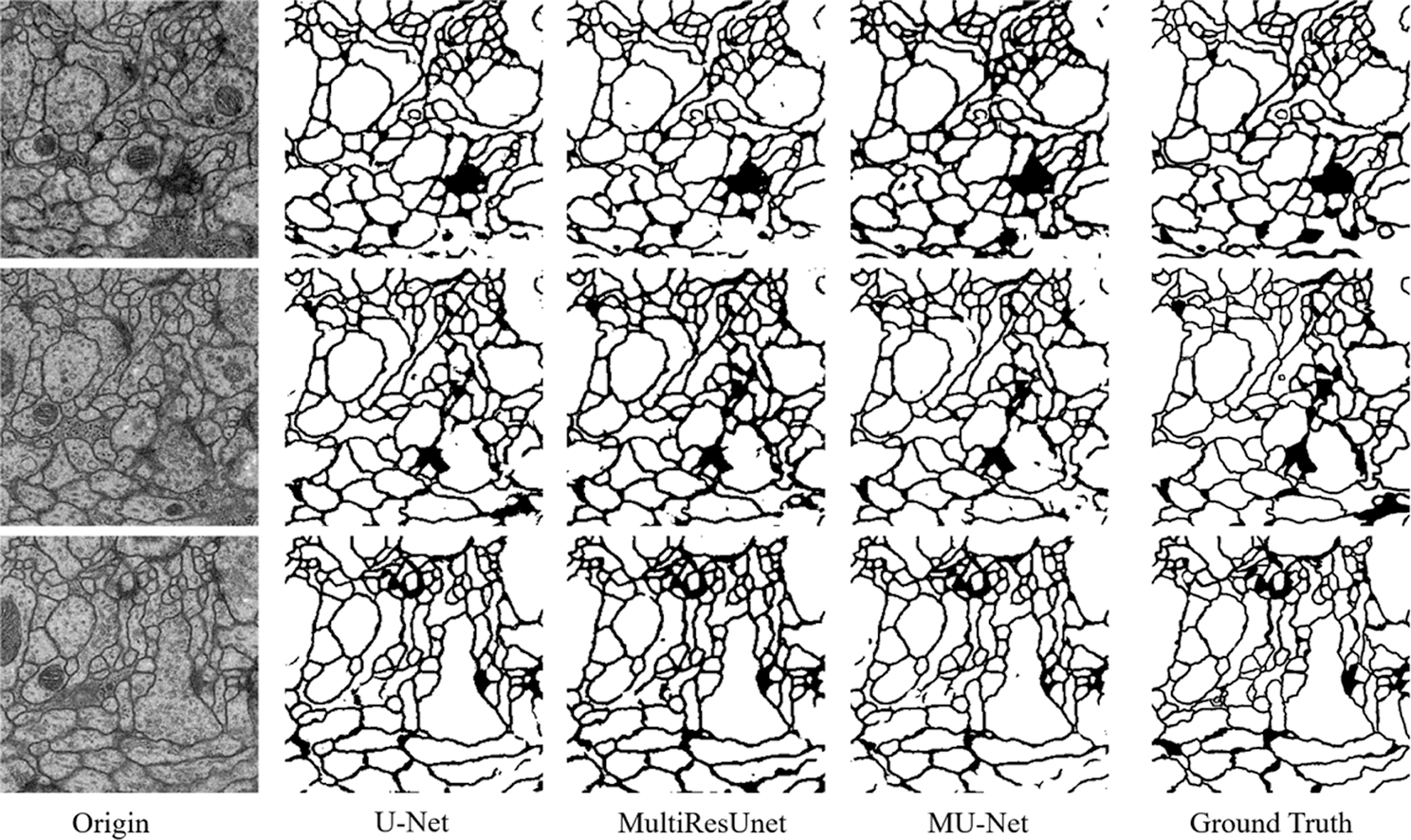
Figure 5: The segmentation results of the EM challenge dataset
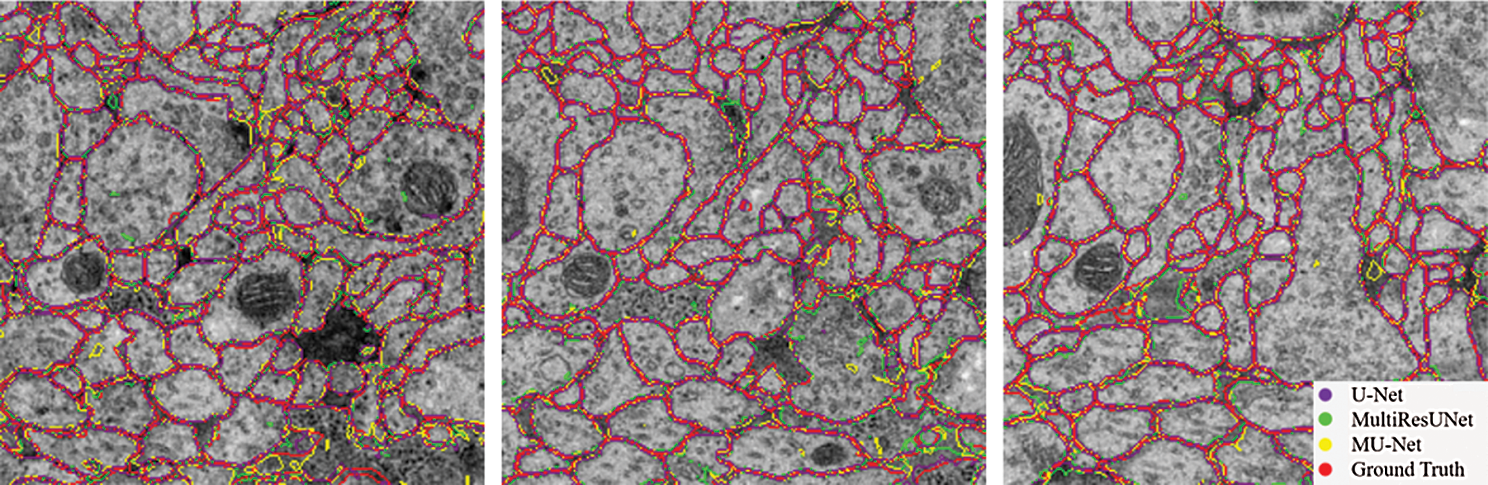
Figure 6: Contour comparison chart of segmentation results
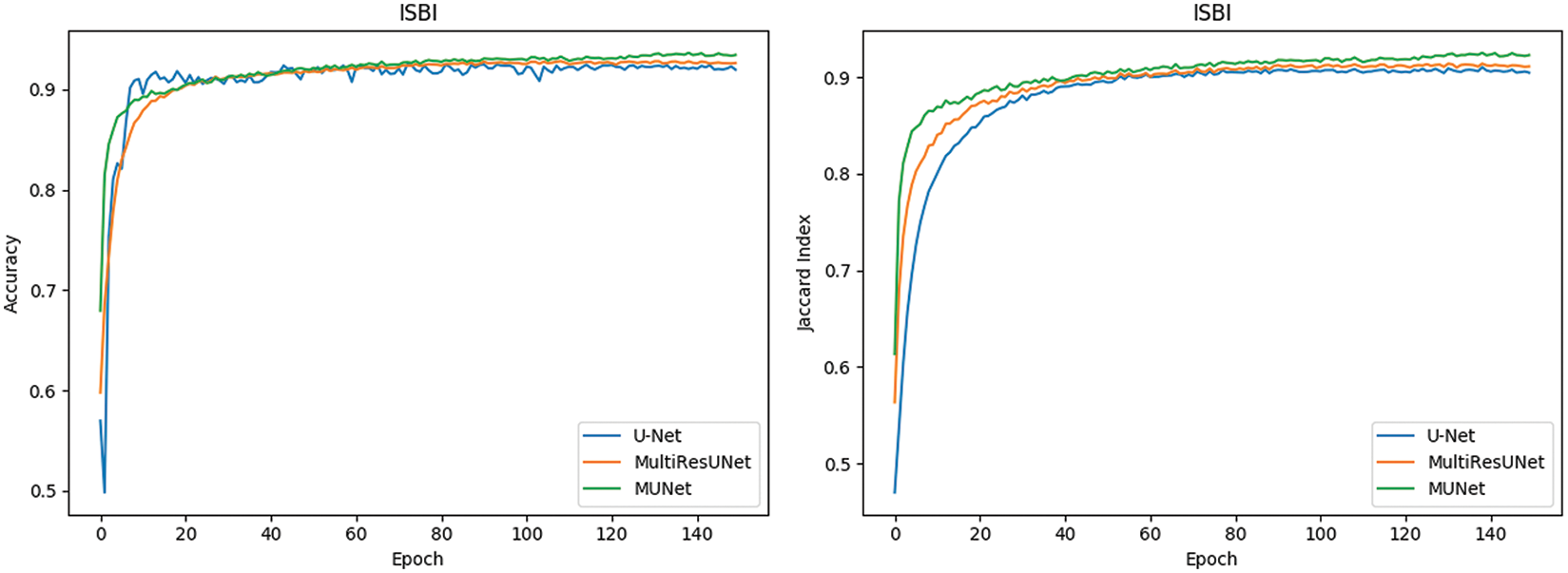
Figure 7: Comparison of segmentation performance of neuron structure
2) Retinal Vessel Segmentation
The second comparative experiment was on retinal vessel segmentation. We use the public DRIVE dataset [28] for retinal vascular segmentation. In DRIVE, there are 40 colour retinal images which are divided into 20 images for training and 20 images for testing. The size of each original image is 565 × 584 pixels, which was rescaled to 448 × 448 for this implementation. Since the dataset is small, we also use data augmentation, with the same strategy used in cell contour segmentation.
The performance comparisons are summarized in Table 1, Compared to U-Net, the parameter quantity decreases from 7,759,585 to 3,445,280 by 55.6%, the accuracy score increases from 0.9575 to 0.9672, the recall score increases from 0.7575 to 0.8099, the Jaccard index increases from 0.7434 to 0.7527, while the Hausdorff distance index dropped from 71.25 to 57.26, which proves the validity of our multiple expansive paths. Compared to MultiResUNet, our model has also improved. We also give examples for visual comparison in Figs. 8 and 9. In Fig. 8, the first column shows the input image, the second column shows the segmentation output using U-Net, the third column shows the segmentation output using MultiResUNet [22], the fourth column shows our architecture MU-Net segmentation result, and the last column is ground truth. Fig. 9 shows that our segmentation results are more accurate in detail. Although the improvement of evaluation metrics is not particularly large, it is better visually. In the process of actual segmentation of retinal vascular cases, the segmentation of the end of the blood vessel is more refined, which is more accurate for some bleeding, exudation, etc. The images of other lesions are less affected when segmented, and are closer to the real label Ground Truth.
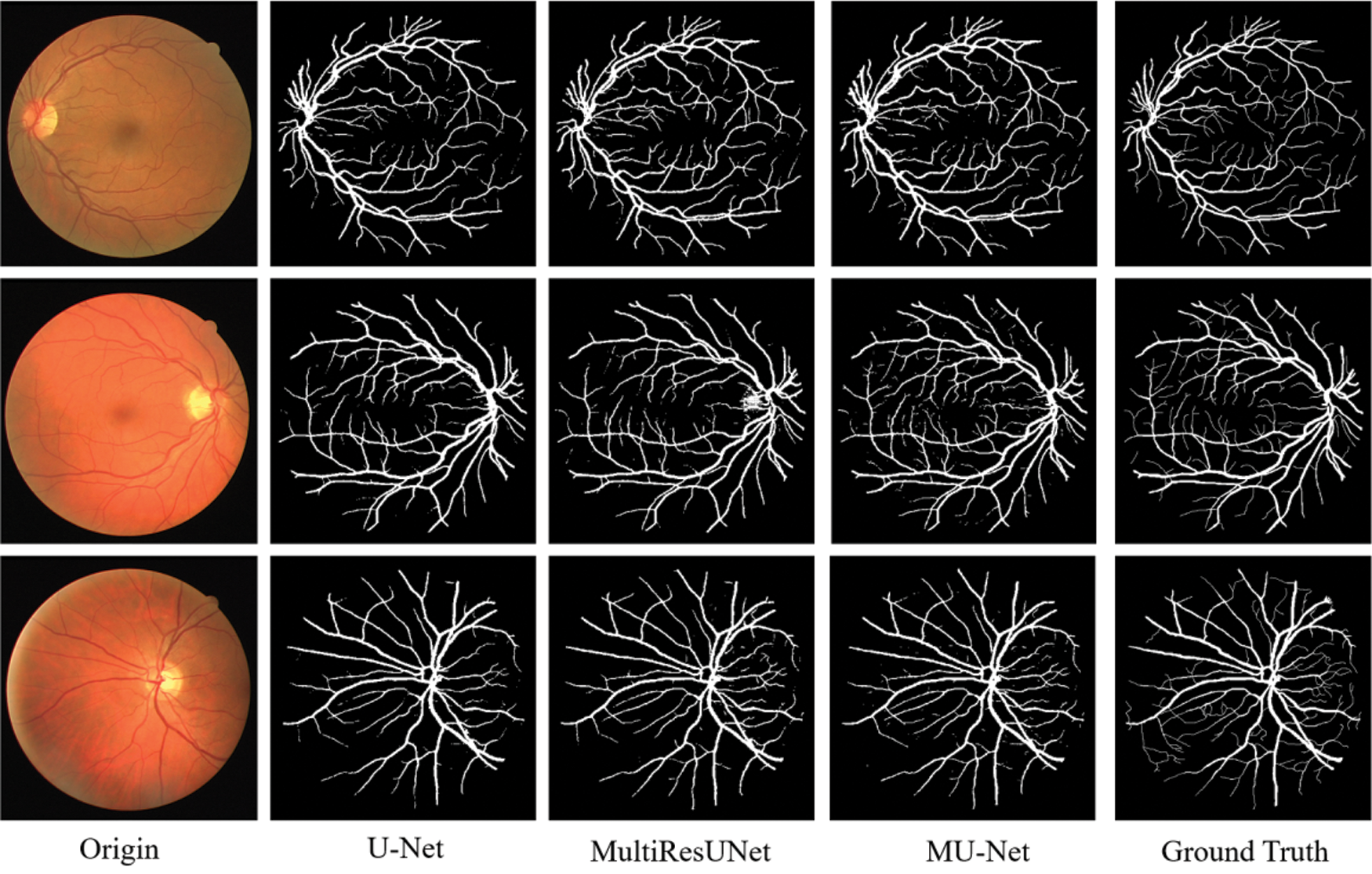
Figure 8: Experimental outputs of DRIVE dataset
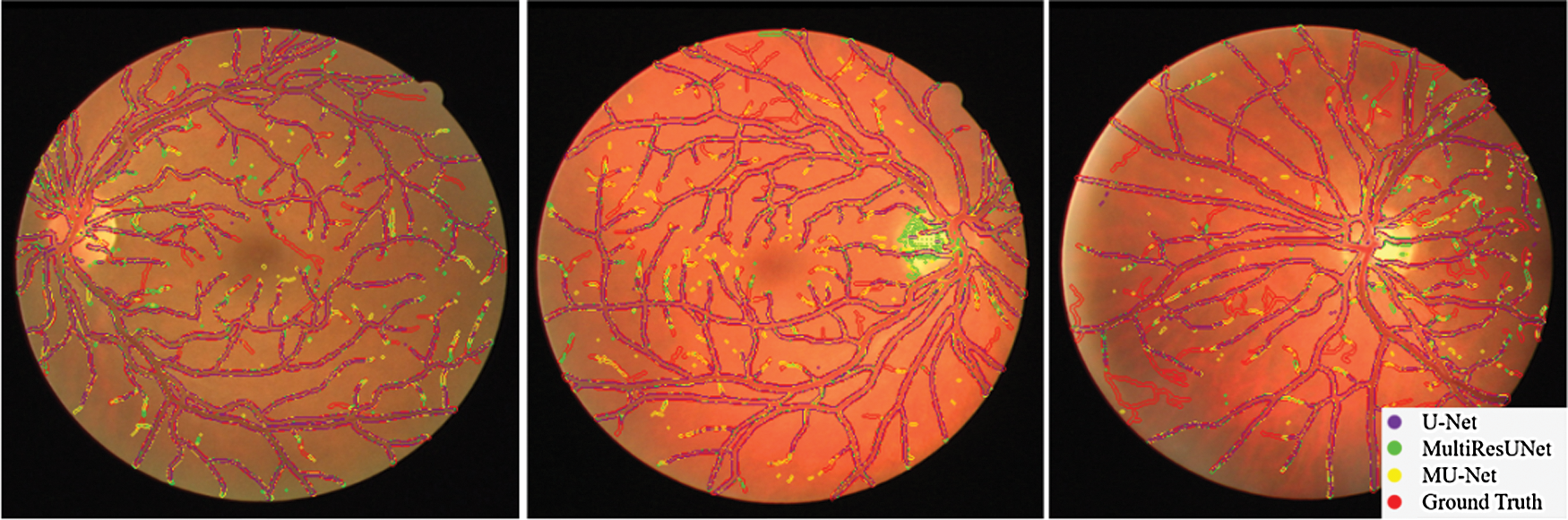
Figure 9: Comparison of segmentation results on DRIVE dataset
In Fig. 10, it can also be seen that the performance of MU-Net is improved, the index convergence is relatively stable, and it has good robustness.
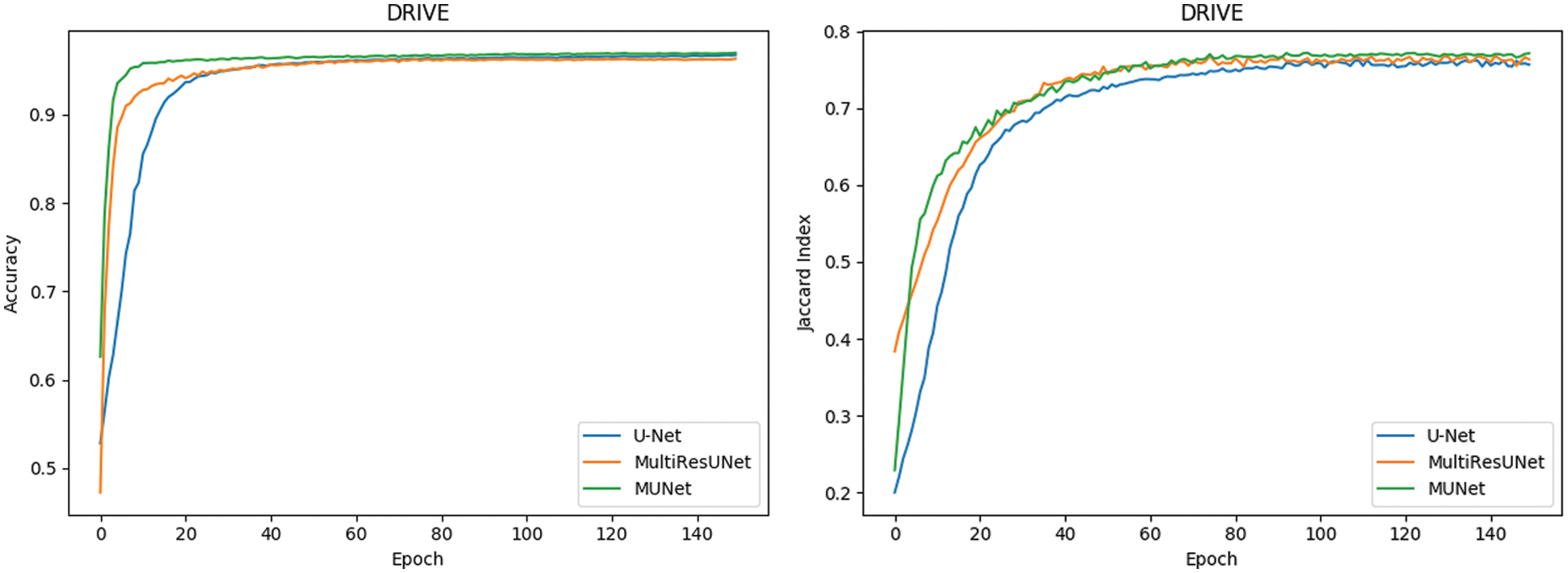
Figure 10: Comparison of retinal blood vessel segmentation
3) Skin Cancer Lesion Segmentation
Finally, the comparison is conducted on the segmentation of skin cancer lesions. The dataset is taken from the Kaggle competition on skin lesion segmentation [29] which contains 2000 samples. The original image size was 192 × 256, we resize the images to 192 × 192 pixels. The original images, as well as corresponding target binary images containing cancer or non-cancer lesions, were included in the samples. We use 80% of the images for training and the rest for testing. In this experiment, no preprocessing is performed on the data.
The metrics for evaluating the segmentation of skin cancer lesions are shown in Table 1. From the comparison, the MU-Net achieves 0.9439, 0.8193, 0.7746, and 52.19 in Accuracy, Recall, Jaccard index, and Hausdorff distance, respectively. Although the recall score is lower than U-Net's 0.8440. MU-Net is better in contour segmentation from the perspective of visual effects, as shown in Fig. 11. In Fig. 11, these five columns are respectively the original image, U-Net segmentation result, MultiResUNet segmentation result, MU-Net segmentation result, and ground truth. The visualization results of the accuracy and Jaccard index indices during segmentation are shown in Fig. 12. Convergence is more stable on the accuracy indices, but on the Jaccard index indices, both U-Net and MUltiResUNet fluctuate for a long time, compared with MU-Net, which is more stable during training.
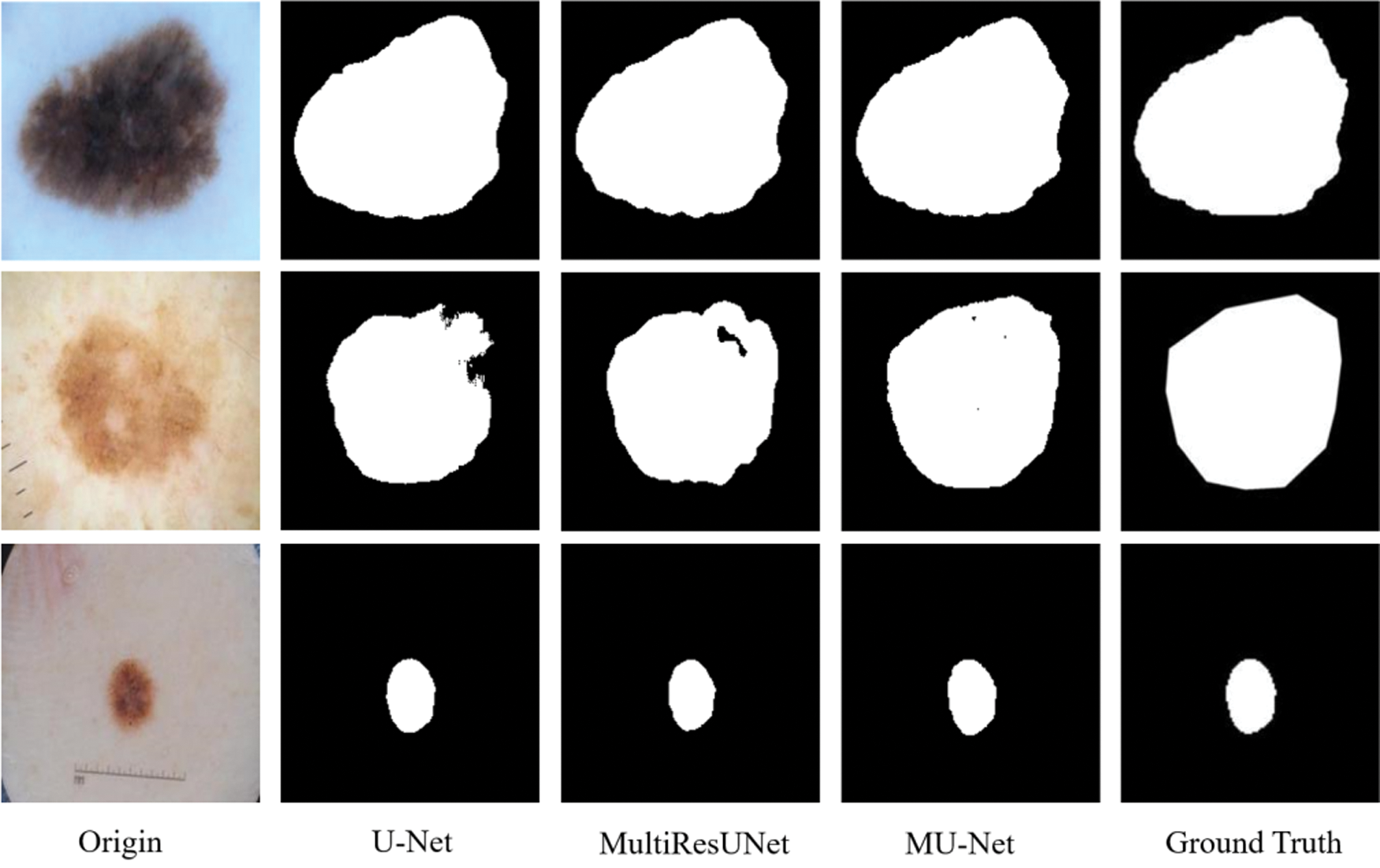
Figure 11: The segmentation of skin cancer lesion image
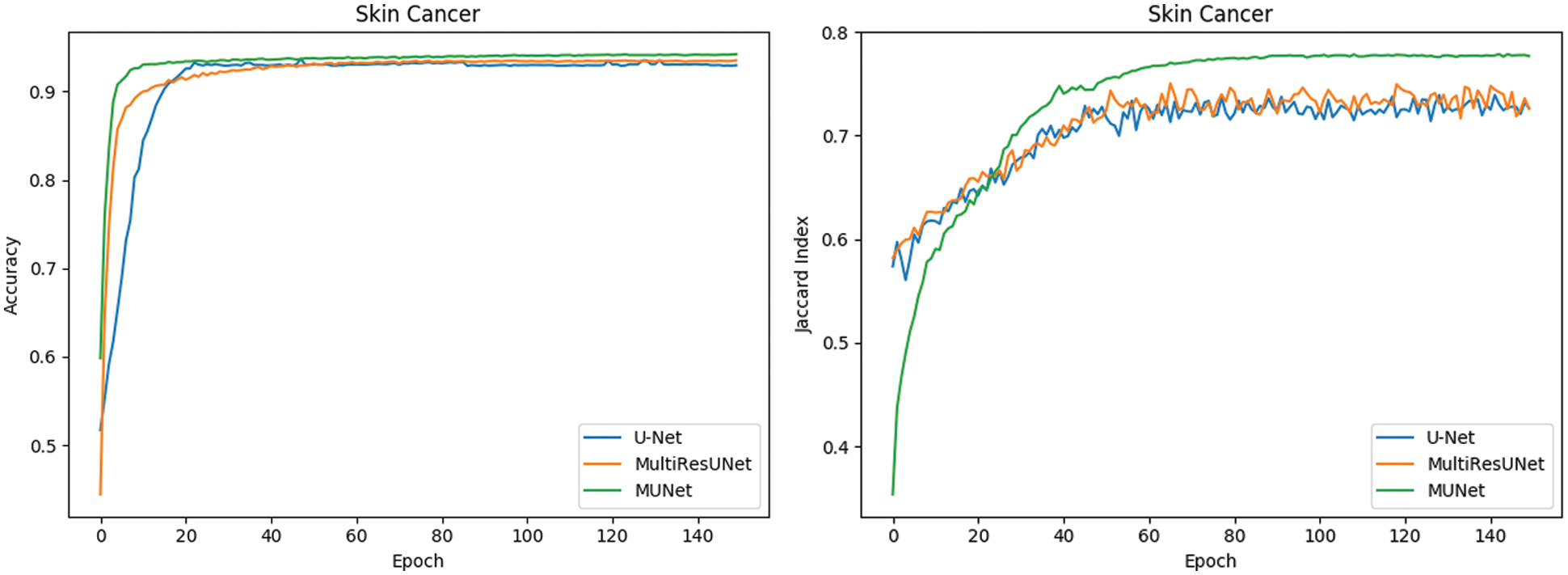
Figure 12: Comparison of segmentation performance of dermoscopy lesions
4) Comparative Experiments
In this work, we also use MU-Net to compare with other more advanced segmentation algorithms that use the same data set. They are the CE-Net model proposed by Gu et al. [20] in 2019, the UNet++ model proposed by Szegedy et al. [23] in 2018 and the SegNet model proposed by Badrinarayanan et al. [30] in 2017. Through the current more advanced algorithm model [31,32] to conduct segmentation comparison experiments on medical data sets, it is proved that the MU-Net proposed in this paper is of great help in improving the segmentation performance of medical images. The specific comparison results are shown in Tables 2 and 3.


Tables 2 and 3 show that our proposed multi-path upsampling convolutional neural network MU-Net has achieved better results in accuracy, Jaccard index, and Hausdorff distance compared with other algorithms. Although the recall score is lower than that of the CE-Net model when training in different data sets, the overall performance of our proposed multi-path upsampling convolutional neural network MU-Net has made progress.
Through comparative experiments on MU-Net on various data sets, the results show that the multi-path upsampling convolutional network MU-Net, which is based on the improvement of the codec architecture, significantly reduces the parameters of the model without affecting the segmentation performance. The MU-Net model is better than the other three models in Accuracy, recall, Jaccard index, and Hausdorff distance, which effectively improves the segmentation performance. And multi-path up-sampling can make more effective use of high-level abstract semantic features while fusing features of different scales. The improved skip connection structure in MU-Net can avoid the problem of the semantic gap to a certain extent.
In this paper, we propose a novel multi-path upsampling convolutional network for medical image segmentation termed MU-Net. The MU-Net directly builds in MultiReUNet (based on U-Net). Of course, our model can also be easily built on other U-Net-based architectures. We redesign the decoder, using a multi-expansive path and remove the convolution operation in the expansive path. Our approach solves the defect that the high-level information is lost in the expansive path of U-Net. Meanwhile, the convolution operation of the expansive path is removed, which greatly reduces the parameter amount of the model, but the segmentation performance is not reduced. Our model is evaluated using three different datasets, including cell contour segmentation, retina blood vessel segmentation, and skin cancer lesion segmentation. The experimental results demonstrate that the proposed MU-Net model shows better performance in segmentation tasks with the half number of network parameters when compared to U-Net and MulitiResUNet on all three datasets. In the future, we would like to explore the same architecture to make more effective use of shallow and high-level features.
Funding Statement: The authors received Sichuan Science and Technology Program (No. 18YYJC1917) funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
REFERENCES
1. Wang, S., Zhou, M., Liu, Z., Gu, D., Zang, Y. et al. (2017). Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Edical Image Analysis, 40, 172–183. DOI 10.1016/j.media.2017.06.014. [Google Scholar] [CrossRef]
2. Cherukuri, V., Ssenyonga, P., Warf, B. C., Kulkarni, A. V., Monga, V. et al. (2018). Learning based segmentation of ct brain images: Application to postoperative hydrocephalic scans. IEEE Transactions on Biomedical Engineering, 65(8), 1871–1884. DOI 10.1109/TBME.2017.2783305. [Google Scholar] [CrossRef]
3. Liskowski, P., Krawiec, K. (2016). Segmenting retinal blood vessels with deep neural networks. IEEE Transactions on Medical Imaging, 35(11), 2369–2380. DOI 10.1109/TMI.2016.2546227. [Google Scholar] [CrossRef]
4. Chen, X., Li, C., Wang, D., Wen, S., Zhang, J. et al. (2020). Surya Nepal, Yang Xiang, and Kui Ren, android HIV: A study of repackaging malware for evading machine-learning detection. IEEE Transactions on Information Forensics and Security, 15(1), 987–1001. DOI 10.1109/TIFS.2019.2932228. [Google Scholar] [CrossRef]
5. Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F. et al. (2017). A survey on deep learning in medical image analysis. Medical Image Analysis, 42, 60–88. DOI 10.1016/j.media.2017.07.005. [Google Scholar] [CrossRef]
6. Roth, H., Lu, R., Lay, L., Harrison, N., Farag, A. P. et al. (2018). Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Medical Image Analysis, 45, 94–107 DOI 10.1016/j.media.2018.01.006. [Google Scholar] [CrossRef]
7. Roth H. R., Oda H., Hayashi, Y., Oda, M., Shimizu, N. et al. (2017). Hierarchical 3D fully convolutional networks for multi-organ segmentation. arXiv:1704.06382. [Google Scholar]
8. Long, J., Shelhamer, E., Darrell, T. (2015). Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440. Boston, USA. [Google Scholar]
9. Ronneberger, O., Fischer, P., Brox, T. (2015). U-Net: Convolutional networks for biomedical image segmentation, vol. 3, pp. 234–241. Cham, Switzerland: Springer International Publishing, DOI 10.1007/978-3-319-24574-4_28. [Google Scholar] [CrossRef]
10. Vladimir, I., Alexey, S. (2018). Ternausnet: U-net with VGG11 encoder pre-trained on ImageNet for image segmentation. arXiv:1801.05746. [Google Scholar]
11. Milletari, F., Navab, N., Ahmadi, S. A. (2016). V-Net: Fully convolutional neural networks for volumetric medical image segmentation. IEEE 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. Stanford, USA. [Google Scholar]
12. Zhou, Z. W., Siddiquee, M. M. R., Tajbakhsh, N., Liang, J. M. (2018). UNet++: A nested U-Net architecture for medical image segmentation. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, 9, 3–11. DOI 10.1007/978-3-030-00889-5. [Google Scholar] [CrossRef]
13. Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich, M. et al. (2018). Attention U-Net: Learning where to look for the pancreas. arXiv:1804.03999. [Google Scholar]
14. Huang, G., Liu, Z., Maaten, L. V. D., Kilian, Q. (2017). Weinberger. densely connected convolutional networks. arXiv:1608.06993. [Google Scholar]
15. Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S. (2015). Going deeper with convolutions. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 1–9. Boston, USA. [Google Scholar]
16. Dolz, J., Ayed, I. B., Desrosiers. C. (2019). Dense multi-path U-Net for ischemic stroke lesion segmentation in multiple image modalities. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, 1, 271–282. DOI 10.1007/978-3-030-11723-8. [Google Scholar] [CrossRef]
17. Guan, S., Khan, A., Sikdar, S., Parag, V. C. (2020). Fully dense UNet for 2D sparse photoacoustic tomography artifact removal. IEEE Journal of Biomedical and Health Informatics, 24(2), 568–576. DOI 10.1109/JBHI.6221020. [Google Scholar] [CrossRef]
18. Zhang, J. W., Jin, Y. Z., Xu, J. L., Xu, X. W., Zhang, Y. C. (2018). MDU-Net: Multi-scale densely connected U-Net for biomedical image segmentation. arXiv:1812.00352. [Google Scholar]
19. Alom, M. Z., Hasan, M., Yakopcic, C., Taha, T. M., Vijayan, K. (2018). Recurrent residual convolutional neural network based on U-Net (R2U-Net) for medical image segmentation. arXiv:1802.06955. [Google Scholar]
20. Gu, Z. W., Cheng, J., Fu, H. Z., Zhou, K., Hao, H. Y. (2019). CE-Net: Context encoder network for 2D medical image segmentation. IEEE Transactions on Medical Imaging, 38(10), 2281–2292. DOI 10.1109/TMI.42. [Google Scholar] [CrossRef]
21. He, K. M., Zhang, X. Y., Ren, S. Q., Sun, J. (2016). Deep residual learning for image recognition. IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778. Las Vegas, USA. [Google Scholar]
22. Ibtehaz, N., Rahman, M. S. (2020). MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Networks, 121, 74–87. DOI 10.1016/j.neunet.2019.08.025. [Google Scholar] [CrossRef]
23. Szegedy, C., Ioffe, S., Vanhoucke, V., Alemi. A. A. (2017). Inception-v4, inception-resnet and the impact of residual connections on learning. arXiv:1602.07261. [Google Scholar]
24. Lin, T. Y., Dollár, P., Girshick, R., He, K. M., Hariharan, B. et al. (2016). Serge belongie. feature pyramid networks for object detection. arXiv:1612.03144. [Google Scholar]
25. He, K., Zhang, X., Ren, S., Sun, J. (2015). Delving deep into rectifiers: Surpassing human-level performance on imageNet classification. IEEE International Conference on Computer Vision, pp. 1026–1034. Santiago, USA. [Google Scholar]
26. Dai, J., Li, Y., He, K., Sun, J., Fan, R. (2016). Object detection via region based fully convolutional networks. Advances in Neural Information Processing Systems, 29, 379–387. [Google Scholar]
27. Zhang, Y., Qiu, Z., Yao, T., Liu, D., Mei, T. (2018). Fully convolutional adaptation networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 6810–6818. Salt Lake City, USA. [Google Scholar]
28. Staal, J., Abramoff, M. D., Niemeijer, M., Viergever, M. A., Ginneken, B. V. (2004). Ridge based vessel segmentation in color images of the retina. IEEE Transactions on Medical Imaging, 23, 501–509. DOI 10.1109/TMI.2004.825627. [Google Scholar] [CrossRef]
29. Codella, N., Gutman, D., Celebi, M. E., Helba, B., Marchetti, M. A. et al. (2017). Skin lesion analysis toward melanoma detection: A challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC). arXiv: 1710.05006. [Google Scholar]
30. Badrinarayanan, V., Kendall, A., Cipolla, R. (2017). Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 39(12), 2481–2495. DOI 10.1109/TPAMI.34. [Google Scholar] [CrossRef]
31. Kingma, D. P., Ba, J. (2014). Adam: A method for stochastic optimization. https://arxiv.org/. [Google Scholar]
32. Ignacio, A. C., Srinivas, C. T., Daniel, R. B., Dan, C., Giusti, A. et al. (2015). Crowdsourcing the creation of image segmentation algorithms for connectomics. Frontiers in Neuroanatomy, 9, 142. DOI 10.3389/fnana.2015.00142. [Google Scholar] [CrossRef]
Appendix







 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |