
 | Computer Modeling in Engineering & Sciences |  |
DOI: 10.32604/cmes.2021.016981
REVIEW
Deep Learning Applications for COVID-19 Analysis: A State-of-the-Art Survey
1School of Computer, Jiangsu University of Science and Technology, Zhenjiang, 212003, China
2School of Automation, Key Laboratory of Measurement and Control for CSE, Ministry of Education, Southeast University, Nanjing, 210096, China
3School of Information Science and Technology, Nantong University, Nantong, 226019, China
*Corresponding Author: Xing Deng. Email: xdeng@just.edu.cn
Received: 16 April 2021; Accepted: 13 July 2021
Abstract: The COVID-19 has resulted in catastrophic situation and the deaths of millions of people all over the world. In this paper, the predictions of epidemiological propagation models, such as SIR and SEIR, are introduced to analyze the earlier COVID-19 propagation. The deep learning methods combined with transfer learning are familiar with classification-detection approaches based on chest X-ray and CT images are presented in detail. Besides, deep learning approaches have also been applied to lung ultrasound (LUS), which has been shown to be more sensitive than chest X-ray and CT images in detecting COVID-19. In the absence of a vaccine, the machine learning-related approaches are applied to analyze vaccine candidates in the realm of biology and medicine. The telehealth system played a major role in combating the pandemic from all aspects and reducing contact with patients during this period. Natural language processing-related methods are utilized to analyze tweets related to the COVID-19 epidemic on social media, and further analyze public sentiment and subject modeling, so as to arrange corresponding measures to appease public sentiment. In particular, this survey is to summarize and analyze the contributions made in various fields during the COVID-19 pandemic by considering both the contribution of deep learning in chest X-ray and CT images, as well as the application of the latest LUS during the COVID-19 pandemic. Telehealth and the importance of public sentiment analysis during a pandemic were also described in detail.
Keywords: COVID-19; epidemiological propagation models; deep learning; transfer learning; classification-detection; lung ultrasound; telehealth system
A pandemic is an occurrence of a disease or wide-scale outbreak of any infectious disease, which can rapidly increase mortality and morbidity rates across a huge area [1]. The pneumonia is caused by a virus known as “severe acute respiratory syndrome coronavirus 2” (SARS-CoV-2), which is dubbed Coronavirus Disease 2019 (COVID-19) by the World Health Organization (WHO) [2–6]. The WHO, on March 11, 2020, then proclaimed the COVID-19 outbreak a global pandemic [7]. A virus is a pathogen consisting of genetic material in a protein coating, such as the common flu, colds, Severe Acquired Immunodeficiency Syndrome (AIDS) and Ebola, which are all infectious diseases of varying severity that are usually caused by viruses [8]. Coronaviruses are enveloped, positive single-stranded large RNA viruses that can infect humans and a variety of animals.
The clinical manifestations of patients with COVID-19 infection are extremely complex. Fever, cough, and shortness of breath are the most prevalent symptoms of patients [9,10], and appear as decreased blood oxygen saturation, blood gas deviation etc. Severe patients will suffer from septic shock, pulmonary edema and acute respiratory distress syndrome (ARDS), heart failure, shock, etc., which may lead to the death of the patient [11–14]. Patients infected with COVID-19 can also experience insomnia [15]. Zhang et al. [16] proposed that melatonin could improve sleep quality in patients. Epidemiological studies have shown that elderly patients are more likely to develop serious diseases, while children tend to have milder symptoms [17]. Although COVID-19 mainly affects the lungs, it usually causes multiple organ involvement. COVID et al. [18] proposed the need for an interdisciplinary approach to address the potential acute post-care needs of recovering COVID-19 patients. The medical clinical phenomena play an important role in detection of COVID-19 and monitoring disease progression. However, some diseases don’t show any obvious symptoms, and even the proportion of individuals infected by SARS-CoV-2 is still uncertain for the whole period of infection [19], which poses a huge threat to public health. Although reverse transcription polymerase chain reaction (RT-PCR) is considered as the gold standard for detecting SARS-CoV-2 in the analysis of respiratory specimens from patients with suspected COVID-19 [20,21], it usually becomes difficult to effectively screen suspicious cases, even with a high false negative rate [22], due to lack of resources and strict testing environment. Ai et al. [20] used RT-PCR as a reference standard to evaluate the performance of CT in the diagnosis of COVID-19. Compared with RT-PCR, the diagnosis of COVID-19 by chest CT images is more sensitive, more stable and more conducive to epidemic control [20].
COVID-19 spread via respiratory and extra-respiratory routes, which may benefit in an explanation of the rapid proliferate of disease [23]. As a result, the use of breathing aid device is very helpful in patients with COVID-19 infection [24]. In addition, in order to limit the number of people infected by means of transmission, simulating the COVID-19 outbreak’s transmission process is critical [25]. Mathematical epidemiology is a venerable and well-respected field [26,27]. For numerous years, mathematical modeling has been employed in epidemiological studies [28], the majority of which are based on the Susceptible-Exposed-Infected-Removed (SEIR) and Susceptible-Infected-Recovered (SIR) models [29]. Menon et al. [30] studied the spread of COVID-19 in a broad population using deterministic mathematical models of disease propagation, such as SIR and its variants. To explain the transmission of infection during the incubation phase, Lopez et al. [31] suggested a modified SEIR model. To track and segregate patients, Levesque et al. [32] devised a parsimonious Crump-Mode-Jagers continuous time branching process of propagation. Modeling epidemics by using mathematical models based on SIR models is of benefit to predicting the spread of epidemics and providing epidemiological parameters [29]. In the meanwhile, it can assist the government in developing COVID-19 prevention and control policies. Deep learning is one of the most representation-learning approaches with multiple levels of representation, and it is capable of explaining complicated problems using learning methods from simple depictions [33]. Deep learning methods are capable of representing the ability and characteristics of learning data accurately in a deep manner [34,35], and it is widely utilized in medical systems such as biomedicine [36,37], smart healthcare [38], drug discovery [39], virus severity and infectivity [40], and medical image analysis [41,42].
Deep learning methods are of great significance for the detection of COVID-19 through the chest X-ray or CT images. Apostolopoulos et al. [43] used Convolutional Neural Networks (CNNs) with transfer learning algorithm to present automatic detection methods for chest X-ray images. The appearance of the chest X-ray image is often difficult to explain because of its non-standard and vague terminology, such as airborne disease, pneumonia, infiltration, patchy opacity, and opacity [20,44–46]. The most common CT findings of COVID-19 infection are the typical histologic pneumonia pattern of lung injury, with ground-glass appearance, consolidation, or both predominating in bilateral, peripheral, and basal areas [44,45,47]. Wang et al. [48] used a fully automatic deep learning approach to diagnose COVID-19 from chest CT images. Loey et al. [49] proposed Conditional Generative Adversarial Nets (CGAN) to detect COVID-19 in chest CT scan images. Deep learning-based methods contributed significantly to the discovery of new drugs, in addition to determining whether a patient’s chest X-ray and CT images are infected with COVID-19. Patankar et al. [50] used the LSTM deep learning method to calculate medications to explore possible drug candidates for the treatment of COVID-19. Zhang et al. [51] established a deep learning model based on a dense fully convolutional neural network (DFCNN) to implement protein-ligand interaction pairs for further discovery of protease medicines related to COVID-19.
Similarly, in fighting against the epidemic, machine learning (ML) methods can also be viewed as a powerful processing method. Sethy et al. [52] used the Support Vector Machine (SVM) method to analyze the features of coronavirus disease and constructed the classification-detection approach for detection. Ong et al. [53] proposed the newly developed machine learning-based Vaxign-ML reverse vaccinology tools to predict COVID-19 vaccine candidates. In this crisis, some people will experience psychopathology, such as depression and anxiety. Due to the large-scale family quarantine, it has greatly restricted direct psychosocial testing of patients [54]. However, an analysis of public comments on social media platforms can provide insight into how people’s mood change during the epidemic. ML has produced promising results in a variety of natural language understanding tasks, including sentiment analysis and text classification. Jelodar et al. [55] presented a systematic framework based on the Natural Language Process (NLP) to extract meaningful comments about COVID-19 on Reddit. Li et al. [56] used NLP and Weibo data to classify the COVID-19-related information. The extraction and classification of social media comments is useful in identifying patient’s emotional changes during an epidemic disease, which can encourage the government to make timely effective arrangements.
The rest of this paper is organized as follows: Mathematical modeling-related approaches were used to predict the spread of the COVID-19 pandemic approaches in Section 2; In Section 3, deep learning-related approaches were used to the chest X-ray and CT images and construct the approaches for the diagnosis of COVID-19 infected patients; and deep learning-based method for detecting COVID-19 in LUS was introduced in detail; and the contribution of ML in biology and medicine was introduced to predict vaccine and antibodies; In Section 4, the application of telehealth systems during the pandemic and the application of NLP-related methods to analyze COVID-19 tweets on social media are introduced. The discussion, future research focus and the conclusion are summarized in Sections 5–7, respectively.
2 The Propagation Analysis of COVID-19
In recent years, the analysis method based on mathematical modeling-related approaches has always been favored by researchers and engineers, and has been widely used in various aspects of the application scenes, such as biology [57], epidemiology [58] and medical sciences [59]. This is due to its precise description of the mechanical model. Traditionally, these approaches involved deterministic models based on algebra, vector calculus, regression, differential equations, etc. to solve real world problems [60]. The standard compartmental model such as SIR, can date back to the mathematical tour de force of Kermack et al. [61]. The SIR model is one of the deterministic models employed to simulate the spread of infectious diseases, which comprises three compartments: susceptible (S), infectious (I) and recovered (R) [30]. The two parameters S and R, as the name implies, represent the susceptible part and the infectious part respectively. In particular, R can be viewed from a macroscopic perspective as the composition of all immune individuals, including individuals who have recovered and died. The overall framework of this survey is shown in Fig. 1.
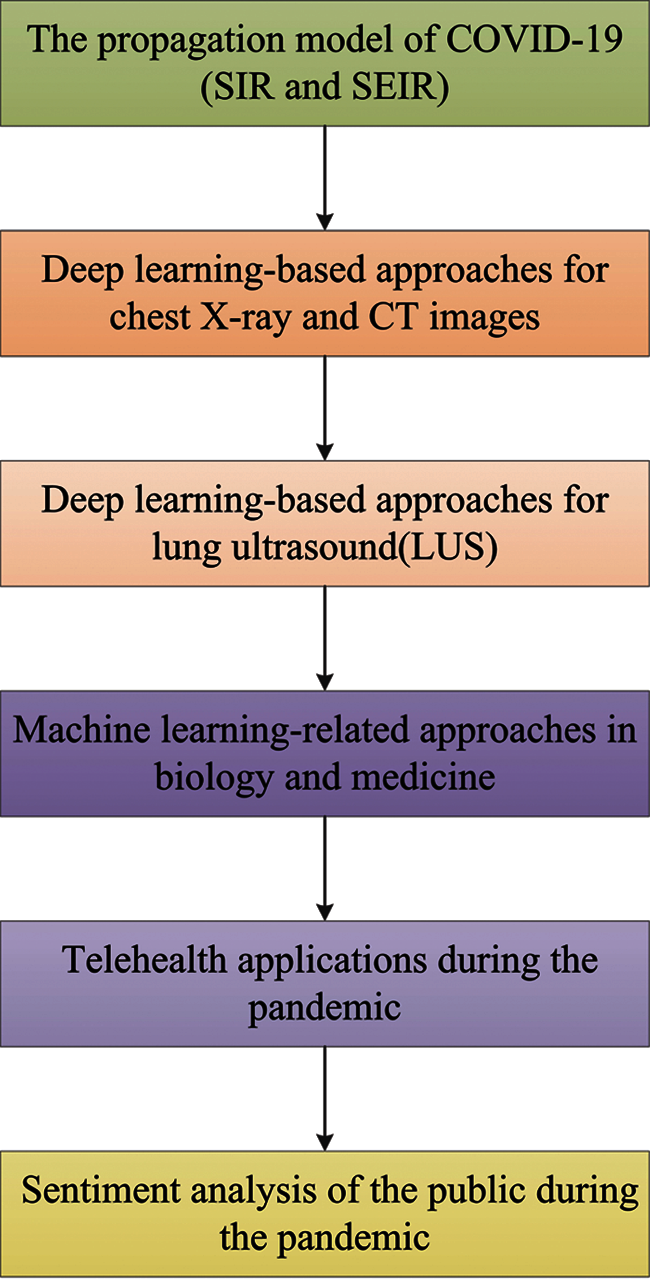
Figure 1: The processing diagram of this paper
Menon et al. [30] used the deterministic mathematical model of the SIR and its variants to study the propagation of COVID-19 in the Indian population. Chen et al. [62] proposed a time-dependent SIR model to address undetectable infections in COVID-19, which tracked the transmission and recovery rate. Wang et al. [63] extended the SIR model to incorporate various types of time-varying quarantine protocols, which contribute to timely analysis and evaluation of the timecourse dynamics of a range of infectious disease epidemics. Compared with the traditional static SIR model, the time-based SIR model proposed is more adaptable and robust. Infectious diseases transmission routes are difficult to determine, and the parameters involved are difficult to estimate [64,65]. Many researchers are always trying to assess the relevant parameters of the emerging coronavirus pandemic, in order to improve prediction accuracy. Heritability, severity, sensitivity and specificity etc. parameters have been evaluated by Shearer et al. [66] and Moss et al. [67]. Biswas et al. [68] studied the law of COVID-19 case data using the SIR model on the Euclidean network and reproduced China’s data with great accuracy under the premise of known relevant parameters, allowing them to plan appropriate measures in advance to limit the number of infections. To reduce the uncertainty of model prediction as much as possible, Zhong et al. [69] developed a simplified SIR model with only two parameters: the infection rate and the removal rate. In particular, Chikina et al. [70] used a simple SIR-like epidemic model to simulate the effect of age-targeted mitigation strategies for a COVID-19-like epidemic, demonstrating that they can effectively reduce mortality and ICU utilization. Singh et al. [71] used an age-structured SIR model to study the transmission process of COVID-19 among age and inter-age social contacts, emphasizing the role of social distance in moderating virus transmission.
Many infectious diseases are in the incubation period, during which individuals are infected with no spread. Therefore, the ‘exposed’ individual compartment is added to the SEIR models to migrate the susceptible individuals. Long incubation periods, different age groups, potential immunity deficiency, asymptomatic and symptomatic carriers, and minimum testing are all included in traditional SEIR epidemiology models [72]. The SEIR model has also traditionally been used to explore epidemic spread with various forms of networks of transmission that define the contact topology [73], such as scalefree networks [74–76], small-world networks [77,78], Oregon graph [79,80] and adaptive networks [81]. Zhan et al. [82] published the dynamics of human migration and the process of disease transmission, and used the classic SEIR model to combine inter-city travel data collected by Baidu migration data [83]. The classic SEIR epidemic model has also been used in [72] and incorporated dynamic features to analyze the spread of different age groups in specific dynamic partitions. Prem et al. [84] used an age-structured SEIR model for several physical distancing measures to simulate the ongoing trajectory of an outbreak in Wuhan. To consider the spread of infection during the incubation phase, a modified SEIR model has been proposed in [85]. Aside from the incubation period and the patient’s age, temperature and humidity have a potential impact on the spread of infectious diseases. Guo et al. [86] studied the link between temperature and humidity by using the equation derived from the SEIR model to analyze the influence of weather on the transmission rate of COVID-19.
Wearing a mask can reduce the likelihood of viral infection, because COVID-19 is highly contagious through a patient’s respiratory tract. Rahman et al. [87] developed systems that automatically detect if pedestrians are wearing masks to limit the growth of COVID-19. Eikenberry et al. [88] developed a compartmental model for assessing the community-wide impact of masks used by the general and asymptomatic public. Self-protection and isolation have effectively controlled the spread of the virus. Peng et al. [89] proposed a generalized SEIR model that improved on the classic SEIR model by accunting for factors such as self-protection mechanism, new isolation status, and preventive measures on the epidemic. The adding of a new quarantined sate is driven by data, which along with the recovery state, constitutes the original R state in the classic SEIR model. The corresponding epidemic model for COVID-19 is shown in Fig. 2.
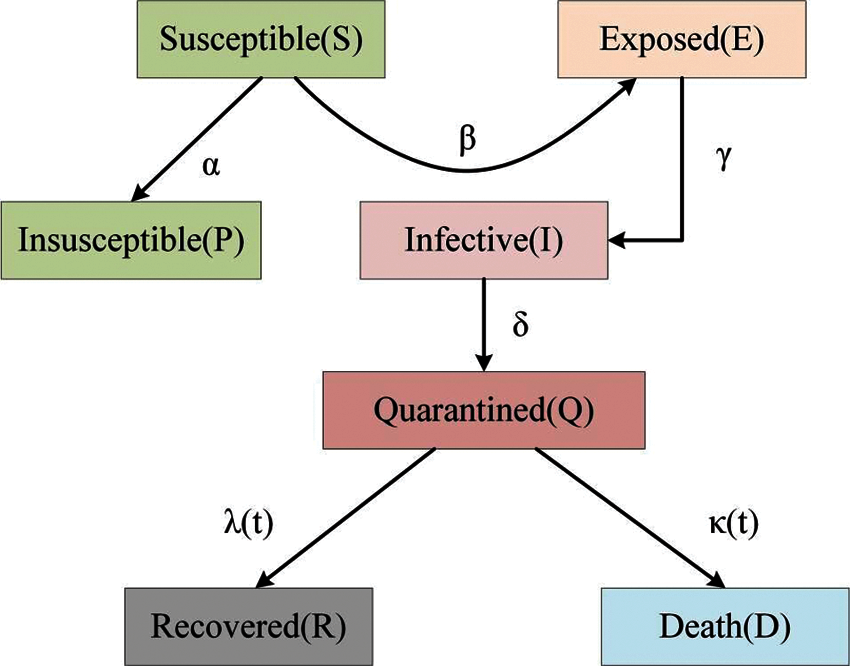
Figure 2: The epidemic model for COVID-19
Many studies have used a hybrid model based on the SEIR model to predict COVID-19 transmission changes more comprehensively and effectively. On the Johns Hopkins COVID-19 dataset, Pandey et al. [90] utilized the SEIR model and the regression model to predict changes in virus’s spread, and the findings showed that the regression model prediction had a higher root mean squared log error (RMSLE) that the SEIR model prediction. Considering the long-term impact of measures taken by the government, such as increasing the duration and intensity of isolation, a 4 + 1 penta-group model was established to predict the future trend of disease outbreaks [91]. A mathematical model of bat-host-reservoir-human transmission was proposed by Chen et al. [92] based on the SEIR model to simulate the potential transmission of bat-to-human infections. Others, meanwhile, use purely mathematical approaches to analyze the virus’s spread. Chen et al. [93] introduced a novel dynamic system with time-delay, which was applied in the differential equation to analyze the COVID-19 incubation duration. A discrete model has been proposed in [94], to simulate the dynamic process of coronavirus transmission between humans or animals in the same or other regions. The model based on the discrete-time Pontryagin maximum principle was applied to achieve optimal control, and it has been shown to be one of the most cost-effective strategies to control the disease.
Aside from the deterministic methods described above, the infectious disease modeling process can also be stochastic [95], which is useful when no relevant epidemiological parameters are available. Hellewell et al. [96] proposed a stochastic transmission model to assess the impact of contact tracing and isolation on the transmission of COVID-19 cases, which is a branch process model of stochastic modeling. In contrast to [96], Levesque et al. [32] paid more attention to the stochastic formulation of COVID-19 and built a parsimonious Crump–Mode–Jagers continuous time branching process of propagation to provide a basis for making decision for contact tracing and isolation. Furthermore, one of the most important properties of epidemics spread is spatial propagation, which is primarily determined by epidemic mechanism, human mobility and control strategy [97]. An in-depth understanding of the spatio-temporal dynamic characteristics of infectious diseases could be helpful for epidemic prevention and control [98]. Arenas et al. [99] provided a mathematical model based on the Microscopic Markov Chain Approach (MMCA) for the spatio-temporal transmission of epidemics, showing that in the current stage of the epidemics, stricter social distance containment measures are required to avoid the health system collapsing.
SIR and SEIR-based mathematical models are deterministic models that are used to predict the propagation of simulated epidemics. Many studies that predicted the spread of COVID-19 epidemics have been based on SIR and SEIR models, which have been analyzed and modified to take into account things such as the effects of social interventions, age, and social distance. However, there are some limitations to using deterministic models to simulate and predict epidemic diseases. To more properly evaluate COVID-19 propagation, a stochastic model was applied, in which random variables and probability distributions were used to characterize parameter interactions. In addition, model parameters are one of the most significant challenges in predicting the propagation of an epidemic. The more complicated the model structure is, the more parameters need to be determined, which makes it difficult to determine all the parameters in the absence of epidemiological data [69]. However, to some extent, the use of stochastic modeling reduces the uncertainty of parameters. According to the findings of the preceding studies, assessing and forecasting the COVID-19 transmission process is critical for raising public awareness and timely mobilizing resources for effective control and treatment.
3 The Application of Neural Network-Based Detection Methods during the Pandemic of COVID-19
Neural network learning is a process. Some sample patterns are successively input to the network under the incentive of its environment, and the weight matrix of each layer of the network is updated according to a certain learning method. The learning process is completed when the weight of each layer of the network converges to a certain value. The resulting neural network can then be used to classify the real data. Wang et al. [100] proposed a method for classifying heartbeats based on a dual fully connected neural network to determine whether the heart rate is abnormal.
CNNs with strong feature extraction capabilities and the ability to automatically learn features from images in a specific field [101–108], which have been widely used in the field of medical imaging [109]. CNNs strong feature extraction capabilities also come in here. Such as, Qin et al. [110] used a deep twin CNN to detect the misfire of diesel engines under strong environmental noise and different working conditions; In order to better mine meaningful deep information and apply it to the prediction of cutter head torque, Qin et al. [111] also utilized CNN and LSTM to extract the implicit features and sequential features of relevant parameters of shield tunneling machines.
3.1 Deep Learning and Transfer Learning-Based Approaches for the Detection of COVID-19 from Chest X-ray and CT Images
The transfer of learnt knowledge from a pre-trained network that completed one task to a new task is a popular strategy for training CNNs [108]. Transfer learning can be accomplished through the three following aspects [43,112,113]: i) ‘shallow tuning’, which adjusts only the last classification layer according to a new task without considering the parameters of other layers; ii) ‘deep tuning’, aiming to retrain all the parameters of the pre-trained network in an end-to-end manner; iii) ‘fine-tuning’, aiming to gradually train more layers by tuning the learning parameters to improve the performance of the network. In recent years, transfer learning has been often used in the field of medical image classification, such as tumor classification [114], skin lesions [115], cancer classification [116], retinal disease diagnosis [117] and pneumonia detection [118].
To reduce the training network time and computational complexity, the transfer learning algorithm has been used on the pre-trained AlexNet model proposed in [119]. By using the pre-trained AlexNet model to transfer the learning weights, bias and features to the approach of diagnosing COVID-19 from chest X-ray and CT images. To better extract lung features, a two-step transfer learning strategy based on COVID-19Net has been proposed in [48]. It uses the large CT-EGFR dataset for auxiliary training, and the pre-trained COVID-19Net is then applied to the COVID-19 dataset. Narin et al. [120] used five pre-trained transfer CNN models (ResNet50, ResNet101, ResNet152, InceptionV3 and Inception ResNetV2) to automatically classify four aspects of healthy, COVID-19, bacterial and viral pneumonia patients. Kassani et al. [121] used CNNs to extract features from images. Ten CNNs were used to distinguish infection of COVID-19 from non-COVID-19 groups, of which Resnet101 differentiated them with an accuracy of 99.51% [122]. Horry et al. [123] also used a pre-trained deep learning model to detect COVID-19 from chest X-ray images. Islam et al. [124] proposed a method of using VGG19, DenseNet121, InceptionV3, InceptionResNetV2 CNNs for transfer learning and combining with a recurrent neural network(RNN) for chest X-ray image diagnosis of COVID-19.
The transfer learning strategy was adopted to directly transfer the weights trained on other large datasets to the trained CNN model, due to the over-fitting problem caused by the limited number of trained images. The aforementioned transfer learning retains the parameters of the pre-trained model, but in order to improve the performance of network, many studies have used a more complex transfer learning strategy, which is applied to the pre-trained model by fine-tuning the parameters. Transfer knowledge via fine-tuning mechanism showed outstanding performance in chest X-ray and CT image classification [125–128]. The transfer learning strategy mentioned in [125–128], there have been many studies on the detection and classification of chest X-ray and CT images using transfer learning and deep learning that have been presented recently. Apostolopoulos et al. [43] proposed state-of-the-art CNNs for transfer learning by tuning the parameters, which shared some hyper-parameters and detected COVID-19 automatically. The COVID-19 detection system, proposed by Razzak et al. [129], used seven pre-trained CNNs model and a fine-tune transfer learning method on top of them to diagnose in healthy, bacterial pneumonia, and viral pneumonia scenarios. The overall accuracy of diagnosis was 98.75%. To improve model classification performance and reduce training time, the three-step technique was used to fine-tune the pre-trained model ResNet-50 and screen COVID-19 patients on the COVIDx dataset [130]. A Bayesian Convolutional Neural Network (BCNN) based on Dropweights, which was introduced in [131] based on a pre-trained ResNet50V2 model and fine-tuning the original model, aims to improve the diagnostic performance of human-machine decisions. In addition, Ucar et al. [132] proposed SqueezeNet a pre-trained model for diagnosing COVID-19 from chest X-ray images that is optimized using Bayesian method. Meanwhile, the SqueezeNet model needed to be fine-tuned and the overall classification accuracy of Bayes-SqueezeNet was 98.3%. Brunese et al. [133] fine-tuned VGG16 to perform transfer learning and distinguish between common lung disease and COVID-19 from chest X-ray images, and then highlighted areas of COVID-like symptoms on chest X-ray images. He et al. [134] proposed an efficient sample deep learning method that combines contrastive self-supervised learning and transfer learning to detect chest CT images. Vaid et al. [135] used CNNs for transfer learning to detect chest X-ray images of patients, with a 96.3% accuracy. Wang et al. [136] proposed a model called CCSHNET for detecting COVID-19 in the chest CT images. The model is based on DCFDCA algorithm of the selected two optimal models, for which they developed a SAPNF algorithm is developed to optimally determine the best pre-training model (PTM) and with a hyper-parameter of number of layers to be removed (NLR, Symbolized as L), and then the proposed L2TfL algorithm realized the feature learning process of the two models.
COVID-19 detection accuracy can be significantly improved by preprocessing the dataset. Toaçar et al. [137] proposed preprocessing the data with fuzzy color technology, followed by training the acquired feature sets with a deep learning model and processing them with the Social Mimic Optimization method, rather than using transfer learning directly. Finally, SVM was used to combine and classify the effective features. A generative adverse network (ACGAN) model based on an auxiliary classifier was used to generate the synthesized chest X-ray images, and it was demonstrated that the synthesized images could be used to improve CNN detection performance [138]. Two data enhancement models were proposed by Sedik et al. [139], the first is to enhance the dataset with simple image transformation to achieve the deep learning model’s effective learning level, and the second is to enhance the data with a GAN network. COVID-19 was detected using CNNs and convolutional long short-term memory (CONVLSTM) after data enhancement. Using a new technique of multi-path data enhancement, Wang et al. [140] extracted features from a self-created CNNs to learn a single image-level representation, and relation-aware representations were learnt from graph convolutional network (GCN). Then they developed Deep feature fusion (DFF), which fuses individual image-level features and relation-aware features from both GCN and CNN, respectively. In [49], five deep transfer learning models combined with classical data augmentation and Conditional Generative Adversarial Nets (CGAN) were proposed to improve the classification performance of the models, with the ResNet50 model combined with classical data augmentation having a classification accuracy of up to 82.91%. The proposed architecture of the classical data augmentation along with the CGAN and DTL models is shown in Fig. 3.
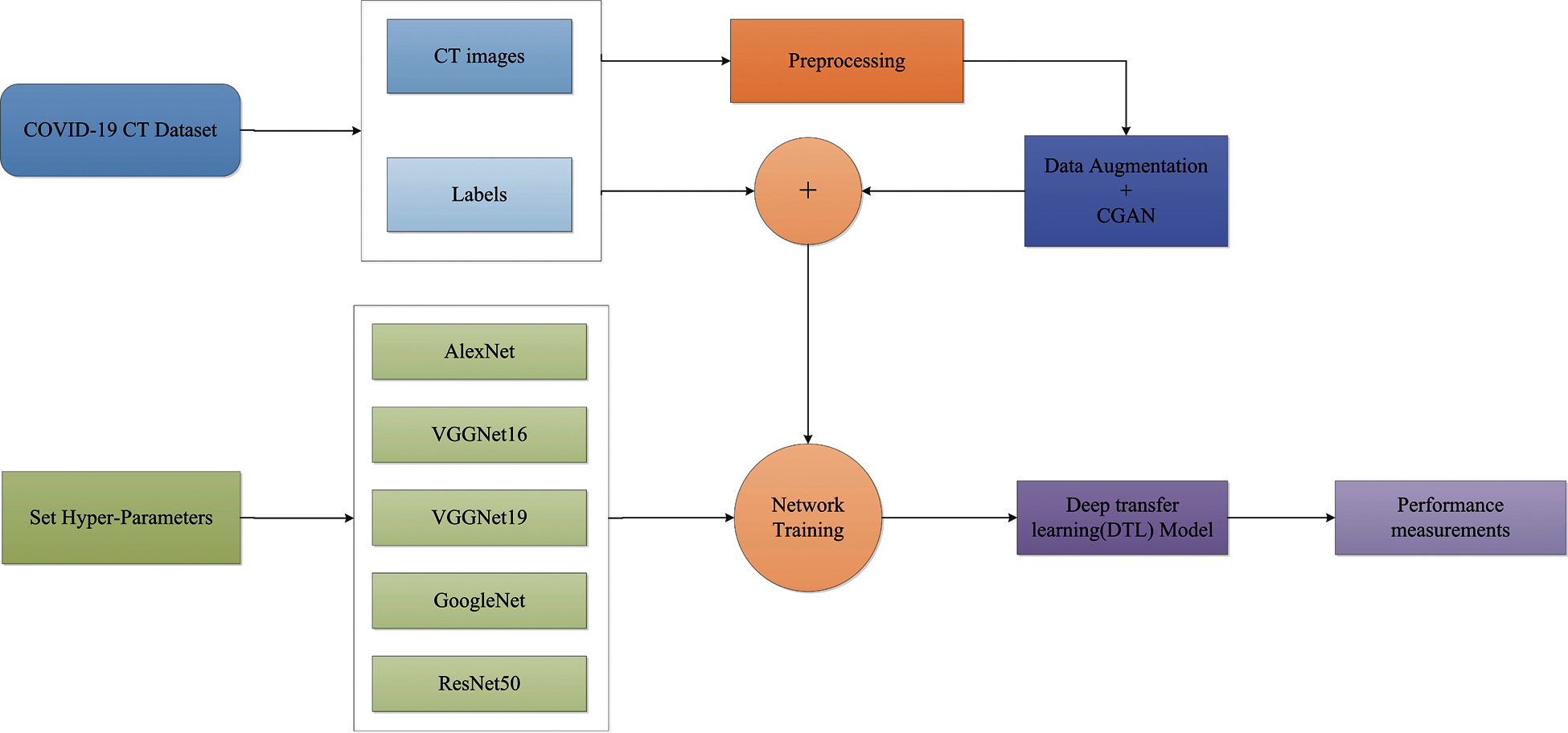
Figure 3: The proposed architecture of the classical data augmentation along with CGAN and DTL models
With an increase in the number of new coronary pneumonia cases worldwide, there are privacy concerns when considering data sharing among hospitals around the world. Ting et al. [141] explored the use of four interrelated digital technologies to enhance COVID-19 response: the Internet of Things, big data analytics, Artificial intelligence (AI), and blockchain. As a result, Kumar et al. [142] proposed using blockchain-based federated learning to train a global deep learning model. Federated learning safeguards the organization’s privacy while training the model on a global scale. They also used Capsule Network-based segmentation and classification to identify COVID-19 patients. The comprehensive experiments show improved performance in detecting COVID-19 patients. Deep learning method can not only detect the state of chest X-ray and CT images, but it can also track the disease, facilitate risk assessment, and arrange corresponding measures. Ye et al. [143] proposed an AI-driven system composed of the Attributed Heterogeneous Information Network (AHIN) and using cGAN to enrich it, namely alpha-Satellite, to automatically provide hierarchical community risk assessments related to COVID-19. Chimmula et al. [144] forecasted the COVID-19 transmission in real time in Canada using the Long short-term memory (LSTM) networks. Alazab et al. [145] also used the Prophet Algorithm (PA), the Autoregressive Moving Average (ARIMA) model integration, and the LSTM neural network to predict the number of confirmed COVID-19 deaths and resuscitation over the next seven days.
3.2 The Hybrid Deep Learning Approaches for the Detection of COVID-19 are Based on Chest X-ray and CT Images
Based on detection of chest X-ray images, Hemdan et al. [146] proposed a deep learning framework, COVIDx-Net, with seven different deep networks to automatically diagnose COVID-19. Song et al. [147] created deep learning-based diagnostic CT systems (Deeppneumonia) in three steps: first, extracting key lung features, then designing a DRE-Net neural network to extract K details from chest CT images and make predictions, and finally, diagnosing at the patient level. This system facilitates rapid identification of patients and the results reveal an AUC of 99% and a sensitivity of 93%. To analyze automated detection and patient monitoring based on chest CT images, Gozes et al. [148] proposed a 3D and 2D deep learning models. The 3D deep learning model analyzed the 3D lungs using existing software [149]. The U-net model was used to segment the lung in the 2D deep model, and pre-trained ResNet-50-2D CNNs were used to detect lung abnormalities. A deep learning system was proposed in [150] to screen patients that consists of two classification models. The first is the ResNet-18 model, and the second is to build a position-attention classification model on top of it to increase overall classification accuracy. The deep learning system also includes a 3D deep learning model to segment lungs from chest CT images. Besides, a 3D deep learning framework, COVNet based on RestNet50, was developed in [151] to extract both 2D local and 3D global representative visual features from volumetric chest CT exams for identifying COVID-19.
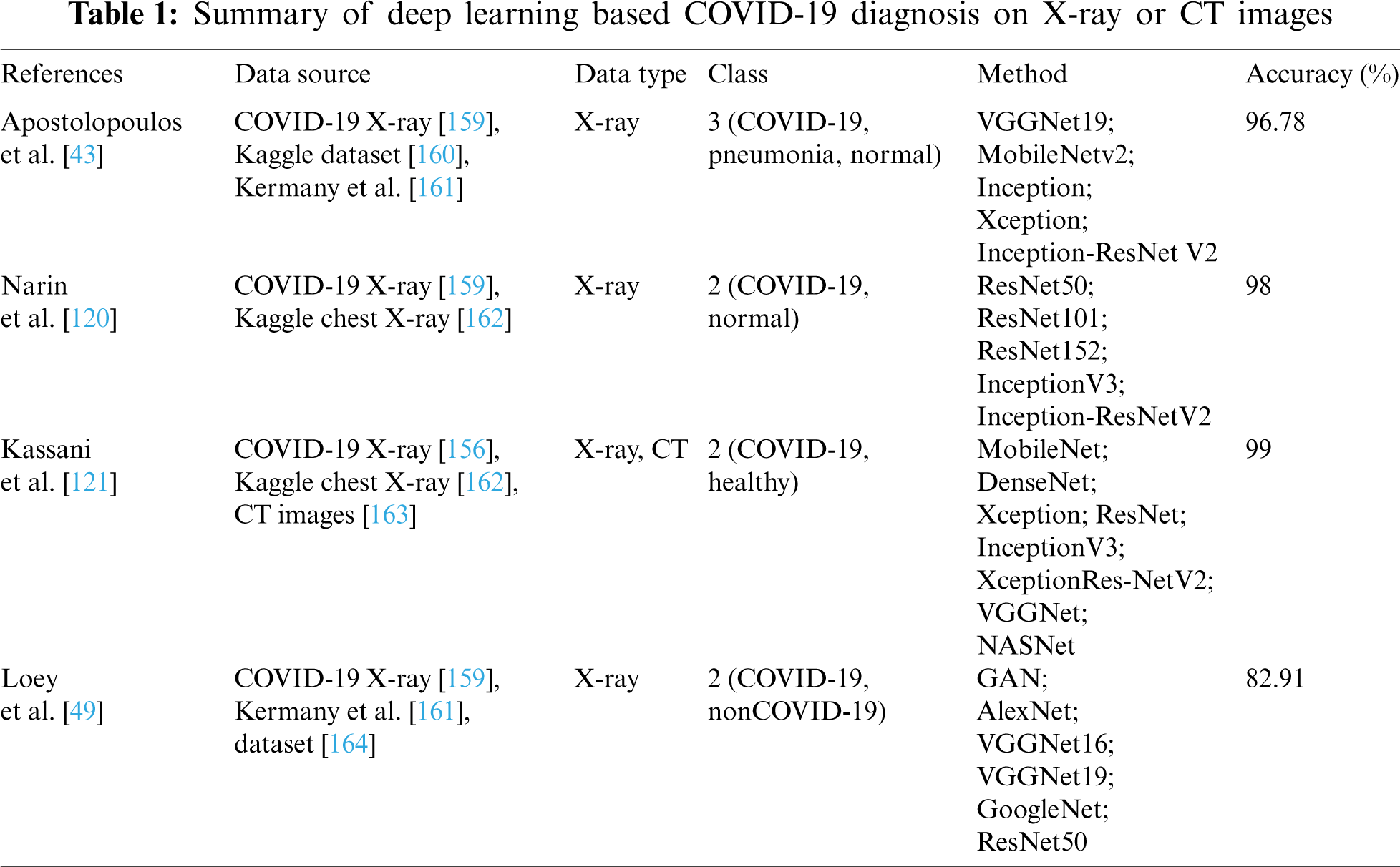
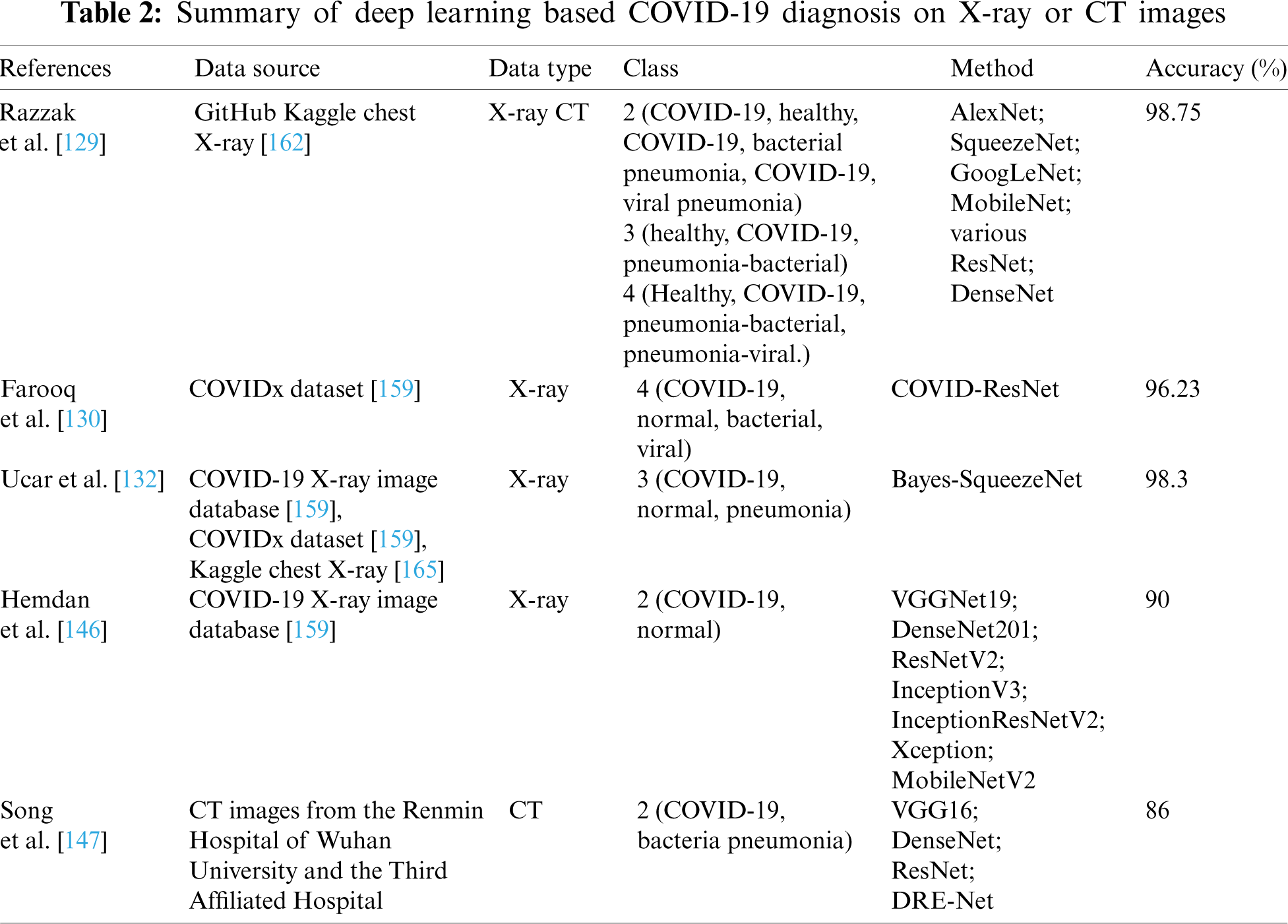
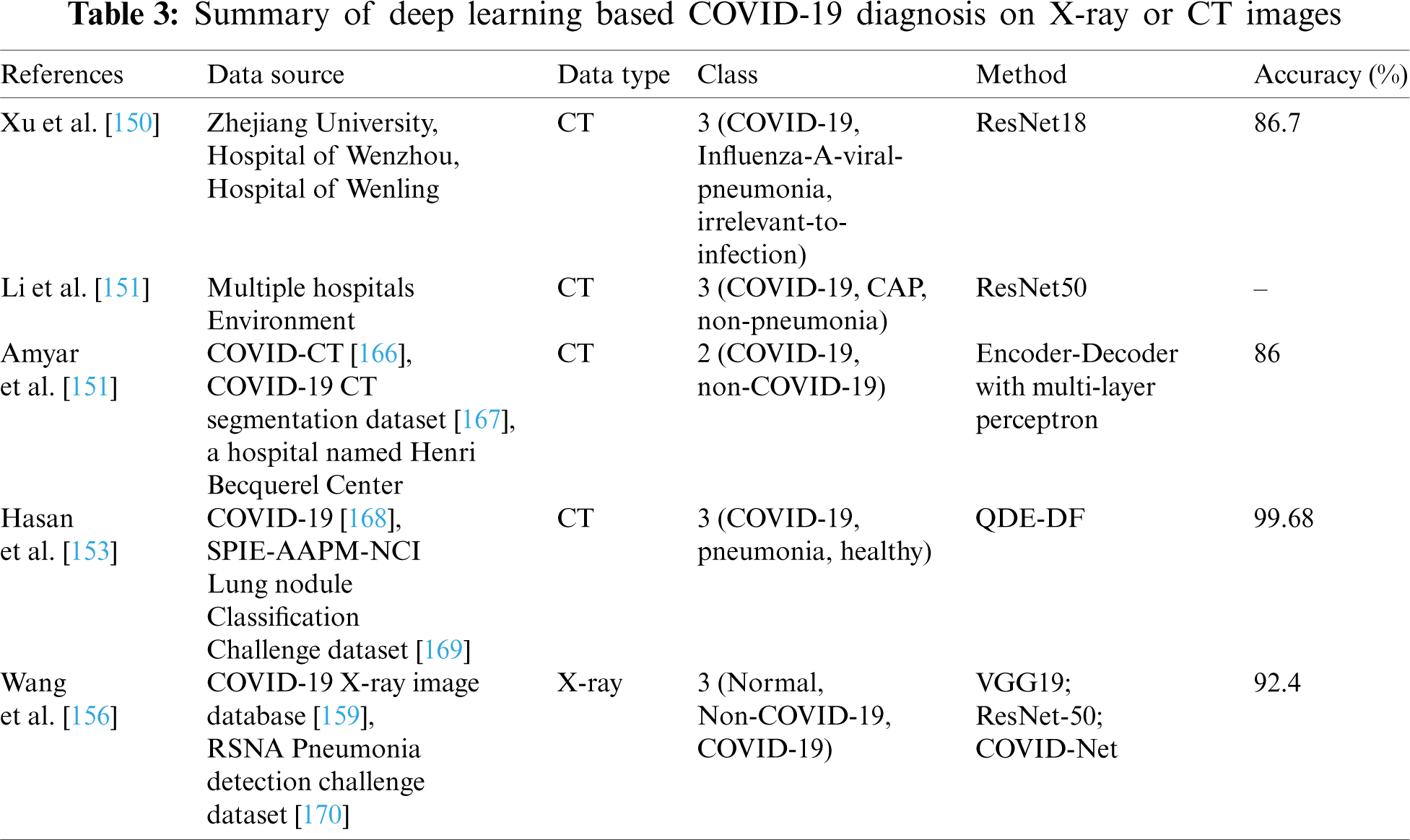
In order to improve model’s generalization capabilities, Amyar et al. [152] proposed a multi-task deep learning model to complete image reconstruction, classification and segmentation to screen for COVID-19 pneumonia. To solve image reconstruction and segmentation, the U-Net architecture was used as encoder and decoder, and CNNs was used to classify images. Hasan et al. [153] provided a method for differentiating between COVID-19 coronavirus, pneumonia, and healthy lungs that combined extracted features from deep networks with Q-deformed entropy handcrafted features. In this study, nine-layer CNNs and the Q-deformed entropy algorithm were used to extract features, which were then given to an LSTM neural network classifier to be classified. Islam et al. [154] proposed a deep learning technology integrating CNN and LSTM to achieve 99.4% accuracy in the automatic diagnosis of COVID-19 from chest X-ray images. Bandyopadhyay et al. [155] adopted LSTM and Gated Recurrent Unit (GRU) models to predict the positive, negative, death, and release of COVID-19 cases, and their predictions matched the clinicians’. The disease may be efficiently predicted and accurately recognized by using deep learning method, allowing appropriate actions to be implemented to effectively prevent the virus’s spread. To detect cases from chest X-ray images, Wang et al. [156] designed a COVID-Net framework. The COVID-Net utilized a lightweight residual projection-extension (PEPX) design pattern to enhance presentation capabilities while reducing computational complexity. Muhammad et al. [157] employed epidemiological data of COVID-19 patients in South Korea and a data mining algorithm to predict the recovery of COVID-19 patients. The results demonstrate that the decision tree’s data mining algorithm has the highest overall accuracy rate in the prediction outcomes, at 99.85%. Saha et al. [158] proposed an automatic detection scheme named EMCNET, which extracted deep features from the chest X-ray images of infected patients using CNN. Four ML methods, namely random forest, support vector machine, decision tree and Adaboost, were used to integrate a binary classifier to detect COVID-19 quickly. Parts of the aforementioned results are tabulated in Tabs. 1–3.
3.3 Deep Learning Approaches for the Detection of COVID-19 Based on LUS
The mixing of air and water in the chest cavity causes the signs of LUS [171]. LUS has become a helpful diagnostic and testing tool in recent years. LUS is frequently used to diagnose lung parenchymal and pleural diseases, as well as in intensive care units [172,173]. LUS offers the advantages of being simple to use, repeatable, radiation-free, and inexpensive [174]. During the current pandemic, LUS has got a lot of attention and has been pushed as a potentially useful tool for real-time detection of COVID-19 [175–177]. LUS, on the other hand, has certain limits. Ultrasonography can not detect lesions in the deep lung, because the inflated lung hinders its conduction. In a nutshell, the anomaly must extend to the pleura’s surface in order to be visible on ultrasound; Otherwise, it needs to be examined by CT [178]. LUS can reduce patient exposure to ionizing radiation and the incidence of cross-contamination by minimizing the need to move the patient, whereas CT requires moving the patient and controlling the long-term risks associated with ionizing radiation [179]. LUS, in particular, is safer for examining pregnant women [180,181].
During the pandemic, Soldati et al. [182] stated that the current clinical evidence strongly suggests that the LUS has the potential to be a diagnostic tool. Soldati et al. [183] also proposed a standardized method for optimizing the LUS examination of COVID-19 patients by using two pocket devices for LUS examination to reduce exposure to medical staff. In developing countries, LUS is an effective tool for diagnosing pneumonia. LUS imaging has emerged as a safe bedside imaging alternative that avoids exposing patients to radiation and minimizes the risk of infection [184]. Bedside LUS has shown better sensitivity than chest X-ray images in the diagnosis of pneumonia in Nepal [185]. Unlike most previous studies that focused on chest X-ray and CT image scanning, Roy et al. [186] studied the use of deep learning approach in the analysis of LUS images. They proposed a deep network called Regularised Spatial Transformer Networks (Reg-STN), which can predict the disease severity score associated with the input frame while also locating pathological artifacts in a weakly supervised manner. To classify COVID-19 from LUS images, Muhammad et al. [187] proposed a light CNN model. Carrer et al. [188] proposed an automatic and unsupervised method for detecting and locating the pleural line in LUS data, as well as using SVM to classify the patients. Controlling the COVID-19 pandemic necessitates the use of a quick and safe diagnostic tool. In comparison to chest X-rays and CT images, LUS has numerous practical advantages [189]. Born et al. [190] developed a frame-based CNN that correctly classifies COVID-19 from LUS datasets. Awasthi et al. [191] established Mini-COVIDNet, a light weight deep neural network, based on COVID-19 detection using LUS video. Dastider et al. [192] proposed an integrated autoencoder-based hybrid CNN-LSTM model that detects COVID-19 severity scores from LUS images very well.
LUS, as previously stated, is effective in detecting COVID-19 pneumonia. In relevant studies in China [178] and Italy [183], the physical basis and LUS pattern of COVID-19 patients were provided, indicating that LUS can be a useful tool for the diagnosis and monitoring of COVID-19 pneumonia. However, no data on the use of LUS in children with COVID-19 pneumonia are currently available. Although COVID-19 infection in children is rarely as severe as in adults, it has been showed in several studies to impact children of all ages, from newborns to adolescents [193–196]. The observation that LUS can detect COVID-19 pneumonia in children is of clinical significance because LUS has been shown to be accurate in diagnosing pediatric pneumonia of any origin [197–201]. Musolino et al. [202] described LUS features of 10 consecutively admitted children with COVID-19 in two tertiary-level pediatric hospitals in Rome, it is mainly manifested as vertical artifacts (70%), pleural irregularities (60%), areas of white lung (10%) and subpleural consolidations (10%). This study is crucial since children with COVID-19 have fewer data on LUS. LUS will be an effective method for detecting children with COVID-19 if physicians have LUS experience recording children with COVID-19.
3.4 Machine Learning-Related Approaches for the Analysis of Biology and Medicine
Deep learning methods have begun to replace traditional methods to develop drugs that effectively combat this epidemic, because traditional methods take a long time for developing a drug. Zhang et al. [203] established a 3D-like protease protein model using homology modeling and detected potential drugs for the 2019-nCoV protease. In order to swiftly and accurately reverse search for the drug targets, a Dense Fully Convolutional Neural Network (DFCNN) deep learning model has been built to accurately sort/identify protein-ligand interaction pairings.
Based on immune profiling data, ML approaches have also been successfully used to predict antigen-specific immunological signatures in vaccines [204,205]. In order to pick the most promising vaccine candidates, Le et al. [206] compiled and shared information on the status of COVID-19 vaccine development activities around the world. Ong et al. [53] applied the Vaxign and the newly developed machine learning-based Vaxign-ML reverse vaccinology tools to predict COVID-19 vaccine candidates. In addition, finding peptides or antibody sequences that can suppress the SARS-CoV-2 virus’s epitope is critical for efficiently inhibiting the virus’s propagation [207]. Magar et al. [207] used a variety of ML approaches, including XGBoost, Random Forest, Multilayer perceptron, SVM and Logistic Regression, to predict the possible inhibitory synthetic antibodies for SARS-CoV-2. ML methods can also be used to complete case diagnosis tasks and prevent the spread of disease. Sethy et al. [52] map the image features collected from CNNs to a higher-dimensional space and implement coronavirus disease classification-detection. COVID-19 infection can be detected using the Routine Blood Exams with ML method, in addition to chest X-rays and CT images. Brinati et al. [208] developed two ML classification models to distinguish between positive and negative SARS-CoV-2 patients, as well as an interpretable Decision Tree model for clinicians interpreting blood tests (even off-line) for COVID-19 suspect cases. Machine learning-related methods for detecting COVID-19 were proposed in [209] based on the clinical text data. Textual clinical reports are divided into four categories using ML methods: COVID, SARS, ARDS, and both (COVID, ARDS). The test accuracy of logistic regression and multinomial Näive Bayes was 96.2%, outperforming other ML approaches. For the purpose of control and treatment of COVID-19 from the root cause, it is necessary to explain the origin of the virus and the viral genomic sequence as soon as possible. Randhawa et al. [210] identified an intrinsic COVID-19 virus genomic signature and related methods together with a machine learning-based alignment-free approach for classification of whole virus genomes.
4 The Telehealth and Sentiment Analysis-Based Analysis Framework of COVID-19 during the Pandemic
4.1 The Telehealth Analysis of COVID-19 during the Pandemic
The implementation of public health measures has a potential effect. In the absence of vaccines, it aims to reduce virus spread by reducing population exposure. For example, Wearable technology can provide real-time remote monitoring to reduce contact between doctors and patients [211]. Multiple interventions stratified together can effectively reduce population exposure and thus viral infections [212]. The concept of telehealth has always existed [213,214]. During the COVID-19 pandemic, scalable telehealth systems were widely used, which were critical to improving the quality of the medical system, ensuring the safety of patients and doctors, and lowering the infection rate [215]. Telemedicine can also support triage and classification of susceptible people before they are referred to the hospital [216].
Smith et al. [217] introduced key requirements for using telehealth, such as flexible funding arrangements, health worker training and accreditation. However, remote systems are still subject to licensing, integration and reimbursement issues in the majority of countries [218]. Wosik et al. [219] describe the role of telehealth in transforming healthcare delivery during the COVID-19 pandemic in the United States (US). Iran offers diagnostic expertise for COVID-19 via teleradiology services delivered via social media platforms [220]. Long-term social isolation may limit physical activity and affect mental health in COVID-19 patients with other conditions, such as cardiovascular diseases, so the use of telehealth health care intervention systems can reduce the morbidity and mortality of acute diseases caused by COVID-19 [221]. Vandekerckhove et al. [222] believe that video counselling in cardiology for further telehealth is essential for a flexible healthcare system for emergency needs outside of the COVID-19 pandemic. Moreover, children and adolescents are using telehealth [223]. Bokolo et al. [224] looked at how telemedicine and virtual software helped patients following the pandemic, as well as the social, organizational, and technical factors that influence patient and physician acceptance of telemedicine and virtual platforms. The social, organizational, and technical aspects of telemedicine and virtual platforms are illustrated in Fig. 4.
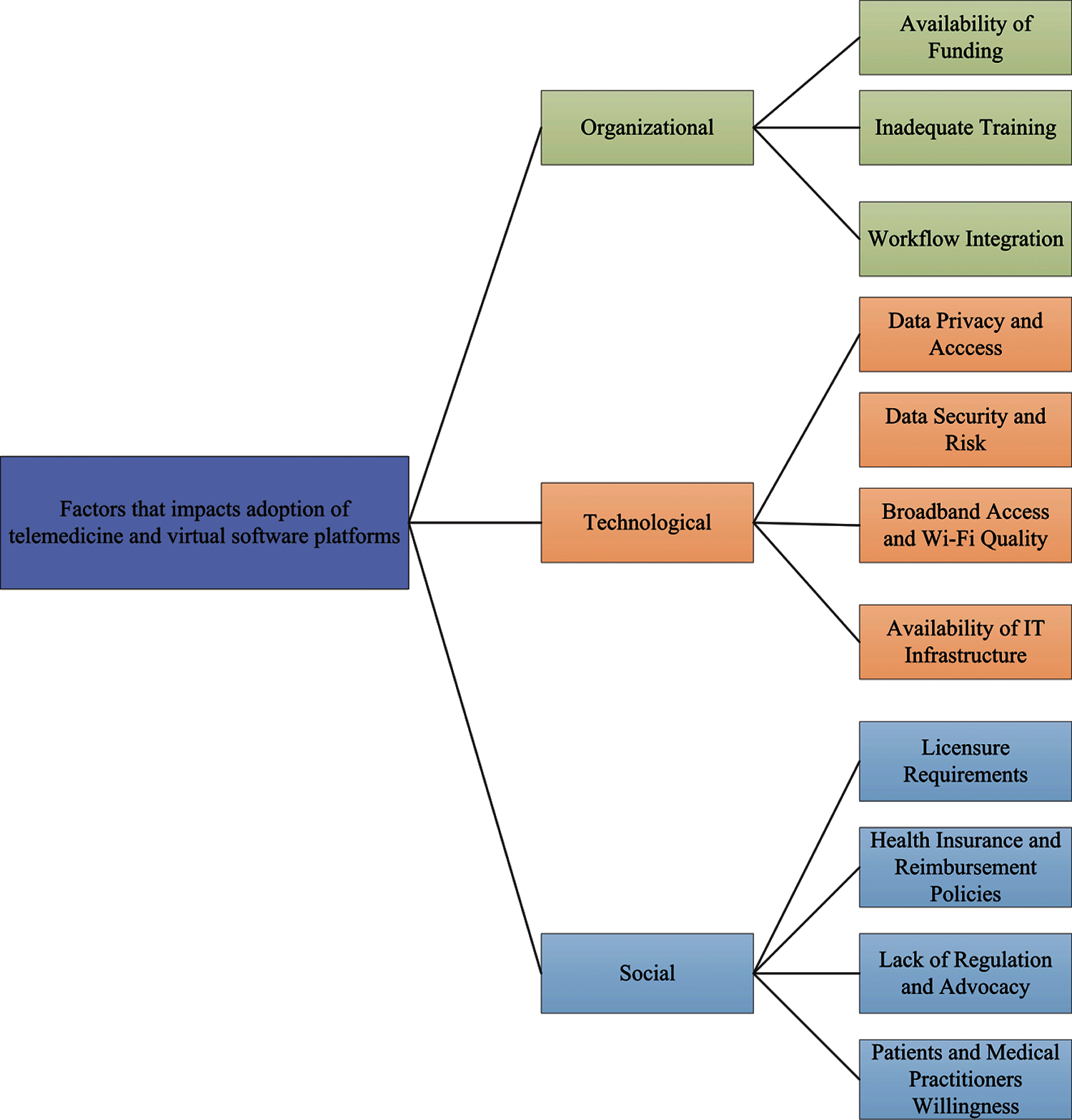
Figure 4: The social, organizational, and technical aspects of telemedicine and virtual platforms
Aslani et al. [225] briefly outlined the needs addressed by telemedicine, as shown in Fig. 5. It is also recommended that all countries respond to COVID-19 using telemedicine capabilities in accordance with their own national measures. Besides, during the COVID-19 pandemic, Jnr et al. [226] introduced practical guide to the use of telemedicine and virtual care. This study fully demonstrates the potential of virtual care solutions.
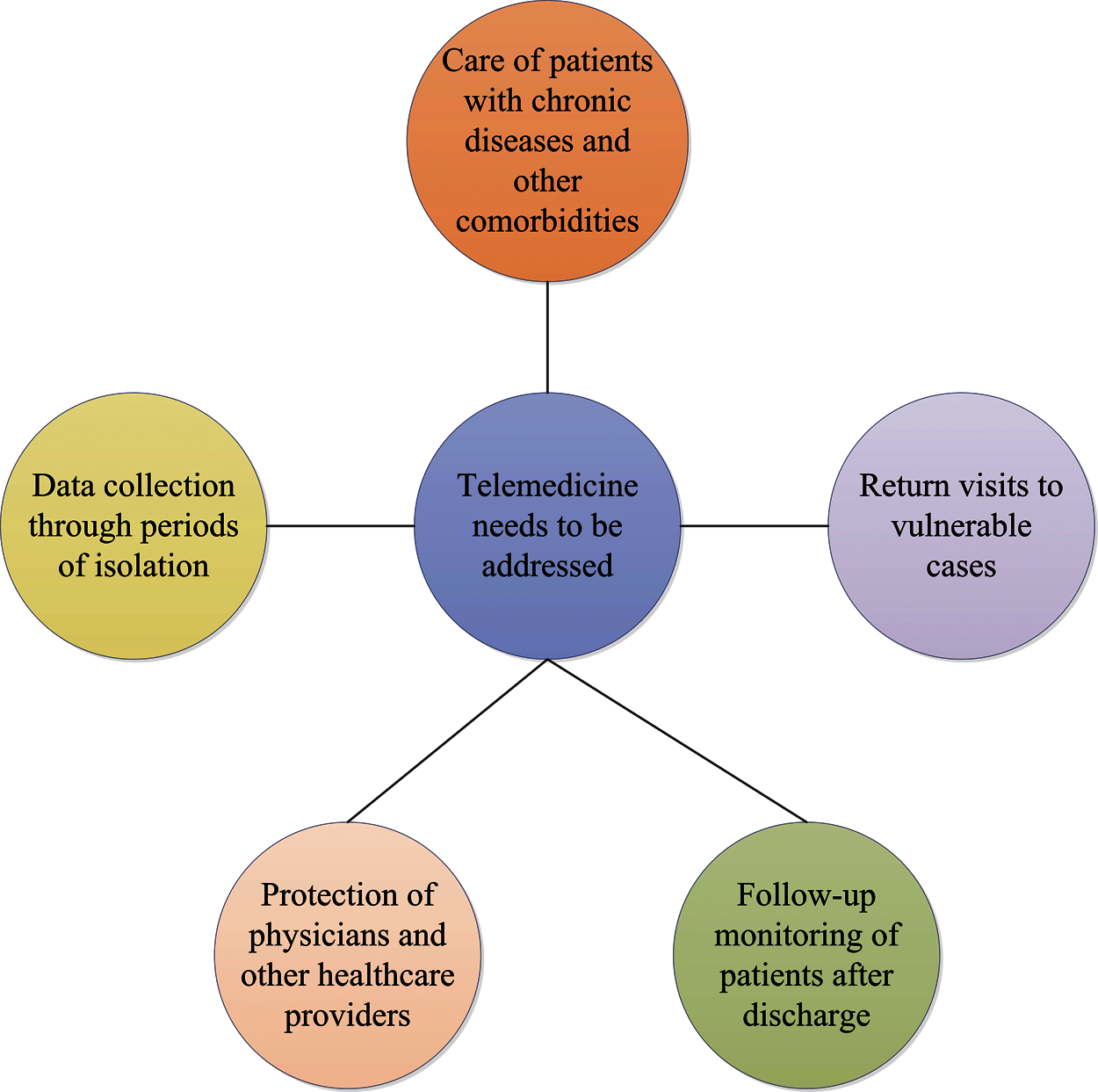
Figure 5: The processing strategy of the telemedicine
4.2 The Sentiment Analysis of the Public during the COVID-19 Pandemic
As we all know, in today’s world, it is common for people to use social media to learn about current events and share their thoughts. During the COVID-19 pandemic, social media played a huge role in keeping people connected and informed. Reports on the epidemic may be viewed online, and people can express their emotions through various social platforms. Hence, analyzing public sentiment has become extremely critical in preventing a series of psychological problems, such as depression.
To better understand the information crisis of COVID-19, it is indispensable to assess public sentiment. Samuel et al. [227] identified public sentiment related to the pandemic using coronavirus-specific tweets, and R statistical software, and its sentiment analysis software package, revealing the growth process of COVID-19 fear and negative sentiment in the US. When short tweets are classified using the Näive Bayesian method, the accuracy rate can reach 91%. Based on RNN, Nemes et al. [228] created an emotion prediction model. They utilized the model to analyze tweets on Twitter, search for words associations, and categorize various types of text into clearer categories of emotional intensity, such as weak positive/negative and strong positive/negative.
As the epidemic worsens, India has adopted blockade measures to prevent the increase in cases. After the lockdown, Indians’ comments on Twitter were studied by Barkur et al. [229]. The full analysis process was carried out using the R software, and a word cloud describing the sentiment of the tweet was generated. According to the study’s findings, the vast majority of Indians are quite in favor of the blockade measures. To assess the sentiment on Twitter in India during the peak of COVID-19 new cases in 2020, Chandra et al. [230] also proposed a deep learning-based LSTM RNN model and a multi-label sentiment classification framework using global vector (Glove) embedding. The results show that the majority of people are pleased with how the authorities handled the epidemic, while a small number are dissatisfied. Chintarapudi et al. [231] used the Bidirectional Encoder Representations from Transformers (BERT) model to analyze tweets from the Indian public during the COVID-19 lockdown and achieved an accuracy rate of 89%. Wang et al. [232] also used the fine-tuned BERT classification model to classify postings linked to the new crown on Chinese social media Weibo, including positive, neutral, and negative, and used the term frequency-inverse document frequency (TF-IDF) topic model to summarize the topics of the posts. This research also uncovered crucial depression-related motifs. Moreover, Fig. 6 depicts the workflow of the sentiment analysis model.
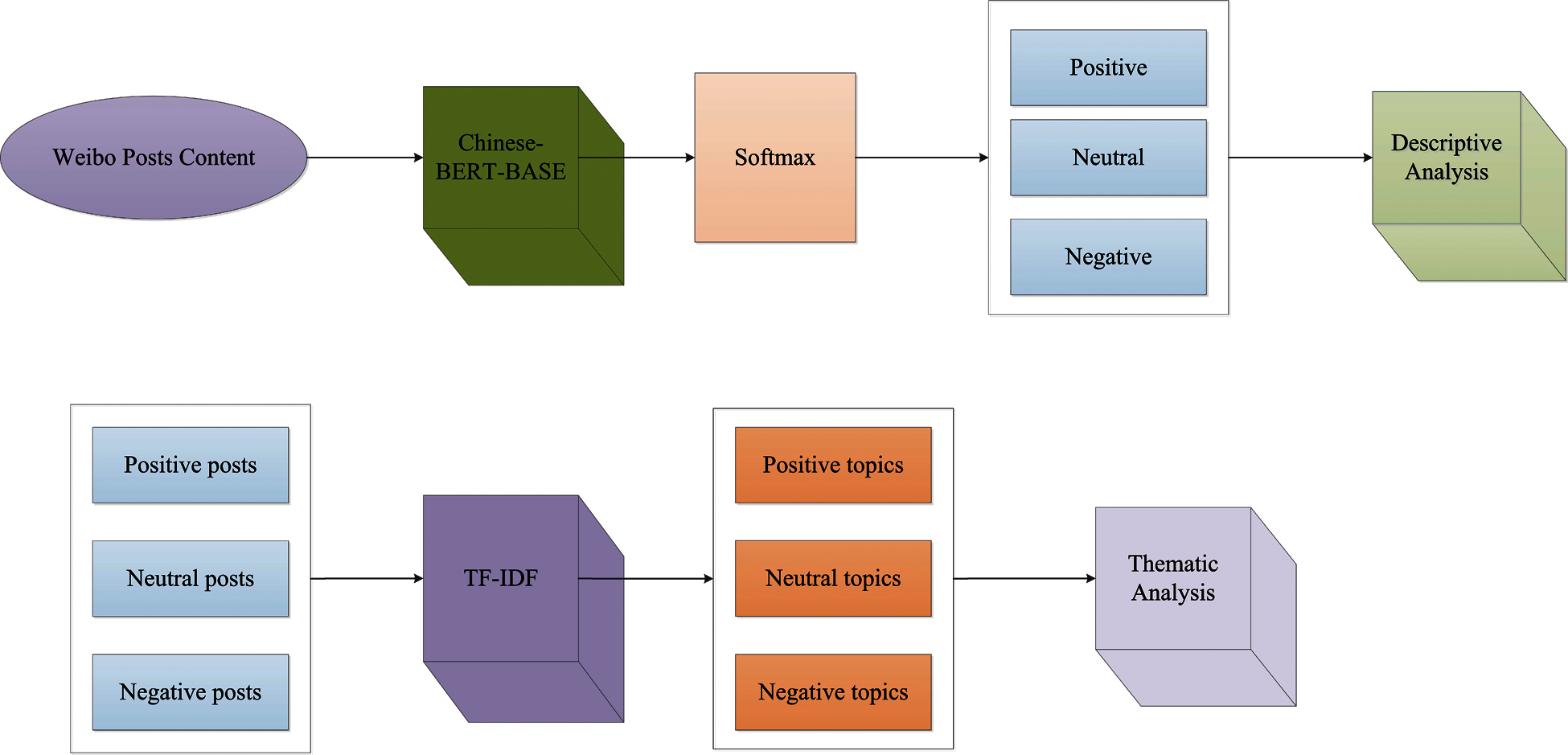
Figure 6: The workflow of the sentiment analysis model
Tclustvid, an intelligent cluster-based classification and topic extraction model, was proposed to categorize COVID-19-related tweets into positive, neutral, and negative emotions, as well as identify the most common topics [233]. The widely unsupervised Latent Dirichlet Allocation (LDA) [234] ML methods have been applied by Xue et al. [235], to analyze tweets concerning epidemics on Twitter. The subjects of 1,597,934 tweets made by 1,299,084 users from 18,413 news media articles and social media were modeled using LDA by de Melo et al. [236]. This research contributes to a deeper understanding of the COVID-19 pandemic’s observations and gives a more meaningful emotional timeline. Furthermore, to see if Twitter and traditional news media cover similar COVID-19 categories and topic kinds, the popular cosine similarity was employed as a measure of topic similarity and defined by Eq. (1),
where ta and tb are essential the vectors used to describe the topics a and b, respectively, and the value of the corresponding similarity range from 0 to 1. The NLP approach can automatically extract COVID-19-related discussions from social media. A systematic framework based on NLP has been presented in [55], which is able to extract meaningful topics about COVID-19 from Reddit. This research has certain significance for the government to promulgate measures related. Lopez et al. [31], like [55], recommend using network analysis techniques, NLP and text mining to analyze a multilanguage Twitter dataset to understand changing policies and common responses to the COVID-19 outbreak across time and countries. Li et al. [56] classified COVID-19-related information into seven types of situational information using NLP and Weibo data. Data classification using SVM, Näive Bayes and random forest classifiers is quite useful in assisting the public in effectively and quickly understanding the novel disease.
During the COVID-19 pandemic, Hung et al. [237] used NLP to determine whether tweets on Twitter were positive, neutral, or negative. Topic modeling using LDA and identifying five general themes. Overall, positive tweets outnumbered negative tweets, according to the study. Sanders et al. [238] built a new pipeline that used state-of-the-art NLP technology to analyze sentiment on topics related to mask wearing on Twitter. The analysis pipeline to extract, label, summarize, and present the themes, topics and sentiment present in tweet corpus. Retrieval and Sampling, Embedding and Sentiment Scoring, Clustering and Subclustering, Cluster and Subcluster Labeling, and Cluster and Subcluster Summarization are the primary sections of the analysis pipeline. Cluster and Subcluster Labeling is the fourth step, in which each word is scored based on its contribution to the Kullback-Leibler divergence, defined by Eq. (2), as it is relates to the word distribution of the cluster and the word distribution of the entire corpus sample,
where Ws and Wc are the word probability distributions for thecorpus sample and sub-sample (cluster), respectively.
Moreover, public sentiment will be influenced by the COVID-19 vaccine’s research and development. To foster public faith in the vaccination, it is vital to analyze public sentiment about the vaccine. From March 1 to November 22, 2020, Hussain et al. [239] extracted social media posts on the COVID-19 vaccine from Facebook and Twitter in the United Kingdom (UK) and the US. And they created a hierarchical hybrid ensemble AI model for sentiment analysis, which consists mostly of the Valence Aware Dictionary for Sentiment Reasoning (VADER), TextBlob and BERT based on deep learning. The study’s findings show that the public’s sentiment towards vaccines in the UK and the US on the two platforms is mostly positive and similar. This is in relation to the country’s recent announcements about vaccine development, testing, and supply, which have allayed public fears and concerns. Different from the method used by Hussain et al. [239], Ritonga et al. [240] employed the Näive Bayes algorithm to analyze the Indonesian public’s remarks on the COVID-19 vaccination in January 2021 in order to better understand people’s perspectives on the vaccine. According to the findings, up to 56% of the public had an unfavorable attitude toward vaccines during this time period.
COVID-19 has wreaked havoc on public health and safety, which have become a global concern [241,242]. Epidemiological propagation models, deep learning and the application of ML methods will aid people understand the transmission process of epidemics and increase our understanding of disease detection so that timely preventive measures may be adopted. Most studies of the COVID-19 propagation process mathematical modeling use the classic SIR and SEIR deterministic models, which take into account the effects of age, temperature, humidity, mitigation measures and other parameters on the epidemic’s transmission. The more complex the structure of the model, the more epidemic parameters are required. The determination of too many unknown parameters will result in a high level of uncertainty in the model’s forecast. To forecast the impact of COVID-19 transmission, a few studies have used stochastic modeling. The goal is to lessen the level of uncertainty caused by parameters. However, thorough stochastic modeling has not yet been solved, and further studies on the application of stochastic modeling methods in epidemiology is required in the future. In particular, insufficient information is always caused by the distribution of different datasets. This is usually because different regions in a dataset may contain irrelevant information as a result of improper data processing and incorrect prediction accuracy of epidemic speed. Furthermore, even with limited samples, the majority of research results indicate that the performance of the residual network is the best among the pre-trained networks. In other fields, for example, Jin et al. [243] used attention residuals network to improve the accuracy of diagnosing actual bearing compound faults. The challenge of detecting COVID-19 can also be exacerbated by the imbalance of the sample in the dataset, which always leads to biases during the deep learning training phase, especially the issues caused by the lack of confidence intervals [33]. In order to address the aforementioned problem, freezing technology was introduced, which resulted in higher performance in the case of fewer COVID-19 data [244,245]. In the setting of modest amounts of data, this evidence shows that ensemble learning [246,247] and multi-tasking learning 152 [248] are more suitable for diagnosing COVID-19. Meanwhile, data preprocessing technology, such as image rotation and clipping, as well as data sample expansion using GAN network, can effectively improve the training test’s accuracy.
During the COVID-19 pandemic, deep learning was used not only for COVID-19 detection on chest X-ray and CT images, but also in the field of LUS. LUS was more sensitive to identifying COVID-19 than chest X-ray and CT images. Current clinical studies implies that LUS has potential accuracy in diagnosing a pandemic. Deep learning and ML approaches have played an extraordinary role in biology and medicine in combating the COVID-19 pandemic, and are now being used to test potential drugs that can successfully target COVID-19. In the biology and medicine fields, ML methods have also been applied, such as using random forest and SVM to analyze and predict the possible inhibitory synthetic antibodies of SARS-CoV-2. Meanwhile, to timely understand the public mood, some studies use NLP-related method to analyze the tweets about COVID-19 on social media, so as to arrange corresponding measures in time to calm the public mood and prevent causing serious public psychological problems.
In this paper, the method proposed has not been applied in real life, because the data results in real data are more easily affected by noise and human factors. As a result, practical considerations should be factored into future design methods. Biology, medicine, AI and other fields have all made great contributions to the global spread of COVID-19. Despite the fact that a vaccine has been developed and is now available for free nationwide vaccination, there are still many problems to be solved in the future, such as risk assessment of human infection and complete resolution of viral infection. The most essential aspect of this paper is full explanation of the epidemiological transmission model and the application of deep learning and transfer learning methods in COVID-19. Because most research on chest X-ray and CT images is based on CNNs, we can pay more attention to its studies in other directions. The study of LUS is less than that of chest X-ray and CT images, and the limitations of LUS can also become a research direction. Finally, the majority of the research is theoretical, and the detection network can be deployed to mobile terminals.
The survey is primarily used to introduce the usage of deep learning and transfer learning detection of COVID-19 in chest X-ray and CT images, as well as deep learning in the detection of viruses in LUS. To effectively combat COVID-19, machine learning-based approaches are used to predict and screen vaccine candidates that can resist the virus, and the importance of the telehealth system throughout the epidemic period, as well as the analysis of public comments on social platforms using NLP and other methods to better understand the public mood and make appropriate arrangements. This paper attempts to provide a comprehensive survey and provides systematic literature reference for subsequent research based on the summary and analysis of the studies during the COVID-19 pandemic period. Some future directions and the facing challenges are shown in Tab. 4.

The rapid spread of COVID-19 has triggered a great crisis to the world. This is an ongoing epidemic that has had a profound impact on all areas of the world, especially public health. Epidemiological propagation models, deep learning, and ML methods have all proven to be powerful tools in the fight against the epidemic. In this survey, we mainly make the following contributions: Firstly, we introduce in detail the prediction of COVID-19 transmission by epidemiological propagation models, including deterministic models (such as SIR and SEIR models) and stochastic models. Mathematical modeling of infectious diseases can effectively predict the intensity, incubation period and duration of infection, as well as the impact of mitigation measures on the spread of COVID-19, allowing appropriate measures to be effectively arranged to control the spread of the virus and reduce the number of infected people to a certain extent, relieving the country’s pressure. Secondly, this paper introduces the application of the combination of deep learning and transfer learning in the classification detection based on chest X-ray and CT images, so as to improve COVID-19 patients’ screening and provide an accurate solution for controlling the epidemic’s spread. Thirdly, unlike most studies in the literature that use chest X-ray and CT images to detetct COVID-19, LUS has a stronger sensitivity in detecting COVID-19. LUS has been demonstrated to have a higher accuracy rate in clinical medicine, and it does not expose patients to radiation when used at the bedside. Fourthly, ML methods have also made great contributions to biology and medicine, including the prediction of potential vaccines and antibodies, etc., aims to prevent the public from becoming infected with diseases from the root. Fifth, in the absence of a vaccine at the time, telehealth systems were used during the pandemic. The adoption of telehealth systems has greatly reduced the rate of contact between health care workers and patients, thereby reducing virus’s spread. Sixth, to interpret the influence of COVID-19 from various aspects, an NLP-related method is introduced to analyze tweets relevant to the COVID-19 epidemic on social media, so that public health departments can timely understand the public mood and prevent serious psychosocial problems from causing human panic. This survey, in particular, tries to provide a detailed description of the research on the diagnostic classification of chest X-ray and CT images during the pandemic period, with a focus on systematically introducing the application of deep learning-based and transfer learning-based methods, as well as highlighting innovations and classification accuracy. More specifically, this survey also presents the employ of LUS in COVID-19, which has been proven to be a safer and more sensitive method. In our future work, the better accurate epidemic spread models that can be applied to accurately simulate the transmission of infectious diseases, and even the clinical diagnosis advice from radiologists in the medical assistance system will be considered.
Acknowledgement: The authors wish to express their appreciation to the reviewers for their helpful suggestions which greatly improved the presentation of this paper.
Funding Statement: This research is supported by National Natural Science Foundation of China (Nos. 61902158, 61806087).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Tumpey, T. M., Garca-Sastre, A., Taubenberger, J. K., Palese, P., Swayne, D. E. et al. (2005). Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: Functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. Journal of Virology, 79(23), 14933–14944. DOI 10.1128/JVI.79.23.14933-14944.2005. [Google Scholar] [CrossRef]
2. Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B. et al. (2020). China novel coronavirus investigating and research team. A novel coronavirus from patients with pneumonia in China. New England Journal of Medicine, 382(8), 727–733. DOI 10.1056/NEJMoa2001017. [Google Scholar] [CrossRef]
3. WHO (2020). World health organization coronavirus disease 2019 (COVID-19) situation report. [Google Scholar]
4. Sohrabi, C., Alsafi, Z., O’Neill, N., Khan, M., Kerwan, A. et al. (2020). World health organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). International Journal of Surgery, 76, 71–76. DOI 10.1016/j.ijsu.2020.02.034. [Google Scholar] [CrossRef]
5. Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W. et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. DOI 10.1038/s41586-020-2008-3. [Google Scholar] [CrossRef]
6. Cucinotta, D., Vanelli, M. (2020). Who declares COVID-19 a pandemic. Acta Bio Medica: Atenei Parmensis, 91(1), 157. DOI 10.23750/abm.v91i1.9397. [Google Scholar] [CrossRef]
7. WHO (2020). Who director-general’s opening remarks at the media briefing on COVID-19-11 march 2020. [Google Scholar]
8. Wilson, D. J., Falush, D., McVean, G. (2005). Germs, genomes and genealogies. Trends in Ecology & Evolution, 20(1), 39–45. DOI 10.1016/j.tree.2004.10.009. [Google Scholar] [CrossRef]
9. Tyrrell, D., Bynoe, M. (1966). Cultivation of viruses from a high proportion of patients with colds. The Lancet, 1(7428), 76–77. DOI 10.1016/S0140-6736(66)92364-6. [Google Scholar] [CrossRef]
10. Lai, C. C., Shih, T. P., Ko, W. C., Tang, H. J., Hsueh, P. R. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19The epidemic and the challenges. International Journal of Antimicrobial Agents, 55(3), 105924. DOI 10.1016/j.ijantimicag.2020.105924. [Google Scholar] [CrossRef]
11. Velavan, T. P., Meyer, C. G. (2020). The COVID-19 epidemic. Tropical Medicine & International Health, 25(3), 278–280. DOI 10.1111/tmi.13383. [Google Scholar] [CrossRef]
12. Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F. et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395(10223), 507–513. DOI 10.1016/S0140-6736(20)30211-7. [Google Scholar] [CrossRef]
13. Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J. et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. DOI 10.1016/S0140-6736(20)30183-5. [Google Scholar] [CrossRef]
14. Woelfel, R., Corman, V. M., Guggemos, W., Seilmaier, M., Zange, S. et al. (2020). Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. DOI 10.1101/2020.03.05.20030502. [Google Scholar] [CrossRef]
15. Klopfenstein, T., Kadiane-Oussou, N., Toko, L., Royer, P. Y., Lepiller, Q. et al. (2020). Features of anosmia in COVID-19. Médecine et Maladies Infectieuses, 50(5), 436–439. DOI 10.1016/j.medmal.2020.04.006. [Google Scholar] [CrossRef]
16. Zhang, R., Wang, X., Ni, L., Di, X., Ma, B. et al. (2020). COVID-19: Melatonin as a potential adjuvant treatment. Life Sciences, 250, 117583. DOI 10.1016/j.lfs.2020.117583. [Google Scholar] [CrossRef]
17. Yuki, K., Fujiogi, M., Koutsogiannaki, S. (2020). COVID-19 pathophysiology: A review. Clinical Immunology, 215, 108427. DOI 10.1016/j.clim.2020.108427. [Google Scholar] [CrossRef]
18. COVID, Gemelli Against and Post-Acute Care Study Group (2020). Post-COVID-19 global health strategies: The need for an interdisciplinary approach. Aging Clinical and Experimental Research, 32(8), 1613–1620. DOI 10.1007/s40520-020-01616-x. [Google Scholar] [CrossRef]
19. Guan, W. J., Ni, Z. Y., Hu, Y., Liang, W. H., Ou, C. Q. et al. (2020). Clinical characteristics of 2019 novel coronavirus infection in China. MedRxiv, 28, 394. DOI 10.1101/2020.02.06.20020974. [Google Scholar] [CrossRef]
20. Ai, T., Yang, Z., Hou, H., Zhan, C., Chen, C. et al. (2020). Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology, 296(2), E32–E40. DOI 10.1148/radiol.2020200642. [Google Scholar] [CrossRef]
21. Dashraath, P., Wong, J. L. J., Lim, M. X. K., Lim, L. M., Li, S. et al. (2020). Coronavirus disease 2019 (COVID-19) pandemic and pregnancy. American Journal of Obstetrics and Gynecology, 222(6), 521–531. DOI 10.1016/j.ajog.2020.03.021. [Google Scholar] [CrossRef]
22. Xie, X., Zhong, Z., Zhao, W., Zheng, C., Wang, F. et al. (2020). Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: Relationship to negative RT-PCR testing. Radiology, 296(2), E41–E45. DOI 10.1148/radiol.2020200343. [Google Scholar] [CrossRef]
23. Murthy, S., Gomersall, C. D., Fowler, R. A. (2020). Care for critically ill patients with COVID-19. JAMA, 323(15), 1499–1500. DOI 10.1001/jama.2020.3633. [Google Scholar] [CrossRef]
24. Islam, M. M., Ullah, S. M. A., Mahmud, S., Raju, S. T. U. (2020). Breathing aid devices to support novel coronavirus (COVID-19) infected patients. SN Computer Science, 1(5), 1–8. DOI 10.1007/s42979-020-00300-1. [Google Scholar] [CrossRef]
25. Rothe, C., Schunk, M., Sothmann, P., Bretzel, G., Froeschl, G. et al. (2020). Transmission of 2019-ncov infection from an asymptomatic contact in Germany. New England Journal of Medicine, 382(10), 970–971. DOI 10.1056/NEJMc2001468. [Google Scholar] [CrossRef]
26. Murray, J. D. (2007Mathematical biology: I. An introduction, vol. 17. Berlin, Germany: Springer Science & Business Media. [Google Scholar]
27. Small, M., Cavanagh, D. (2020). Modelling strong control measures for epidemic propagation with networks-a COVID-19 case study. IEEE Access, 8, 109719–109731. DOI 10.1109/ACCESS.2020.3001298. [Google Scholar] [CrossRef]
28. Nokes, D., Anderson, R. (1988). The use of mathematical models in the epidemiological study of infectious diseases and in the design of mass immunization programmes. Epidemiology & Infection, 101(1), 1–20. DOI 10.1017/S0950268800029186. [Google Scholar] [CrossRef]
29. Kruijshaar, M. E., Barendregt, J. J., Hoeymans, N. (2002). The use of models in the estimation of disease epidemiology. Bulletin of the World Health Organization, 80, 622–628. DOI 10.1590/S0042-96862002000800005. [Google Scholar] [CrossRef]
30. Menon, A., Rajendran, N. K., Chandrachud, A., Setlur, G. (2020). Modelling and simulation of COVID-19 propagation in a large population with specific reference to India. DOI 10.1101/2020.04.30.20086306. [Google Scholar] [CrossRef]
31. Lopez, C. E., Vasu, M., Gallemore, C. (2020). Understanding the perception of COVID-19 policies by mining a multilanguage twitter dataset. arXiv preprint arXiv:2003.10359. [Google Scholar]
32. Levesque, J., Maybury, D. W., Shaw, R. D. (2021). A model of COVID-19 propagation based on a gamma subordinated negative binomial branching process. Journal of Theoretical Biology, 512(2), 110536. DOI 10.1016/j.jtbi.2020.110536. [Google Scholar] [CrossRef]
33. Islam, M. M., Karray, F., Alhajj, R., Zeng, J. (2021). A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19). IEEE Access, 9, 30551–30572. DOI 10.1109/ACCESS.2021.3058537. [Google Scholar] [CrossRef]
34. LeCun, Y., Bengio, Y., Hinton, G. (2015). Deep learning. Nature, 521(7553), 436–444. DOI 10.1038/nature14539. [Google Scholar] [CrossRef]
35. Goodfellow, I., Bengio, Y., Courville, A., Bengio, Y. (2016). Deep learning, vol. 1. Cambridge: MIT Press. [Google Scholar]
36. Mamoshina, P., Vieira, A., Putin, E., Zhavoronkov, A. (2016). Applications of deep learning in biomedicine. Molecular Pharmaceutics, 13(5), 1445–1454. DOI 10.1021/acs.molpharmaceut.5b00982. [Google Scholar] [CrossRef]
37. Wainberg, M., Merico, D., Delong, A., Frey, B. J. (2018). Deep learning in biomedicine. Nature Biotechnology, 36(9), 829–838. DOI 10.1038/nbt.4233. [Google Scholar] [CrossRef]
38. Esteva, A., Robicquet, A., Ramsundar, B., Kuleshov, V., DePristo, M. et al. (2019). A guide to deep learning in healthcare. Nature Medicine, 25(1), 24–29. DOI 10.1038/s41591-018-0316-z. [Google Scholar] [CrossRef]
39. Chen, H., Engkvist, O., Wang, Y., Olivecrona, M., Blaschke, T. (2018). The rise of deep learning in drug discovery. Drug Discovery Today, 23(6), 1241–1250. DOI 10.1016/j.drudis.2018.01.039. [Google Scholar] [CrossRef]
40. Asraf, A., Islam, M. Z., Haque, M. R., Islam, M. M. (2020). Deep learning applications to combat novel coronavirus (COVID-19) pandemic. SN Computer Science, 1(6), 1–7. DOI 10.1007/s42979-020-00383-w. [Google Scholar] [CrossRef]
41. Shen, D., Wu, G., Suk, H. I. (2017). Deep learning in medical image analysis. Annual Review of Biomedical Engineering, 19(1), 221–248. DOI 10.1146/annurev-bioeng-071516-044442. [Google Scholar] [CrossRef]
42. Deng, X., Shao, H., Shi, L., Wang, X., Xie, T. (2020). A classification-detection approach of COVID-19 based on chest X-ray and CT by using keras pre-trained deep learning models. Computer Modeling in Engineering & Sciences, 125(2), 579–596. DOI 10.32604/cmes.2020.011920. [Google Scholar] [CrossRef]
43. Apostolopoulos, I. D., Mpesiana, T. A. (2020). COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine, 43(2), 635–640. DOI 10.1007/s13246-020-00865-4. [Google Scholar] [CrossRef]
44. Kanne, J. P., Little, B. P., Chung, J. H., Elicker, B. M., Ketai, L. H. (2020). Essentials for radiologists on COVID-19: An update-radiology scientific expert panel. Radiology, 2, 296. DOI 10.1148/radiol.2020200527. [Google Scholar] [CrossRef]
45. Bernheim, A., Mei, X., Huang, M., Yang, Y., Fayad, Z. A. et al. (2020). Chest CT findings in coronavirus disease-19 (COVID-19Relationship to duration of infection. Radiology, 295(3), 200463. DOI 10.1148/radiol.2020200463. [Google Scholar] [CrossRef]
46. Chang, D., Lin, M., Wei, L., Xie, L., Zhu, G. et al. (2020). Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. JAMA, 323(11), 1092–1093. DOI 10.1001/jama.2020.1623. [Google Scholar] [CrossRef]
47. Pan, F., Ye, T., Sun, P., Gui, S., Liang, B. et al. (2020). Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology, 295(3), 715–721. DOI 10.1148/radiol.2020200370. [Google Scholar] [CrossRef]
48. Wang, S., Zha, Y., Li, W., Wu, Q., Li, X. et al. (2020). A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. European Respiratory Journal, 56(2). DOI 10.1183/13993003.00775-2020. [Google Scholar] [CrossRef]
49. Loey, M., Manogaran, G., Khalifa, N. E. M. (2020). A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images. Neural Computing and Applications, 1–13(4), 281. DOI 10.1007/s00521-020-05437-x. [Google Scholar] [CrossRef]
50. Patankar, S. (2020). Deep learning-based computational drug discovery to inhibit the RNA dependent RNA polymerase: Application to SARS-CoV and COVID-19. https://osf.io/6kpbg. [Google Scholar]
51. Zhang, H., Saravanan, K. M., Yang, Y., Hossain, M. T., Li, J. et al. (2020). Deep learning based drug screening for novel coronavirus 2019-nCoV. Interdisciplinary Sciences: Computational Life Sciences, 12, 368–376. DOI 10.20944/preprints202002.0061.v1. [Google Scholar] [CrossRef]
52. Sethy, P. K., Behera, S. K., Ratha, P. K., Biswas, P. (2020). Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. https://pdfs.semanticscholar.org/9da0/35f1d7372cfe52167ff301bc12d5f415caf1.pdf. [Google Scholar]
53. Ong, E., Wong, M. U., Huffman, A., He, Y. (2020). COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Frontiers in Immunology, 11, 1581. DOI 10.3389/fimmu.2020.01581. [Google Scholar] [CrossRef]
54. Pfefferbaum, B., North, C. S. (2020). Mental health and the COVID-19 pandemic. New England Journal of Medicine, 383(6), 510–512. DOI 10.1056/NEJMp2008017. [Google Scholar] [CrossRef]
55. Jelodar, H., Wang, Y., Orji, R., Huang, S. (2020). Deep sentiment classification and topic discovery on novel coronavirus or COVID-19 online discussions: NLP using LSTM recurrent neural network approach. IEEE Journal of Biomedical and Health Informatics, 24(10), 2733–2742. DOI 10.1109/JBHI.2020.3001216. [Google Scholar] [CrossRef]
56. Li, L., Zhang, Q., Wang, X., Zhang, J., Wang, T. et al. (2020). Characterizing the propagation of situational information in social media during COVID-19 epidemic: A case study on weibo. IEEE Transactions on Computational Social Systems, 7(2), 556–562. DOI 10.1109/TCSS.2020.2980007. [Google Scholar] [CrossRef]
57. Ingalls, B. P. (2013). Mathematical modeling in systems biology: An introduction. Cambridge: MIT Press. [Google Scholar]
58. Siettos, C. I., Russo, L. (2013). Mathematical modeling of infectious disease dynamics. Virulence, 4(4), 295–306. DOI 10.4161/viru.24041. [Google Scholar] [CrossRef]
59. Marchetti, C., Bianchi, A., Bassi, M., Gori, R., Lamberti, C. et al. (2006). Mathematical modeling and numerical simulation in maxillo-facial virtual surgery (VISU). Journal of Craniofacial Surgery, 17(4), 661–667. DOI 10.1097/00001665-200607000-00009. [Google Scholar] [CrossRef]
60. Bissell, C., Dillon, C. (2000). Telling tales: Models, stories and meanings. For the Learning of Mathematics, 20(3), 3–11. https://www.jstor.org/stable/40248331. [Google Scholar]
61. Kermack, W. O., McKendrick, A. G. (1927). A contribution to the mathematical theory of epidemics. Proceedings of the Royal Society of London. Series A, Containing Papers of a Mathematical and Physical Character, 115(772), 700–721. DOI 10.1098/rspa.1927.0118. [Google Scholar] [CrossRef]
62. Chen, Y. C., Lu, P. E., Chang, C. S., Liu, T. H. (2020). A time-dependent sir model for COVID-19 with undetectable infected persons. IEEE Transactions on Network Science and Engineering, 7(4), 3279–3294. DOI 10.1109/TNSE.2020.3024723. [Google Scholar] [CrossRef]
63. Wang, L., Zhou, Y., He, J., Zhu, B., Wang, F. et al. (2020). An epidemiological forecast model and software assessing interventions on the COVID-19 epidemic in China. Journal of Data Science, 18(3), 409–432. DOI 10.6339/JDS.202007_18(3). 0003. [Google Scholar] [CrossRef]
64. Read, J. M., Bridgen, J. R., Cummings, D. A., Ho, A., Jewell, C. P. (2020). Novel coronavirus 2019-nCoV: Early estimation of epidemiological parameters and epidemic predictions. MedRxiv, 10(7), 1258. DOI 10.1101/2020.01.23.20018549. [Google Scholar] [CrossRef]
65. Wang, H., Wang, Z., Dong, Y., Chang, R., Xu, C. et al. (2020). Phase-adjusted estimation of the number of coronavirus disease 2019 cases in Wuhan, China. Cell Discovery, 6(1), 1–8. DOI 10.1038/s41421-020-0148-0. [Google Scholar] [CrossRef]
66. Shearer, F., Walker, J., Tellioglu, N., McCaw, J. M., McVernon, J. et al. (2020). Assessing the risk of spread of COVID-19 to the asia pacific region. DOI 10.1101/2020.04.09.20057257. [Google Scholar] [CrossRef]
67. Moss, R., Wood, J., Brown, D., Shearer, F., Black, A. J. et al. (2020). Modelling the impact of COVID-19 in Australia to inform transmission reducing measures and health system preparedness. DOI 10.1101/2020.04.07.20056184. [Google Scholar] [CrossRef]
68. Biswas, K., Khaleque, A., Sen, P. (2020). COVID-19 spread: Reproduction of data and prediction using a sir model on euclidean network. arXiv preprint arXiv:2003.07063. [Google Scholar]
69. Zhong, L., Mu, L., Li, J., Wang, J., Yin, Z. et al. (2020). Early prediction of the 2019 novel coronavirus outbreak in the mainland china based on simple mathematical model. IEEE Access, 8, 51761–51769. DOI 10.1109/ACCESS.2020.2979599. [Google Scholar] [CrossRef]
70. Chikina, M., Pegden, W. (2020). Modeling strict age-targeted mitigation strategies for COVID-19. PLoS One, 15(7), e0236237. DOI 10.1371/journal.pone.0236237. [Google Scholar] [CrossRef]
71. Singh, R., Adhikari, R. (2020). Age-structured impact of social distancing on the COVID-19 epidemic in India. arXiv preprint arXiv:2003.12055. [Google Scholar]
72. Radulescu, A., Williams, C., Cavanagh, K. (2020). Management strategies in a SEIR-type model of COVID 19 community spread. Scientific Reports, 10(1), 1–16. DOI 10.1038/s41598-020-77628-4. [Google Scholar] [CrossRef]
73. Diekmann, O., Heesterbeek, H., Britton, T. (2012). Mathematical tools for understanding infectious disease dynamics, vol. 7. New Jersey: Princeton University Press. [Google Scholar]
74. Pastor-Satorras, R., Vespignani, A. (2001). Epidemic spreading in scale-free networks. Physical Review Letters, 86(14), 3200–3203. DOI 10.1103/PhysRevLett.86.3200. [Google Scholar] [CrossRef]
75. Boguná, M., Pastor-Satorras, R., Vespignani, A. (2003). Absence of epidemic threshold in scale-free networks with degree correlations. Physical Review Letters, 90(2), 28701. DOI 10.1103/PhysRevLett.90.028701. [Google Scholar] [CrossRef]
76. Small, M., Tse, C. K., Walker, D. M. (2006). Super-spreaders and the rate of transmission of the sars virus. Physica D: Nonlinear Phenomena, 215(2), 146–158. DOI 10.1016/j.physd.2006.01.021. [Google Scholar] [CrossRef]
77. Small, M., Tse, C. K. (2005). Small world and scale free model of transmission of sars. International Journal of Bifurcation and Chaos, 15(5), 1745–1755. DOI 10.1142/S0218127405012776. [Google Scholar] [CrossRef]
78. Small, M., Tse, C. K. (2005). Clustering model for transmission of the sars virus: Application to epidemic control and risk assessment. Physica A: Statistical Mechanics and its Applications, 351(2–4), 499–511. DOI 10.1016/j.physa.2005.01.009. [Google Scholar] [CrossRef]
79. Wang, Y., Chakrabarti, D., Wang, C., Faloutsos, C. (2003). Epidemic spreading in real networks: An eigenvalue viewpoint. 22nd International Symposium on Reliable Distributed Systems. IEEE. [Google Scholar]
80. Chakrabarti, D., Wang, Y., Wang, C., Leskovec, J., Faloutsos, C. (2008). Epidemic thresholds in real networks. ACM Transactions on Information and System Security, 10(4), 1–26. DOI 10.1145/1284680.1284681. [Google Scholar] [CrossRef]
81. Gross, T., Dima, C. J. D., Blasius, B. (2006). Epidemic dynamics on an adaptive network. Physical Review Letters, 96(20), 208701. DOI 10.1103/PhysRevLett.96.208701. [Google Scholar] [CrossRef]
82. Zhan, C., Tse, C. K., Fu, Y., Lai, Z., Zhang, H. (2020). Modeling and prediction of the 2019 coronavirus disease spreading in China incorporating human migration data. PLoS One, 15(10), e0241171. DOI 10.1371/journal.pone.0241171. [Google Scholar] [CrossRef]
83. Migration, B. (2020). https://qianxi.baidu.com. [Google Scholar]
84. Prem, K., Liu, Y., Russell, T. W., Kucharski, A. J., Eggo, R. M. et al. (2020). The effect of control strategies to reduce social mixing on outcomes of the COVID-19 epidemic in Wuhan, China: A modelling study. The Lancet Public Health, 5(5), e261–e270. DOI 10.1016/S2468-2667(20)30073-6. [Google Scholar] [CrossRef]
85. López, L., Rodo, X. (2021). A modified seir model to predict the COVID-19 outbreak in Spain and Italy: Simulating control scenarios and multi-scale epidemics. Results in Physics, 21(1274), 103746. DOI 10.1016/j.rinp.2020.103746. [Google Scholar] [CrossRef]
86. Guo, X. J., Zhang, H., Zeng, Y. P. (2020). Transmissibility of COVID-19 and its association with temperature and humidity. Infectious Diseases of Poverty, 9(1). [Google Scholar]
87. Rahman, M. M., Manik, M. M. H., Islam, M. M., Mahmud, S., Kim, J. H. (2020). An automated system to limit COVID-19 using facial mask detection in smart city network. 2020 IEEE International IOT, Electronics and Mechatronics Conference(IEMTRONICS). IEEE. DOI 10.1109/IEMTRONICS51293.2020.9216386. [Google Scholar] [CrossRef]
88. Eikenberry, S. E., Mancuso, M., Iboi, E., Phan, T., Eikenberry, K. et al. (2020). To mask or not to mask: Modeling the potential for face mask use by the general public to curtail the COVID-19 pandemic. Infectious Disease Modelling, 5(8), 293–308. DOI 10.1016/j.idm.2020.04.001. [Google Scholar] [CrossRef]
89. Peng, L., Yang, W., Zhang, D., Zhuge, C., Hong, L. (2020). Epidemic analysis of COVID-19 in China by dynamical modeling. arXiv preprint arXiv:2002.06563. [Google Scholar]
90. Pandey, G., Chaudhary, P., Gupta, R., Pal, S. (2020). Seir and regression model based COVID-19 outbreak predictions in India. arXiv preprint arXiv:2004.00958. [Google Scholar]
91. Huang, G., Pan, Q., Zhao, S., Gao, Y., Gao, X. (2020). Prediction of COVID-19 outbreak in China and optimal return date for university students based on propagation dynamics. Journal of Shanghai Jiaotong University (Science), 25(2), 140–146. DOI 10.1007/s12204-020-2167-2. [Google Scholar] [CrossRef]
92. Chen, T. M., Rui, J., Wang, Q. P., Zhao, Z. Y., Cui, J. A. et al. (2020). A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infectious Diseases of Poverty, 9(1), 1–8. DOI 10.1186/s40249-020-00640-3. [Google Scholar] [CrossRef]
93. Chen, Y., Cheng, J., Jiang, Y., Liu, K. (2020). A time delay dynamical model for outbreak of 2019-nCoV and the parameter identification. Journal of Inverse and Ill-Posed Problems, 28(2), 243–250. DOI 10.1515/jiip-2020-0010. [Google Scholar] [CrossRef]
94. Khajji, B., Kada, D., Balatif, O., Rachik, M. (2020). A multi-region discrete time mathematical modeling of the dynamics of COVID-19 virus propagation using optimal control. Journal of Applied Mathematics and Computing, 64(1–2), 255–281. DOI 10.1007/s12190-020-01354-3. [Google Scholar] [CrossRef]
95. Allen, L. J. (2017). A primer on stochastic epidemic models: Formulation, numerical simulation, and analysis. Infectious Disease Modelling, 2(2), 128–142. DOI 10.1016/j.idm.2017.03.001. [Google Scholar] [CrossRef]
96. Hellewell, J., Abbott, S., Gimma, A., Bosse, N. I., Jarvis, C. I. et al. (2020). Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. The Lancet Global Health, 8(4), e488–e496. DOI 10.1016/S2214-109X(20)30074-7. [Google Scholar] [CrossRef]
97. Gross, B., Zheng, Z., Liu, S., Chen, X., Sela, A. et al. (2020). Spatio-temporal propagation of COVID-19 pandemics. EPL (Europhysics Letters), 131(5), 58003. DOI 10.1209/0295-5075/131/58003. [Google Scholar] [CrossRef]
98. Xiong, Y., Wang, Y., Chen, F., Zhu, M. (2020). Spatial statistics and influencing factors of the novel coronavirus pneumonia 2019 epidemic in Hubei Province, China. DOI 10.21203/rs.3.rs-16858/v2. [Google Scholar] [CrossRef]
99. Arenas, A., Cota, W., Gómez-Gardenes, J., Gómez, S., Granell, C. et al. (2020). A mathematical model for the spatiotemporal epidemic spreading of COVID 19. DOI 10.1101/2020.03.21.20040022. [Google Scholar] [CrossRef]
100. Wang, H., Shi, H., Lin, K., Qin, C., Zhao, L. et al. (2020). A high-precision arrhythmia classification method based on dual fully connected neural network. Biomedical Signal Processing and Control, 58(2), 101874. DOI 10.1016/j.bspc.2020.101874. [Google Scholar] [CrossRef]
101. Yan, Q., Zhang, L., Liu, Y., Zhu, Y., Sun, J. et al. (2020). Deep HDR imaging via a non-local network. IEEE Transactions on Image Processing, 29, 4308–4322. DOI 10.1109/TIP.2020.2971346. [Google Scholar] [CrossRef]
102. Yan, Q., Gong, D., Zhang, Y. (2018). Two-stream convolutional networks for blind image quality assessment. IEEE Transactions on Image Processing, 28(5), 2200–2211. DOI 10.1109/TIP.2018.2883741. [Google Scholar] [CrossRef]
103. Yan, Q., Gong, D., Zhang, P., Shi, Q., Sun, J. et al. (2019). Multi-scale dense networks for deep high dynamic range imaging. 2019 IEEE Winter Conference on Applications of Computer Vision. IEEE. DOI 10.1109/WACV.2019.00012. [Google Scholar] [CrossRef]
104. Yan, Q., Gong, D., Shi, Q., van den Hengel, A., Shen, C. et al. (2019). Attention-guided network for ghost-free high dynamic range imaging. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 1751–1760. [Google Scholar]
105. Gong, D., Yang, J., Liu, L., Zhang, Y., Reid, I. et al. (2017). From motion blur to motion flow: A deep learning solution for removing heterogeneous motion blur. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2319–2328. [Google Scholar]
106. He, T., Shen, C., Tian, Z., Gong, D., Sun, C. et al. (2019). Knowledge adaptation for efficient semantic segmentation. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. [Google Scholar]
107. Gong, D., Liu, L., Le, V., Saha, B., Mansour, M. R. et al. (2019). Memorizing normality to detect anomaly: Memory-augmented deep autoencoder for unsupervised anomaly detection. Proceedings of the IEEE/CVF International Conference on Computer Vision. [Google Scholar]
108. Pan, S. J., Yang, Q. (2009). A survey on transfer learning. IEEE Transactions on Knowledge and Data Engineering, 22(10), 1345–1359. DOI 10.1109/TKDE.2009.191. [Google Scholar] [CrossRef]
109. Lee, J. G., Jun, S., Cho, Y. W. (2017). Deep learning in medical imaging: General overview. Korean Journal of Radiology, 18(4), 570–584. [Google Scholar]
110. Qin, C., Jin, Y., Tao, J., Xiao, D., Yu, H. et al. (2021). Dtcnnmi: A deep twin convolutional neural networks with multi-domain inputs for strongly noisy diesel engine misfire detection. Measurement, 180(14), 109548. DOI 10.1016/j.measurement.2021.109548. [Google Scholar] [CrossRef]
111. Qin, C., Shi, G., Tao, J., Yu, H., Jin, Y. et al. (2021). Precise cutterhead torque prediction for shield tunneling machines using a novel hybrid deep neural network. Mechanical Systems and Signal Processing, 151(6), 107386. DOI 10.1016/j.ymssp.2020.107386. [Google Scholar] [CrossRef]
112. Li, Q., Cai, W., Wang, X., Zhou, Y., Feng, D. D. et al. (2014). Medical image classification with convolutional neural network. 2014 13th International Conference on Control Automation Robotics & Vision. IEEE. DOI 10.1109/ICARCV.2014.7064414. [Google Scholar] [CrossRef]
113. Abbas, A., Abdelsamea, M. M., Gaber, M. M. (2021). Classification of COVID-19 in chest X-ray images using detrac deep convolutional neural network. Applied Intelligence, 51(2), 854–864. DOI 10.1007/s10489-020-01829-7. [Google Scholar] [CrossRef]
114. Huynh, B. Q., Li, H., Giger, M. L. (2016). Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. Journal of Medical Imaging, 3(3), 34501. DOI 10.1117/1.JMI.3.3.034501. DOI 10.1109/BHI.2017.7897283. [Google Scholar] [CrossRef]
115. Elmahdy, M. S., Abdeldayem, S. S., Yassine, I. A. (2017). Low quality dermal image classification using transfer learning. 2017 IEEE EMBS International Conference on Biomedical & Health Informatics. IEEE. DOI 10.1109/BHI.2017.7897283. [Google Scholar] [CrossRef]
116. Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M. et al. (2017). Dermatologist-level classification of skin cancer with deep neural networks. Nature, 542(7639), 115–118. DOI 10.1038/nature21056. [Google Scholar] [CrossRef]
117. Raghu, M., Zhang, C., Kleinberg, J., Bengio, S. (2019). Transfusion: Understanding transfer learning for medical imaging. arXiv preprint arXiv:1902.07208. [Google Scholar]
118. Chouhan, V., Singh, S. K., Khamparia, A., Gupta, D., Tiwari, P. et al. (2020). A novel transfer learning based approach for pneumonia detection in chest X-ray images. Applied Sciences, 10(2), 559. DOI 10.3390/app10020559. [Google Scholar] [CrossRef]
119. Maghdid, H. S., Asaad, A. T., Ghafoor, K. Z., Sadiq, A. S., Khan, M. K. (2021). Diagnosing COVID-19 pneumonia from X-ray and ct images using deep learning and transfer learning algorithms. arXiv preprint arXiv:2004.00038. [Google Scholar]
120. Narin, A., Kaya, C., Pamuk, Z. (2021). Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849. DOI 10.1007/s10044-021-00984-y. [Google Scholar] [CrossRef]
121. Kassani, S. H., Kassasni, P. H., Wesolowski, M. J., Schneider, K. A., Deters, R. (2020). Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: A machine learning-based approach. arXiv preprint arXiv:2004.10641. [Google Scholar]
122. Ardakani, A. A., Kanafi, A. R., Acharya, U. R., Khadem, N., Mohammadi, A. (2020). Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Computers in Biology and Medicine, 121(10229), 103795. DOI 10.1016/j.compbiomed.2020.103795. [Google Scholar] [CrossRef]
123. Horry, M. J., Chakraborty, S., Paul, M., Ulhaq, A., Pradhan, B. et al. (2020). X-ray image based COVID-19 detection using pre-trained deep learning models. DOI 10.31224/osf.io/wx89s. [Google Scholar] [CrossRef]
124. Islam, M. M., Islam, M. Z., Asraf, A., Ding, W. (2020). Diagnosis of COVID-19 from X-rays using combined CNN-RNN architecture with transfer learning. DOI 10.1101/2020.08.24.20181339. [Google Scholar] [CrossRef]
125. Anthimopoulos, M., Christodoulidis, S., Ebner, L., Christe, A., Mougiakakou, S. (2016). Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Transactions on Medical Imaging, 35(5), 1207–1216. DOI 10.1109/TMI.2016.2535865. [Google Scholar] [CrossRef]
126. Gao, M., Bagci, U., Lu, L., Wu, A., Buty, M. et al. (2018). Holistic classification of CT attenuation patterns for interstitial lung diseases via deep convolutional neural networks. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, 6(1), 1–6. DOI 10.1080/21681163.2015.1124249. [Google Scholar] [CrossRef]
127. Shin, H. C., Roth, H. R., Gao, M., Lu, L., Xu, Z. et al. (2016). Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Transactions on Medical Imaging, 35(5), 1285–1298. DOI 10.1109/TMI.2016.2528162. [Google Scholar] [CrossRef]
128. Weiss, K., Khoshgoftaar, T. M., Wang, D. (2016). A survey of transfer learning. Journal of Big data, 3(1), 1–40. DOI 10.1186/s40537-016-0043-6. [Google Scholar] [CrossRef]
129. Razzak, I., Naz, S., Rehman, A., Khan, A., Zaib, A. (2020). Improving coronavirus (COVID-19) diagnosis using deep transfer learning. DOI 10.1101/2020.04.11.20054643. [Google Scholar] [CrossRef]
130. Farooq, M., Hafeez, A. (2020). Covid-resnet: A deep learning framework for screening of COVID-19 from radiographs. arXiv preprint arXiv:2003.14395. [Google Scholar]
131. Ghoshal, B., Tucker, A. (2020). Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv preprint arXiv:2003.10769. [Google Scholar]
132. Ucar, F., Korkmaz, D. (2020). Covidiagnosis-net: Deep bayes-squeezenet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Medical Hypotheses, 140(1122–1131), 109761. DOI 10.1016/j.mehy.2020.109761. [Google Scholar] [CrossRef]
133. Brunese, L., Mercaldo, F., Reginelli, A., Santone, A. (2020). Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Computer Methods and Programs in Biomedicine, 196(20), 105608. DOI 10.1016/j.cmpb.2020.105608. [Google Scholar] [CrossRef]
134. He, X., Yang, X., Zhang, S., Zhao, J., Zhang, Y. et al. (2020). Sample-efficient deep learning for COVID-19 diagnosis based on CT scans. MedRxiv, 3(3), 34501. DOI 10.1101/2020.04.13.20063941. [Google Scholar] [CrossRef]
135. Vaid, S., Kalantar, R., Bhandari, M. (2020). Deep learning COVID-19 detection bias: Accuracy through artificial intelligence. International Orthopaedics, 44(8), 1539–1542. DOI 10.1007/s00264-020-04609-7. [Google Scholar] [CrossRef]
136. Wang, S. H., Nayak, D. R., Guttery, D. S., Zhang, X., Zhang, Y. D. (2021). COVID-19 classification by ccshnet with deep fusion using transfer learning and discriminant correlation analysis. Information Fusion, 68(2), 131–148. DOI 10.1016/j.inffus.2020.11.005. [Google Scholar] [CrossRef]
137. Toaçar, M., Ergen, B., Cömert, Z. (2020). COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Computers in Biology and Medicine, 121, 103805. DOI 10.1016/j.compbiomed.2020.103805. [Google Scholar] [CrossRef]
138. Waheed, A., Goyal, M., Gupta, D., Khanna, A., Al-Turjman, F. et al. (2020). COVIDGAN: Data augmentation using auxiliary classifier gan for improved COVID-19 detection. IEEE Access, 8, 91916–91923. DOI 10.1109/ACCESS.2020.2994762. [Google Scholar] [CrossRef]
139. Sedik, A., Iliyasu, A. M., El-Rahiem, A., Abdel Samea, M. E., Abdel-Raheem, A. et al. (2020). Deploying machine and deep learning models for efficient data-augmented detection of COVID-19 infections. Viruses, 12(7), 769. DOI 10.3390/v12070769. [Google Scholar] [CrossRef]
140. Wang, S. H., Govindaraj, V. V., Górriz, J. M., Zhang, X., Zhang, Y. D. (2021). COVID-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Information Fusion, 67, 208–229. DOI 10.1016/j.inffus.2020.10.004. [Google Scholar] [CrossRef]
141. Ting, D. S. W., Carin, L., Dzau, V., Wong, T. Y. (2020). Digital technology and COVID-19. Nature Medicine, 26(4), 459–461. DOI 10.1038/s41591-020-0824-5. [Google Scholar] [CrossRef]
142. Kumar, R., Khan, A. A., Kumar, J., Zakria, A., Golilarz, N. A. et al. (2021). Blockchain-federated-learning and deep learning models for COVID-19 detection using CT imaging. IEEE Sensors Journal, 1. DOI 10.1109/JSEN.2021.3076767. [Google Scholar] [CrossRef]
143. Ye, Y., Hou, S., Fan, Y., Qian, Y., Zhang, Y. et al. (2020). Alpha-satellite: An AI-driven system and benchmark datasets for hierarchical community-level risk assessment to help combat COVID-19. arXiv preprint arXiv:2003.12232. [Google Scholar]
144. Chimmula, V. K. R., Zhang, L. (2020). Time series forecasting of COVID-19 transmission in canada using LSTM networks. Chaos, Solitons & Fractals, 135(1), 109864. DOI 10.1016/j.chaos.2020.109864. [Google Scholar] [CrossRef]
145. Alazab, M., Awajan, A., Mesleh, A., Abraham, A., Jatana, V. et al. (2020). COVID-19 prediction and detection using deep learning. International Journal of Computer Information Systems and Industrial Management Applications, 12, 168–181. http://www.softcomputing.net/ijcisim_1.pdf. [Google Scholar]
146. Hemdan, E. E. D., Shouman, M. A., Karar, M. E. (2020). Covidx-net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv preprint arXiv:2003.11055. [Google Scholar]
147. Song, Y., Zheng, S., Li, L., Zhang, X., Zhang, X. et al. (2021). Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 1. DOI 10.1109/TCBB.2021.3065361. [Google Scholar] [CrossRef]
148. Gozes, O., Frid-Adar, M., Greenspan, H., Browning, P. D., Zhang, H. et al. (2020). Rapid ai development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning CT image analysis. arXiv preprint arXiv:2003.05037. [Google Scholar]
149. Inc., R. (2021). https://www.radlogics.com. [Google Scholar]
150. Xu, X., Jiang, X., Ma, C., Du, P., Li, X. et al. (2020). A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering, 6(10), 1122–1129. DOI 10.1016/j.eng.2020.04.010. [Google Scholar] [CrossRef]
151. Li, L., Qin, L., Xu, Z., Yin, Y., Wang, X. et al. (2020). Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology, 296(2), E65–E71. DOI 10.1148/radiol.2020200905. [Google Scholar] [CrossRef]
152. Amyar, A., Modzelewski, R., Li, H., Ruan, S. (2020). Multi-task deep learning based ct imaging analysis for COVID-19 pneumonia: Classification and segmentation. Computers in Biology and Medicine, 126(8), 104037. DOI 10.1016/j.compbiomed.2020.104037. [Google Scholar] [CrossRef]
153. Hasan, A. M., Al-Jawad, M. M., Jalab, H. A., Shaiba, H., Ibrahim, R. W. et al. (2020). Classification of COVID-19 coronavirus, pneumonia and healthy lungs in CT scans using q-deformed entropy and deep learning features. Entropy, 22(5), 517. DOI 10.3390/e22050517. [Google Scholar] [CrossRef]
154. Islam, M. Z., Islam, M. M., Asraf, A. (2020). A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Informatics in Medicine Unlocked, 20, 100412. DOI 10.1016/j.imu.2020.100412. [Google Scholar] [CrossRef]
155. Bandyopadhyay, S. K., Dutta, S. (2020). Machine learning approach for confirmation of COVID-19 cases: Positive, negative, death and release. DOI 10.1101/2020.03.25.20043505. [Google Scholar] [CrossRef]
156. Wang, L., Lin, Z. Q., Wong, A. (2020). COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Scientific Reports, 10(1), 1–12. DOI 10.1038/s41598-020-76550-z. [Google Scholar] [CrossRef]
157. Muhammad, L., Islam, M. M., Usman, S. S., Ayon, S. I. (2020). Predictive data mining models for novel coronavirus (COVID-19) infected patientsecovery. SN Computer Science, 1(4), 1–7. DOI 10.1007/s42979-020-00216-w. [Google Scholar] [CrossRef]
158. Saha, P., Sadi, M. S., Islam, M. M. (2021). Emcnet: Automated COVID-19 diagnosis from X-ray images using convolutional neural network and ensemble of machine learning classifiers. Informatics in Medicine Unlocked, 22(3), 100505. DOI 10.1016/j.imu.2020.100505. [Google Scholar] [CrossRef]
159. Cohen, J. P., Morrison, P., Dao, L., Roth, K., Duong, T. Q. et al. (2020). COVID-19 image data collection: Prospective predictions are the future. arXiv preprint arXiv:2006.11988. [Google Scholar]
160. Dataset, K. (2020). https://www.kaggle.com/andrewmvd/convid19-X-rays. [Google Scholar]
161. Kermany, D. S., Goldbaum, M., Cai, W., Valentim, C. C., Liang, H. et al. (2018). Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell, 172(5), 1122–1131. DOI 10.1016/j.cell.2018.02.010. [Google Scholar] [CrossRef]
162. Chest X-ray Repository, K (2020). https://www.kaggle.com/paultimothymooney/chest-xray-Pneumonia. [Google Scholar]
163. Kaggle, R. P. D. C. (2020). https://www.kaggle.com/c/rsna-pneumonia-detection-challenge. [Google Scholar]
164. Dataset Accessed (2020). https://drive.google.com/uc?id=1coM7x3378fOu2l6Pg2wldaOI7Dntu1a. [Google Scholar]
165. Kermany, D., Zhang, K., Goldbaum, M. et al. (2018). Labeled optical coherence tomography (OCT) and chest X-ray images for classification. Mendeley Data, 2(2). DOI 10.17632/rscbjbr9sj.3. [Google Scholar] [CrossRef]
166. Yang, X., He, X., Zhao, J., Zhang, Y., Zhang, S. et al. (2020). Covid-CT-dataset: A CT scan dataset about COVID-19. arXiv preprint arXiv:2003.13865. [Google Scholar]
167. Segmentation Dataset, C. C. (2020). http://medicalsegmentation.com/covid19/. [Google Scholar]
168. Archive, C. S. A. N. L. N. C. C. D (2020). https://www.cancerimagingarchive.net/. [Google Scholar]
169. COVID-19 (2020). https://radiopaedia.org/. [Google Scholar]
170. Radiological Society of North America (2020). RSNA pneumo-nia detection challenge. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data. [Google Scholar]
171. Mojoli, F., Bouhemad, B., Mongodi, S., Lichtenstein, D. (2019). Lung ultrasound for critically ill patients. American Journal of Respiratory and Critical Care Medicine, 199(6), 701–714. DOI 10.1164/rccm.201802-0236CI. [Google Scholar] [CrossRef]
172. Raheja, R., Brahmavar, M., Joshi, D., Raman, D. (2019). Application of lung ultrasound in critical care setting: A review. Cureus, 11(7), DOI 10.7759/cureus.5233. [Google Scholar] [CrossRef]
173. Tutino, L., Cianchi, G., Barbani, F., Batacchi, S., Cammelli, R. et al. (2010). Time needed to achieve completeness and accuracy in bedside lung ultrasound reporting in intensive care unit. Scandinavian Journal of Trauma, Resuscitation and Emergency Medicine, 18(1), 1–4. DOI 10.1186/1757-7241-18-44. [Google Scholar] [CrossRef]
174. Mayo, P., Copetti, R., Feller-Kopman, D., Mathis, G., Maury, E. et al. (2019). Thoracic ultrasonography: A narrative review. Intensive Care Medicine, 45(9), 1200–1211. DOI 10.1007/s00134-019-05725-8. [Google Scholar] [CrossRef]
175. Demi, L. (2020). Lung ultrasound: The future ahead and the lessons learned from COVID-19. The Journal of the Acoustical Society of America, 148(4), 2146–2150. DOI 10.1121/10.0002183. [Google Scholar] [CrossRef]
176. Smargiassi, A., Soldati, G., Borghetti, A., Scoppettuolo, G., Tamburrini, E. et al. (2020). Lung ultrasonography for early management of patients with respiratory symptoms during COVID-19 pandemic. Journal of Ultrasound, 1–8. DOI 10.1007/s40477-020-00501-7. [Google Scholar] [CrossRef]
177. McDermott, C., Lacki, M. B., Sainsbury, B., Henry, J., Filippov, M. et al. (2021). Sonographic diagnosis of COVID-19: A review of image processing for lung ultrasound. Frontiers in Big Data, 4, 2. DOI 10.3389/fdata.2021.612561. [Google Scholar] [CrossRef]
178. Peng, Q. Y., Wang, X. T., Zhang, L. N. (2020). Findings of lung ultrasonography of novel corona virus pneumonia during the 2019–2020 epidemic. Intensive Care Medicine, 46(5), 849–850. DOI 10.1007/s00134-020-05996-6. [Google Scholar] [CrossRef]
179. Gargani, L., Picano, E. (2015). The risk of cumulative radiation exposure in chest imaging and the advantage of bedside ultrasound. Critical Ultrasound Journal, 7(1), 1–4. DOI 10.1186/s13089-015-0020-x. [Google Scholar] [CrossRef]
180. Buonsenso, D., Raffaelli, F., Tamburrini, E., Biasucci, D., Salvi, S. et al. (2020). Clinical role of lung ultrasound for diagnosis and monitoring of COVID-19 pneumonia in pregnant women. Ultrasound in Obstetrics & Gynecology, 56(1), 106–109. DOI 10.1002/uog.22055. [Google Scholar] [CrossRef]
181. Kalafat, E., Yaprak, E., Cinar, G., Varli, B., Ozisik, S. et al. (2020). Lung ultrasound and computed tomographic findings in pregnant woman with COVID-19. Ultrasound in Obstetrics & Gynecology, 55(6), 835–837. DOI 10.1002/uog.22034. [Google Scholar] [CrossRef]
182. Soldati, G., Smargiassi, A., Inchingolo, R., Buonsenso, D., Perrone, T. et al. (2020). Is there a role for lung ultrasound during the COVID-19 pandemic? Journal of Ultrasound in Medicine, 39(7), 1459–1462. DOI 10.1002/jum.15284. [Google Scholar] [CrossRef]
183. Soldati, G., Smargiassi, A., Inchingolo, R., Buonsenso, D., Perrone, T. et al. (2020). Proposal for international standardization of the use of lung ultrasound for patients with COVID-19: A simple, quantitative, reproducible method. Journal of Ultrasound in Medicine, 39(7), 1413–1419. DOI 10.1002/jum.15285. [Google Scholar] [CrossRef]
184. Sultan, L. R., Sehgal, C. M. (2020). A review of early experience in lung ultrasound in the diagnosis and management of COVID-19. Ultrasound in Medicine & Biology, 46(9), 2530–2545. DOI 10.1016/j.ultrasmedbio.2020.05.012. [Google Scholar] [CrossRef]
185. Amatya, Y., Rupp, J., Russell, F. M., Saunders, J., Bales, B. et al. (2018). Diagnostic use of lung ultrasound compared to chest radiograph for suspected pneumonia in a resource-limited setting. International Journal of Emergency Medicine, 11(1), 1–5. DOI 10.1186/s12245-018-0170-2. [Google Scholar] [CrossRef]
186. Roy, S., Menapace, W., Oei, S., Luijten, B., Fini, E. et al. (2020). Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Transactions on Medical Imaging, 39(8), 2676–2687. DOI 10.1109/TMI.2020.2994459. [Google Scholar] [CrossRef]
187. Muhammad, G., Hossain, M. S. (2021). COVID-19 and non-COVID-19 classification using multi-layers fusion from lung ultrasound images. Information Fusion, 72(7), 80–88. DOI 10.1016/j.inffus.2021.02.013. [Google Scholar] [CrossRef]
188. Carrer, L., Donini, E., Marinelli, D., Zanetti, M., Mento, F. et al. (2020). Automatic pleural line extraction and COVID-19 scoring from lung ultrasound data. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 67(11), 2207–2217. DOI 10.1109/TUFFC.2020.3005512. [Google Scholar] [CrossRef]
189. Trauer, M. M., Matthies, A., Mani, N., McDermott, C., Jarman, R. (2020). The utility of lung ultrasound in COVID-19: A systematic scoping review. Ultrasound, 28(4), 208–222. DOI 10.1177/1742271X20950779. [Google Scholar] [CrossRef]
190. Born, J., Wiedemann, N., Brändle, G., Buhre, C., Rieck, B. et al. (2020). Accelerating COVID-19 differential diagnosis with explainable ultrasound image analysis. arXiv preprint arXiv:2009.06116. [Google Scholar]
191. Awasthi, N., Dayal, A., Cenkeramaddi, L. R., Yalavarthy, P. K. (2021). Mini-covidnet: Efficient light weight deep neural network for ultrasound based point-of-care detection of COVID-19. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 68(6), 2023–2037. DOI 10.1109/TUFFC.2021.3068190. [Google Scholar] [CrossRef]
192. Dastider, A. G., Sadik, F., Fattah, S. A. (2021). An integrated autoencoder-based hybrid CNN-LSTM model for COVID-19 severity prediction from lung ultrasound. Computers in Biology and Medicine, 132(2), 104296. DOI 10.1016/j.compbiomed.2021.104296. [Google Scholar] [CrossRef]
193. Dong, Y., Mo, X., Hu, Y., Qi, X., Jiang, F. et al. (2020). Epidemiology of COVID-19 among children in China. Pediatrics, 145(6). DOI 10.1542/peds.2020-0702. [Google Scholar] [CrossRef]
194. Lu, W., Zhang, S., Chen, B., Chen, J., Xian, J. et al. (2020). A clinical study of noninvasive assessment of lung lesions inpatients with coronavirus disease-19 (COVID-19) by bedside ultrasound. Ultraschall in der Medizin (Stuttgart, Germany: 1980). [Google Scholar]
195. Qiu, H., Wu, J., Hong, L., Luo, Y., Song, Q. et al. (2020). Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: An observational cohort study. The Lancet Infectious Diseases, 20(6), 689–696. DOI 10.1016/S1473-3099(20)30198-5. [Google Scholar] [CrossRef]
196. Tagarro, A., Epalza, C., Santos, M., Sanz-Santaeufemia, F. J., Otheo, E. et al. (2021). Screening and severity of coronavirus disease 2019 (COVID-19) in children in Madrid, Spain. JAMA Pediatrics, 175(3), 316–317. DOI 10.1001/jamapediatrics.2020.1346. [Google Scholar] [CrossRef]
197. Pereda, M. A., Chavez, M. A., Hooper-Miele, C. C., Gilman, R. H., Steinhoff, M. C. et al. (2015). Lung ultrasound for the diagnosis of pneumonia in children: A meta-analysis. Pediatrics, 135(4), 714–722. DOI 10.1542/peds.2014-2833. [Google Scholar] [CrossRef]
198. Berce, V., Tomazin, M., Gorenjak, M., Berce, T., Lovreni, B. (2019). The usefulness of lung ultrasound for the aetiological diagnosis of community-acquired pneumonia in children. Scientific Reports, 9(1), 1–10. DOI 10.1038/s41598-019-54499-y. [Google Scholar] [CrossRef]
199. Musolino, A. M., Tomà, P., Supino, M. C., Scialanga, B., Mesturino, A. et al. (2019). Lung ultrasound features of children with complicated and noncomplicated community acquired pneumonia: A prospective study. Pediatric Pulmonology, 54(9), 1479–1486. DOI 10.1002/ppul.24426. [Google Scholar] [CrossRef]
200. Supino, M. C., Buonsenso, D., Scateni, S., Scialanga, B., Mesturino, M. A. et al. (2019). Point-of-care lung ultrasound in infants with bronchiolitis in the pediatric emergency department: A prospective study. European Journal of Pediatrics, 178(5), 623–632. DOI 10.1007/s00431-019-03335-6. [Google Scholar] [CrossRef]
201. Buonsenso, D., Brancato, F., Valentini, P., Curatola, A., Supino, M. et al. (2020). The use of lung ultrasound to monitor the antibiotic response of community-acquired pneumonia in children: A preliminary hypothesis. Journal of Ultrasound in Medicine, 39(4), 817–826. DOI 10.1002/jum.15147. [Google Scholar] [CrossRef]
202. Musolino, A. M., Supino, M. C., Buonsenso, D., Ferro, V., Valentini, P. et al. (2020). Lung ultrasound in children with COVID-19: Preliminary findings. Ultrasound in Medicine & Biology, 46(8), 2094–2098. DOI 10.1016/j.ultrasmedbio.2020.04.026. [Google Scholar] [CrossRef]
203. Zhang, H., Saravanan, K. M., Yang, Y., Hossain, M. T., Li, J. et al. (2020). Deep learning based drug screening for novel coronavirus 2019-NCoV. Interdisciplinary Sciences: Computational Life Sciences, 12, 368–376. DOI 10.20944/preprints202002.0061.v1. [Google Scholar] [CrossRef]
204. Chaudhury, S., Duncan, E. H., Atre, T., Storme, C. K., Beck, K. et al. (2018). Identification of immune signatures of novel adjuvant formulations using machine learning. Scientific Reports, 8(1), 1–11. DOI 10.1038/s41598-018-35452-x. [Google Scholar] [CrossRef]
205. Chaudhury, S., Duncan, E. H., Atre, T., Dutta, S., Spring, M. D. et al. (2020). Combining immunoprofiling with machine learning to assess the effects of adjuvant formulation on human vaccine-induced immunity. Human Vaccines & Immunotherapeutics, 16(2), 400–411. DOI 10.1080/21645515.2019.1654807. [Google Scholar] [CrossRef]
206. Le, T. T., Andreadakis, Z., Kumar, A., Román, R. G., Tollefsen, S. et al. (2020). The COVID-19 vaccine development landscape. Nature Reviews Drug Discovery, 19(5), 305–306. DOI 10.1038/d41573-020-00073-5. [Google Scholar] [CrossRef]
207. Magar, R., Yadav, P., Farimani, A. B. (2021). Potential neutralizing antibodies discovered for novel corona virus using machine learning. Scientific Reports, 11(1), 1–11. DOI 10.1038/s41598-021-84637-4. [Google Scholar] [CrossRef]
208. Brinati, D., Campagner, A., Ferrari, D., Locatelli, M., Banfi, G. et al. (2020). Detection of COVID-19 infection from routine blood exams with machine learning: A feasibility study. Journal of Medical Systems, 44(8), 1–12. DOI 10.1007/s10916-020-01597-4. [Google Scholar] [CrossRef]
209. Khanday, A. M. U. D., Rabani, S. T., Khan, Q. R., Rouf, N., Din, M. M. U. (2020). Machine learning based approaches for detecting COVID-19 using clinical text data. International Journal of Information Technology, 12(3), 731–739. DOI 10.1007/s41870-020-00495-9. [Google Scholar] [CrossRef]
210. Randhawa, G. S., Soltysiak, M. P., El Roz, H., de Souza, C. P., Hill, K. A. et al. (2020). Machine learning using intrinsic genomic signatures for rapid classification of novel pathogens: COVID-19 case study. PLoS One, 15(4), e0232391. DOI 10.1371/journal.pone.0232391. [Google Scholar] [CrossRef]
211. Islam, M. M., Mahmud, S., Muhammad, L., Islam, M. R., Nooruddin, S. et al. (2020). Wearable technology to assist the patients infected with novel coronavirus (COVID-19). SN Computer Science, 1(6), 1–9. DOI 10.1007/s42979-020-00335-4. [Google Scholar] [CrossRef]
212. Ferguson, N., Laydon, D., Nedjati Gilani, G., Imai, N., Ainslie, K. et al. (2020). Report 9: Impact of non-pharmaceutical interventions (NPIS) to reduce COVID 19 mortality and healthcare demand. DOI 10.25561/77482. [Google Scholar] [CrossRef]
213. Suganthi, J., Umareddy, N., Awasthi, N. (2012). Medical alert systems with telehealth & telemedicine monitoring using GSM and GPS technology. 2012 Third International Conference on Computing, Communication and Networking Technologies. IEEE. DOI 10.1109/ICCCNT.2012.6396073. [Google Scholar] [CrossRef]
214. Ade, M., Doulamis, N., Wagle, S. S., Ullah, M. G. (2011). Telehealth: Healthcare technologies and telehealth emergency (the) system. 2011 2nd International Conference on Wireless Communication, Vehicular Technology, Information Theory and Aerospace & Electronic Systems Technology. IEEE. [Google Scholar]
215. Ullah, S. M. A., Islam, M. M., Mahmud, S., Nooruddin, S., Raju, S. T. U. et al. (2021). Scalable telehealth services to combat novel coronavirus (COVID-19) pandemic. SN Computer Science, 2(1), 1–8. DOI 10.1007/s42979-020-00401-x. [Google Scholar] [CrossRef]
216. Hollander, J. E., Carr, B. G. (2020). Virtually perfect? Telemedicine for COVID-19. New England Journal of Medicine, 382(18), 1679–1681. DOI 10.1056/NEJMp2003539. [Google Scholar] [CrossRef]
217. Smith, A. C., Thomas, E., Snoswell, C. L., Haydon, H., Mehrotra, A. et al. (2020). Telehealth for global emergencies: Implications for coronavirus disease 2019 (COVID-19). Journal of Telemedicine and Telecare, 26(5), 309–313. DOI 10.1177/1357633X20916567. [Google Scholar] [CrossRef]
218. Ohannessian, R., Duong, T. A., Odone, A. (2020). Global telemedicine implementation and integration within health systems to fight the COVID-19 pandemic: A call to action. JMIR Public Health and Surveillance, 6(2), e18810. DOI 10.2196/18810. [Google Scholar] [CrossRef]
219. Wosik, J., Fudim, M., Cameron, B., Gellad, Z. F., Cho, A. et al. (2020). Telehealth transformation: COVID-19 and the rise of virtual care. Journal of the American Medical Informatics Association, 27(6), 957–962. DOI 10.1093/jamia/ocaa067. [Google Scholar] [CrossRef]
220. Davarpanah, A. H., Mahdavi, A., Sabri, A., Langroudi, T. F., Kahkouee, S. et al. (2020). Novel screening and triage strategy in iran during deadly coronavirus disease 2019 (COVID-19) epidemic: Value of humanitarian teleconsultation service. Journal of the American College of Radiology, 17(6), 734–738. DOI 10.1016/j.jacr.2020.03.015. [Google Scholar] [CrossRef]
221. Neubeck, L., Hansen, T., Jaarsma, T., Klompstra, L., Gallagher, R. (2020). Delivering healthcare remotely to cardiovascular patients during COVID-19: A rapid review of the evidence. European Journal of Cardiovascular Nursing, 19(6), 486–494. DOI 10.1177/1474515120924530. [Google Scholar] [CrossRef]
222. Vandekerckhove, P., Vandekerckhove, Y., Tavernier, R., de Jaegher, K., de Mul, M. (2020). Leveraging user experience to improve video consultations in a cardiology practice during the COVID-19 pandemic: Initial insights. Journal of Medical Internet Research, 22(6), e19771. DOI 10.2196/19771. [Google Scholar] [CrossRef]
223. Patel, P. D., Cobb, J., Wright, D., Turer, R. W., Jordan, T. et al. (2020). Rapid development of telehealth capabilities within pediatric patient portal infrastructure for COVID-19 care: Barriers, solutions, results. Journal of the American Medical Informatics Association, 27(7), 1116–1120. DOI 10.1093/jamia/ocaa065. [Google Scholar] [CrossRef]
224. Bokolo, A. J. (2020). Exploring the adoption of telemedicine and virtual software for care of outpatients during and after COVID-19 pandemic. Irish Journal of Medical Science, 1971, 1–10. DOI 10.1007/s11845-020-02299-z. [Google Scholar] [CrossRef]
225. Aslani, N., Garavand, A. (2020). The role of telemedicine to control COVID-19. http://eprints.iums.ac.ir/23785/1/The%20role%20of%20telemedicine%20to%20control%20COVID-19.pdf. [Google Scholar]
226. Jnr, B. A. (2020). Use of telemedicine and virtual care for remote treatment in response to COVID-19 pandemic. Journal of Medical Systems, 44(7), 1–9. DOI 10.1007/s10916-020-01596-5. [Google Scholar] [CrossRef]
227. Samuel, J., Ali, G., Rahman, M., Esawi, E., Samuel, Y. et al. (2020). COVID-19 public sentiment insights and machine learning for tweets classification. Information–An International Interdisciplinary Journal, 11(6), 314. [Google Scholar]
228. Nemes, L., Kiss, A. (2020). Social media sentiment analysis based on COVID-19. Journal of Information and Telecommunication, 5(1), 1–15. DOI 10.1080/24751839.2020.1790793. [Google Scholar] [CrossRef]
229. Barkur, G., Vibha, G. B. K. (2020). Sentiment analysis of nationwide lockdown due to COVID 19 outbreak: Evidence from india. Asian Journal of Psychiatry, 51(2), 102089. DOI 10.1016/j.ajp.2020.102089. [Google Scholar] [CrossRef]
230. Chandra, R., Krishna, A. (2021). COVID-19 sentiment analysis via deep learning during the rise of novel cases. arXiv preprint arXiv:2104.10662. [Google Scholar]
231. Chintalapudi, N., Battineni, G., Amenta, F. (2021). Sentimental analysis of COVID-19 tweets using deep learning models. Infectious Disease Reports, 13(2), 329–339. DOI 10.3390/idr13020032. [Google Scholar] [CrossRef]
232. Wang, T., Lu, K., Chow, K. P., Zhu, Q. (2020). COVID-19 sensing: Negative sentiment analysis on social media in China via bert model. IEEE Access, 8, 138162–138169. DOI 10.1109/ACCESS.2020.3012595. [Google Scholar] [CrossRef]
233. Satu, M. S., Khan, M. I., Mahmud, M., Uddin, S., Summers, M. A. et al. (2021). Tclustvid: A novel machine learning classification model to investigate topics and sentiment in COVID-19 tweets. Knowledge-Based Systems. DOI 10.1016/j.knosys.2021.107126. [Google Scholar] [CrossRef]
234. Blei, D. M., Ng, A. Y., Jordan, M. I. (2003). Latent dirichlet allocation. The Journal of Machine Learning Research, 3, 993–1022. https://www.jmlr.org/papers/volume3/blei03a/blei03a.pdf?TB_iframe=true&width=370.8&height=658.8. [Google Scholar]
235. Xue, J., Chen, J., Hu, R., Chen, C., Zheng, C. et al. (2020). Twitter discussions and emotions about the COVID-19 pandemic: Machine learning approach. Journal of Medical Internet Research, 22(11), e20550. DOI 10.2196/20550. [Google Scholar] [CrossRef]
236. de Melo, T., Figueiredo, C. M. (2021). Comparing news articles and tweets about COVID-19 in brazil: Sentiment analysis and topic modeling approach. JMIR Public Health and Surveillance, 7(2), e24585. DOI 10.2196/24585. [Google Scholar] [CrossRef]
237. Hung, M., Lauren, E., Hon, E. S., Birmingham, W. C., Xu, J. et al. (2020). Social network analysis of COVID-19 sentiments: Application of artificial intelligence. Journal of Medical Internet Research, 22(8), e22590. DOI 10.2196/22590. [Google Scholar] [CrossRef]
238. Sanders, A., White, R., Severson, L., Ma, R., McQueen, R. et al. (2020). Unmasking the conversation on masks: Natural language processing for topical sentiment analysis of COVID-19 twitter discourse. DOI 10.1101/2020.08.28.20183863. [Google Scholar] [CrossRef]
239. Hussain, A., Tahir, A., Hussain, Z., Sheikh, Z., Gogate, M. et al. (2021). Artificial intelligence-enabled analysis of public attitudes on facebook and twitter toward COVID-19 vaccines in the united kingdom and the united states: Observational study. Journal of Medical Internet Research, 23(4), e26627. DOI 10.2196/26627. [Google Scholar] [CrossRef]
240. Ritonga, M., Al Ihsan, M. A., Anjar, A., Rambe, F. H. (2021). Sentiment analysis of COVID-19 vaccine in indonesia using nave bayes algorithm. IOP Conference Series: Materials Science and Engineering. vol. 1088, https://iopscience.iop.org/article/10.1088/1757-899X/1088/1/012045/pdf. [Google Scholar]
241. Wang, C., Horby, P. W., Hayden, F. G., Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. The Lancet, 395(10223), 470–473. DOI 10.1016/S0140-6736(20)30185-9. [Google Scholar] [CrossRef]
242. Holshue, M. L., DeBolt, C., Lindquist, S., Lofy, K. H., Wiesman, J. et al. (2020). First case of 2019 novel coronavirus in the united states. New England Journal of Medicine, 382(10), 929–936. DOI 10.1056/NEJMoa2001191. [Google Scholar] [CrossRef]
243. Jin, Y., Qin, C., Huang, Y., Liu, C. (2021). Actual bearing compound fault diagnosis based on active learning and decoupling attentional residual network. Measurement, 173(13), 108500. DOI 10.1016/j.measurement.2020.108500. [Google Scholar] [CrossRef]
244. Tsiknakis, N., Trivizakis, E., Vassalou, E. E., Papadakis, G. Z., Spandidos, D. A. et al. (2020). Interpretable artificial intelligence framework for COVID-19 screening on chest X-rays. Experimental and Therapeutic Medicine, 20(2), 727–735. DOI 10.3892/etm.2020.8797. [Google Scholar] [CrossRef]
245. Ezzat, D., Ella, H. A., Ella, H. A. (2020). GSA-densenet121-COVID-19: A hybrid deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization algorithm. arXiv preprint arXiv: 2004.05084. [Google Scholar]
246. Rajaraman, S., Siegelman, J., Alderson, P. O., Folio, L. S., Folio, L. R. et al. (2020). Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access, 8, 115041–115050. DOI 10.1109/ACCESS.2020.3003810. [Google Scholar] [CrossRef]
247. Singh, M., Bansal, S., Ahuja, S., Dubey, R. K., Panigrahi, B. K. et al. (2021). Transfer learning-based ensemble support vector machine model for automated COVID-19 detection using lung computerized tomography scan data. Medical & Biological Engineering & Computing, 59, 1–15. DOI 10.1007/s11517-020-02299-2. [Google Scholar] [CrossRef]
248. Alom, M. Z., Rahman, M., Nasrin, M. S., Taha, T. M., Asari, V. K. (2020). Covid_mtnet: COVID-19 detection with multi-task deep learning approaches. arXiv preprint arXiv:2004.03747. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |