
 | Computer Modeling in Engineering & Sciences |  |
DOI: 10.32604/cmes.2021.016728
REVIEW
Multi-Disease Prediction Based on Deep Learning: A Survey
College of Data Science Software Engineering, Qingdao University, Qingdao, 2660712, China
*Corresponding Author: Zhihan Lv. Email: lvzhihan@qdu.edu.cn
Received: 20 March 2021; Accepted: 06 May 2021
Abstract: In recent years, the development of artificial intelligence (AI) and the gradual beginning of AI's research in the medical field have allowed people to see the excellent prospects of the integration of AI and healthcare. Among them, the hot deep learning field has shown greater potential in applications such as disease prediction and drug response prediction. From the initial logistic regression model to the machine learning model, and then to the deep learning model today, the accuracy of medical disease prediction has been continuously improved, and the performance in all aspects has also been significantly improved. This article introduces some basic deep learning frameworks and some common diseases, and summarizes the deep learning prediction methods corresponding to different diseases. Point out a series of problems in the current disease prediction, and make a prospect for the future development. It aims to clarify the effectiveness of deep learning in disease prediction, and demonstrates the high correlation between deep learning and the medical field in future development. The unique feature extraction methods of deep learning methods can still play an important role in future medical research.
Keywords: Deep learning; disease prediction; Internet of Things; COVID-19; precision medicine
With the progress of society, people's living habits and environmental conditions are gradually changing, which invisibly increases people's hidden dangers of various diseases. Major diseases such as Brain Health, Cardiovascular Diseases, Vision Impairment, Diabetes, Cancer, etc., have a serious impact all over the world. Looking at diabetes alone, there are 422 million people in the world who are troubled, and Type 2 diabetes patients account for more than 90% [1]. With the increase of age, the heart senescence and loss of function make the risk of heart diseases increase. Heart disease deaths account for more than 30% of global deaths [2]. These major diseases seriously affect human health and reduce personal productivity. At the same time, it will aggravate social pressure and increase medical and health care expenditures. The main purpose of disease prediction is to predict the risk probability of an individual suffering from a certain disease in the future. There are a large number of influencing factors that need to be taken into account for diverse diseases in different populations. Including a set of features with extremely wide dimensions that need to be detected, as well as complex and variable individual and disease differences need to be distinguished. Simply wanting to accomplish these tasks manually is not only thorny, but also consumes a lot of human and financial resources.
The development of society has promoted the progress of technology. It makes the acquisition of medical data sets easier and easier. Medical institutions all over the world are collecting various health-related data, and the scale is constantly expanding. The types of medical data include basic patient information, electronic health records (EHR) [3], electronic medical records (EMR) [4], image data, medical instrument data, etc. Although they belong to one type of basic data, they are extremely vulnerable to impact. For example, incomplete or redundant data caused by its timeliness, or personal security reasons make the data untrue and privacy exposure issues. In addition, medical data is different from ordinary data in complexity, high-dimensionality, heterogeneity, irregularity, and involves more unstructured data, which is difficult to achieve by simply handling these data manually. Moreover, medical diagnosis in reality is still limited by many factors, such as medical conditions, the level of the diagnostician, and the differences between different patients. Based on the above problems, people urgently need some auxiliary methods to help predict and diagnose diseases.
Among the many methods of studying complex data, machine learning has developed rapidly, and the deep learning part of it has suddenly emerged and has become the most eye-catching branch in the field of machine learning. The ability of deep learning to process complex data, the method of extracting the main features of multi-dimensional data, the efficient response to unstructured data, and the classification strategy with higher accuracy are all superior to the previous technical methods. This also allows more people to begin to understand and step into the field of deep learning. In recent years, the development of deep learning technology has become more and more mature. It has brought disruptive changes in many fields such as image recognition, speech recognition, and natural language processing. At the same time, the application of deep learning to disease prediction has gradually attracted people's attention and has achieved many impressive results [5]. The characteristics of medical data are highly compatible with the deep learning model, which can better complete various tasks.
The foundation of deep learning is artificial neural network (ANN), which is a structure composed of multiple neurons according to a certain level, also known as a perceptron. With the changes in people's demand for models, the expansion of data scale and the improvement of computing power, the neural network has gradually evolved from a shallow model to a deep model, and structurally becomes a multilayer perceptron, it also called deep neural network (DNN). A deep neural network includes an input layer, a hidden layer, and an output layer. The core of the traditional multi-layer perceptron neural network is the back propagation algorithm during training, as shown in Fig. 1.
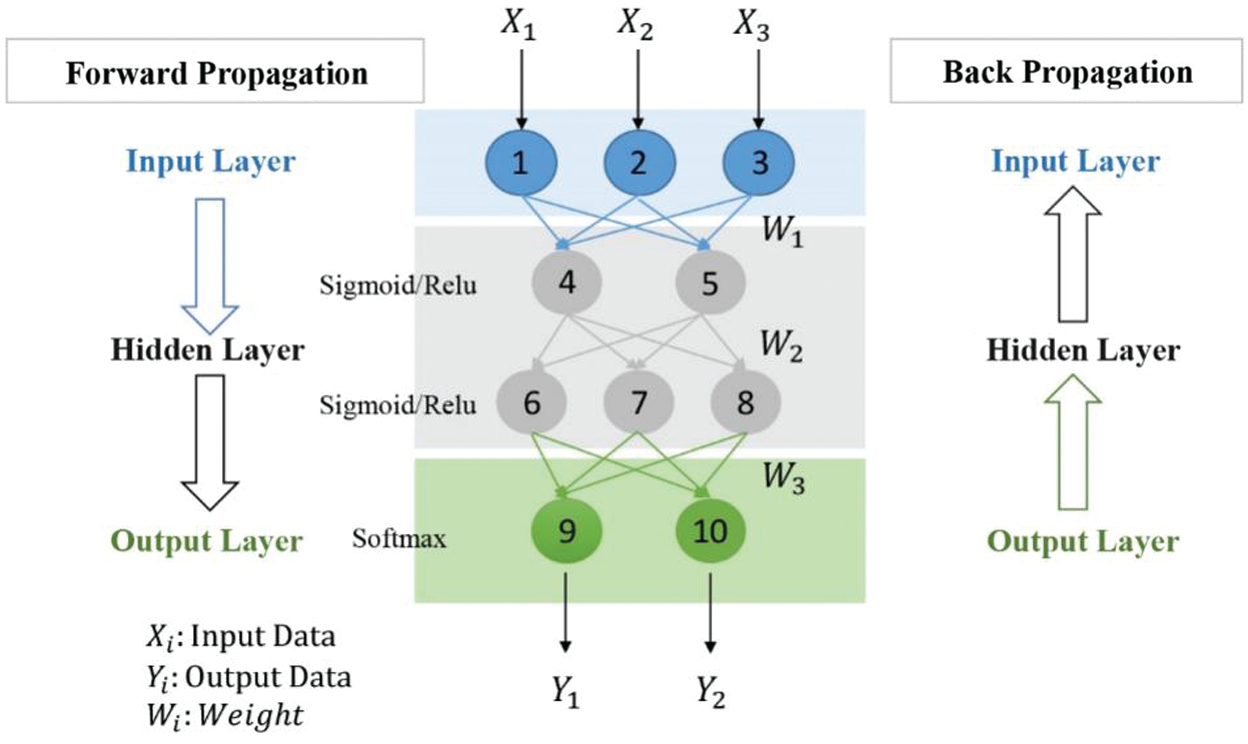
Figure 1: Deep neural network
It includes the procedure of forward propagation of information and the process of optimizing loss function through back propagation. Visually speaking, it is like letting a computer build a series of programs similar to the human brain, exercising the computer has human intelligence, so that it can simulate the principles of human vision, hearing, and other intelligent behaviors, and evolve in the process of continuous practice. Similarly, when predicting diseases, the computer can be used to imitate medical experts to diagnose diseases and accumulate prior experience in the continuous practice process to improve the accuracy of prediction, thereby making the model more robust. For a variety of medical data and problems, DNN has gradually evolved a variety of network structure models to deal with [6], which will be explained in the next section.
In the follow-up content of this article, the second part first introduces some popular deep learning models and methods, and briefly introduces some improved model frameworks, leading to some basic medical problem solving strategies. For different diseases, the third part briefly demonstrates the concept of diseases, overviews the deep learning methods used in predicting diseases, and summarizes the experiences and improvements. The fourth part will point out some of the problems and challenges that exist when the current deep learning methods are combined with disease prediction. The fifth part introduces the extension of the intelligent fusion of the two under the current technological development conditions, and prospects for the future application prospects. The sixth part summarizes the content of the full text.
When a neural network containing only a fully connected layer reaches a deeper level, the amount of parameters will gradually increase, which will increase the memory footprint and increase the computational cost. In order to overcome the problems caused by some fully connected layers and promote the development of neural networks to a deeper level, a solution has been gradually explored. By using the idea of local correlation and weight sharing, a convolutional neural network (CNN) is designed, which not only greatly reduces the number of parameters, but also greatly improves the training efficiency. The proposal of LeNet in 1998 marked the birth of CNN and defined the basic framework of CNN. Fig. 2 shows the basic structure, including convolutional layer, pooling layer, fully connected layer. And the architectural changes after convolution and pooling, at the end, get output after fc.
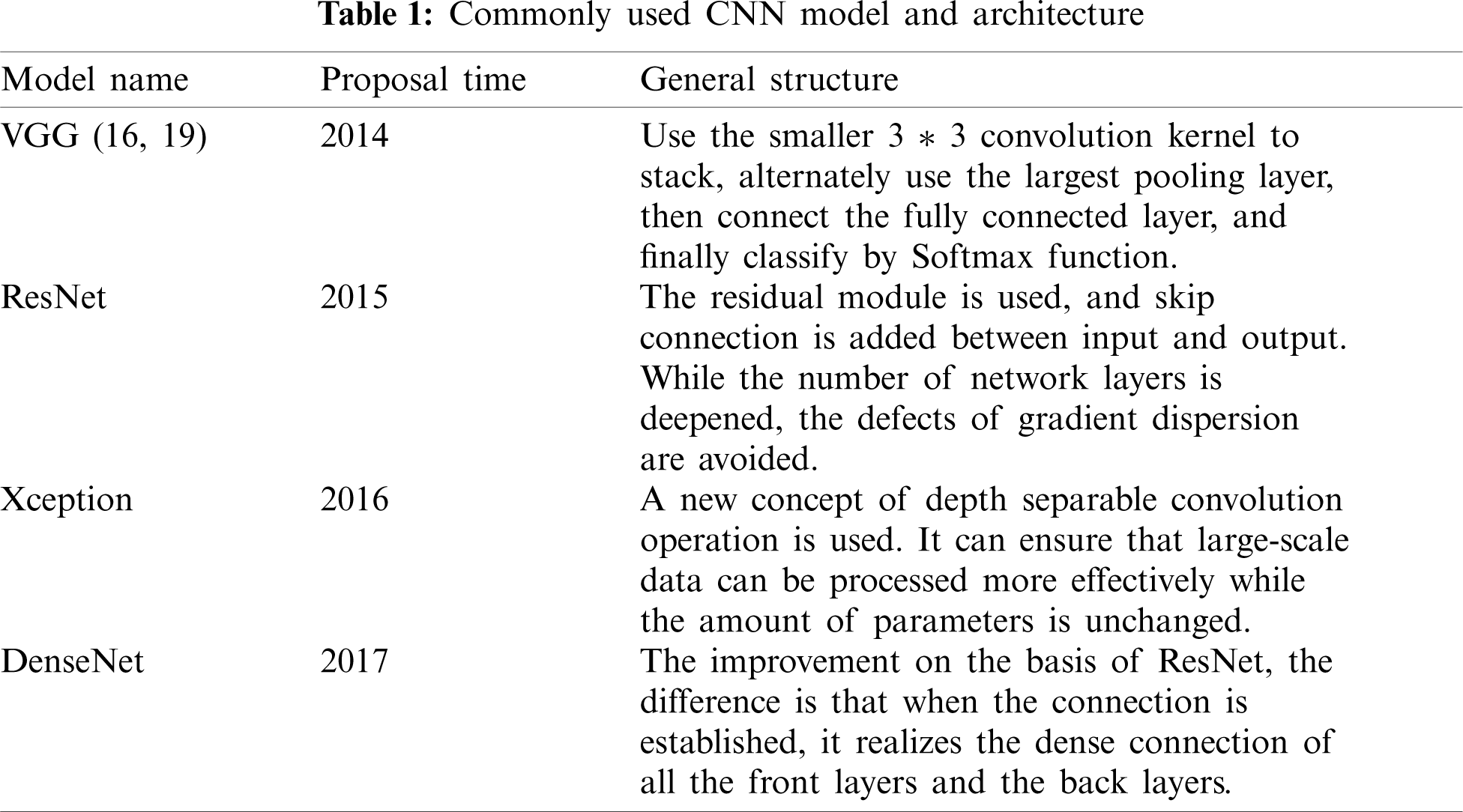
However, due to the influence of computer computing power and data volume at that time, it has not been developed to a greater extent. Until 2012, the overcoming of some objective factors, coupled with the excellent results of CNN in some international competitions, CNN quickly became the new favorite method in the field of computer vision. After that, improved CNN models and CNN variant models starting occurrence, such as VGG net [7], ResNet [8], DenseNet [9], Xception [10], etc. As shown in Tab. 1, the basic information of several frameworks is introduced. Although CNN was originally invented for computer vision, in subsequent studies it has gradually proved that CNN is also quite effective in other fields [11,12], and the prominent directions include medicine. Imaging classification, disease prediction [13,14], medical image visualization [15], etc. CNN can process many types of data, the most prominent is the processing of pictures with spatial local correlation data, such as medical field chest X-ray, CT and other image data. Regardless of whether it is a single image for disease diagnosis or a combination of multiple images for auxiliary prediction [16], the CNN model can realize the processing of medical images through the feature extraction and fusion technology of complex data. Then through the fully connected layer to achieve disease classification, its performance is better than traditional machine learning models [17].
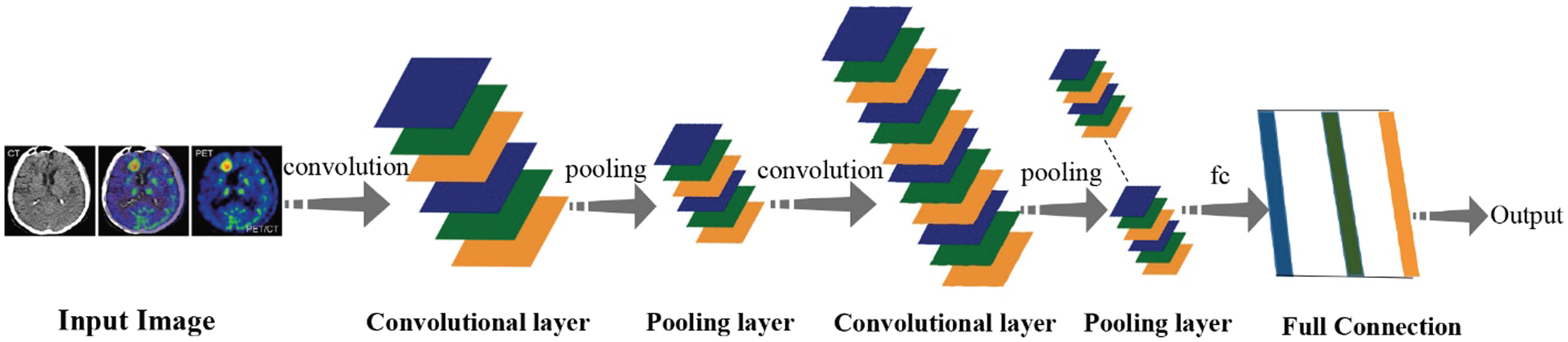
Figure 2: Convolutional neural networks (CNN)
Time series forecasting, task-based dialogue, etc. all require models to learn features from the sequence. The neurons in each layer of the neural network including the fully connected layer cannot be connected to each other, which leads to the problem of not being able to effectively manage the time series. CNN can solve some problems in the spatial dimension, but it cannot handle data in the temporal dimension. In response to some of the problems mentioned above, recurrent neural networks (RNN) came into being.
RNN is composed of neurons and feedback loops. For scenarios where the previous input and the next input have a dependency relationship, RNN shows unique advantages [18]. RNN performs better than most machine learning algorithms when processing big data with delays and abnormal noise. However, RNN faces the possibility of gradient explosion and gradient disappearance during the training process, as well as the short-term memory defects of RNN [19], which derived improved models LSTM and GRU. Compared with the basic RNN, LSTM adds a state vector and a gate mechanism. As shown in Fig. 3, the main structure of LSTM includes: Input Gate, Forget Gate, Output Gate. The GRU only contains Reset Gate and Update Gate. Through the gating mechanism to achieve the control of information refresh and forgetting. In this figure,
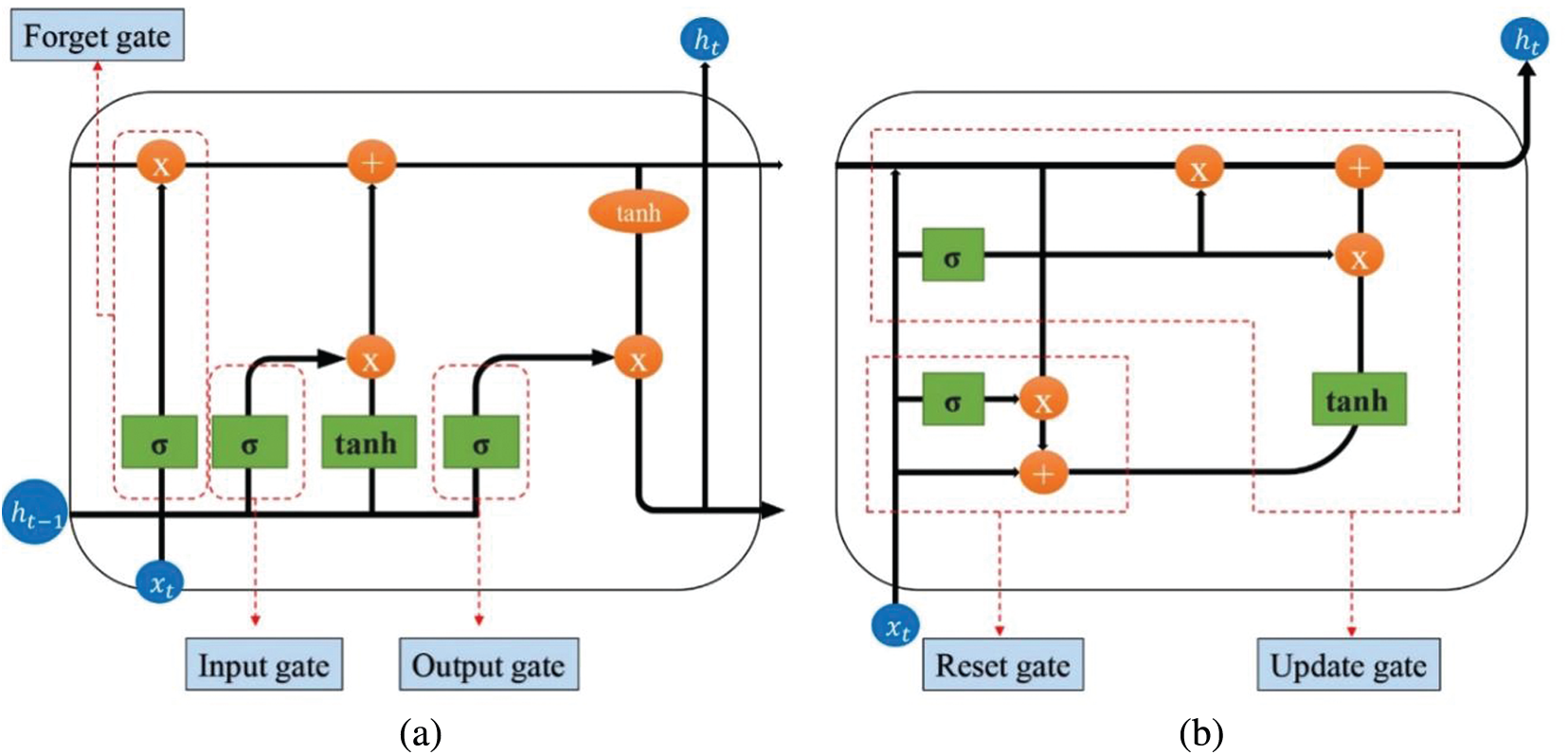
Figure 3: Improved RNN structures. (a) LSTM structure diagram (b) GRU structure diagram
The existence of a large number of unlabeled data in reality. It promotes the creation of unsupervised learning algorithms. It is difficult to label some exact labels on the data collected in actual medical scenarios. This requires some technology to automatically classify or aggregate the data to discover more further potential value of the data. Furthermore, there are many ways to obtain medical data, and it is related to the actual medical environment. Some data will be lost occasionally, and the information will be incomplete or distorted [27]. As a neural network model, autoencoder perform efficient feature extraction on unlabeled data through unsupervised learning, and perform well in feature dimensionality reduction, noise removal and restoration of original data [28].
The classic AE model consists of two modules: encoding and decoding, as shown in Fig. 4. Since the input data itself has no label information, only the input data x itself, AE uses x as the output at the same time, constructing a mapping of
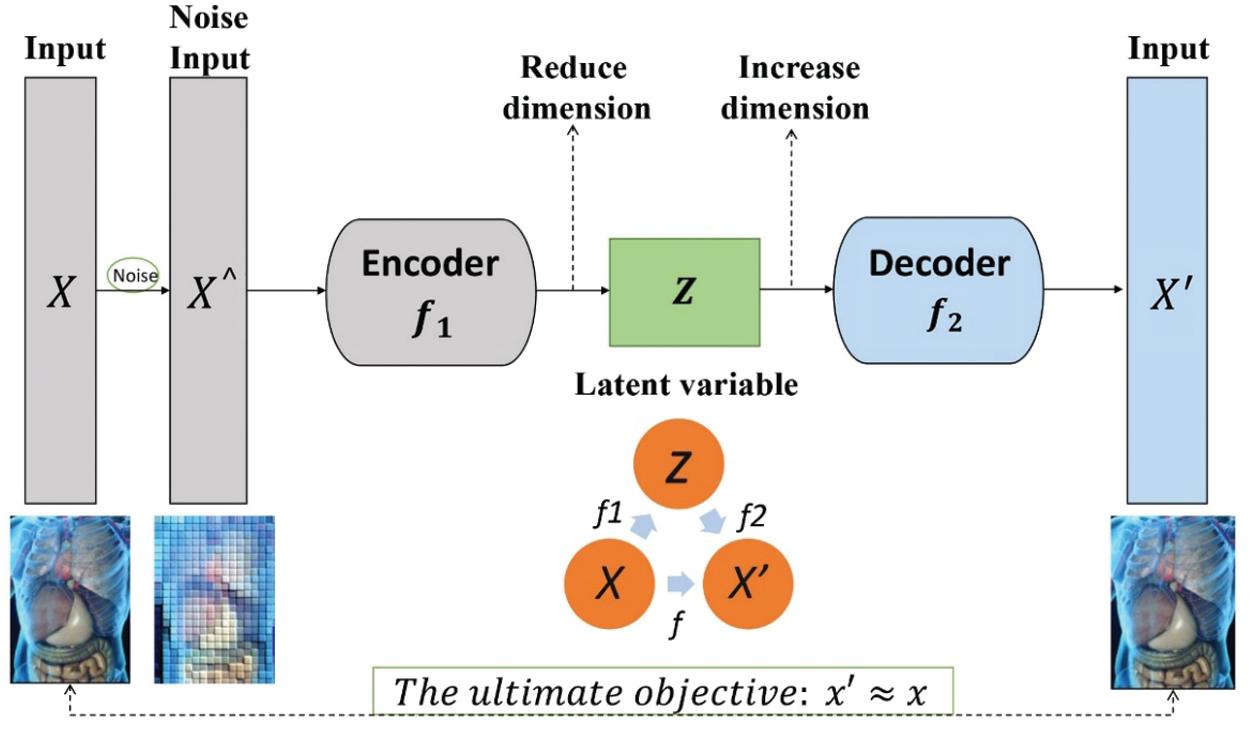
Figure 4: Autoencoder architecture
2.4 Generative Adversarial Networks (GAN)
GAN is a kind of model which is used to train synthetic image based on game theory. The idea of game theory is mainly embodied in the way of network training. Two sub-networks are set up respectively: generation network and discriminant network. They use a process similar to a competition, then apply the backpropagation algorithm to continuously optimize the generation network and the discriminant network. After that, obtain mutual improvement in turn. The generating network is mainly responsible for learning the true distribution of samples, and the discriminating network is mainly responsible for correctly distinguishing the samples generated by the generating network from the real samples. The essence of the generative network is a deconvolutional neural network, and the discrimination network is a convolutional neural network.
Since Gan was first proposed in 2014 to the present, regardless of the image size or image fidelity, the performance of GAN has been constantly improving and breakthrough. The main application of GAN is in image processing, including image-to-image translation, using GAN for image restoration [32,33], improving image resolution [34], medical image fusion [35,36] etc. Recently, the application of Gan in medical imaging has gradually increased and achieved remarkable results [37]. Traditional Gan often encounters instability problems such as gradient disappearance and mode collapse during training. Most of the improved methods introduced for these problems have good performance [38–40]. Although GAN is still in the early stages of medical imaging, it has already demonstrated its advantages in the medical field. With the further resolution of these deficiencies, GAN will certainly provide effective help to the medical field to the greatest extent.
Except for the models mentioned above, Deep Belief Networks (DBN) has also appeared in medical-related research many times in recent years. For example, when processing medical images, it helps to achieve better image fusion [41]. Through DBN processing of voice information, help Parkinson's disease diagnosis [42] and so on. DBN can be used in both supervised learning and unsupervised learning, it can play the role of classifier in supervised learning, and it have the same effects as AE on unsupervised learning, assist doctor process medical information more comprehensively.
In handling practical applications, research methods that combine multiple models perform better. Different models are used to deal with problems from different angles. The method of combining CNN and RNN proposed by [43] uses CNN to process horizontal data, RNN solves vertical problems, and then combines with Bagging, the accuracy is significantly better than independent models.
Besides, transfer learning has become more and more popular for the past few years, and the trend of combining with deep learning has become more obvious. One of the drawbacks of deep learning is that data dependence is relatively serious and requires the help of a large amount of data. Transfer learning can be completed with a small amount of data, which is similar to a “polymorphic” idea, which provides ideas for model reuse, which not only saves time, it is also conducive to the further improvement of the model. More transfer learning should be combined with a variety of DL models [44]. Talo et al. [45] use deep transfer learning to recognize magnetic resonance images, which can effectively assist radiologists in their diagnostic work. Transfer learning is applied to medical research, and new problems are dealt with through the trained model, and the initial model is slightly modified to be used in cancer image lesion recognition, all have good performance [46]. With the maturity of medical imaging technology, transfer learning can help the medical field achieve more far-reaching research.
3 Disease Classification and Related Prediction
Pneumonia is an inflammation of multiple organs in the lungs caused by a variety of factors and pathogens. Bacterial pneumonia is the most common. Common symptoms include difficulty breathing, fever, and chest pain, which have a greater impact on the health of children and the elderly. According to statistics, the ratio of pneumonia patients to deaths is huge across the world. Although antibiotics and vaccines currently exist, there are still major hidden dangers to health and even life threatening. COVID-19, which is raging around the world, is a mild to moderate respiratory disease. It is essentially pneumonia caused by a coronavirus [47]. COVID-19 not only causes damage to the lungs, but also affects other human systems. The harm including cardiovascular system, cranial nervous system [48,49], etc. Since February 19, new coronary pneumonia has gradually developed into a global health problem, which is extremely important for the early diagnosis and prediction of pneumonia.
Obstruction of the respiratory tract caused by pneumonia is more common, and patients with severe lung damage can have symptoms of breathing difficulties. Fan et al. [50] designed a portable respiration measurement device, combined with the algorithm of BP neural network, which effectively improved the performance of existing ventilators. The detection of pneumonia disease is generally through X-ray image or chest radiography, and the efficient processing of images by deep learning can help radiologists to screen abnormal images of the lungs. Voroshnina et al. [51] used X-ray images to test the CNN model. The data set used contains 5863 X images, with clear image classification of diseased lungs and normal lungs. The author determined the most suitable CNN model by setting different layers, optimizers, training parameters and epochs, and finally came up with a 4-layer alternating Conv2D layer and the structure of the MaxPooling2D layer has the highest pneumonia detection accuracy rate of 89.24%. Rajpurkar et al. [52] improved on the traditional CNN model and constructed the CheXNet network, which contains 121 layers. Also tested on the chest X-ray image data set, the index exceeded the previous optimal model, and the results of detecting pneumonia were better than the level of professional radiologists. Qjidaa's team [53] applied the pre-trained and improved VGG16 network model to pneumonia prediction. The advantage of this network is that it is simple in design and can extract better features in the image. The article shows that this model has the same performance as radiologists in the location of lesions. When using this model to predict chest radiographs, the external (different data sets) AUC obtained is as high as 95% and the accuracy rate is 87.5%. In order to better monitor COVID-19, [54] evaluated three pre-trained models Xception, VGG16, and InceptionResnet-V2. In the end, the CNN model using Xception has the highest accuracy, with an accuracy rate of 97.19% higher than the others model's 95.42% and 93.87%.
At the same time, the application of transfer learning can provide new ideas for predicting pneumonia [55]. First build a deep learning framework, and then use transfer learning to apply the parameters that have been trained in other data sets to your own model, which can avoid the situation where the optimal parameters cannot be obtained due to insufficient data. Liang et al. [56] proposed a novel way to predict pneumonia. The underlying framework is a convolutional network containing a residual structure, where the residual structure reduces overfitting, and the convolutional layer avoids the loss of feature information caused by the increase in model depth. Combined with transfer learning, the final accuracy rate is 90.5%, which is better than Xception's 87.8%. Other indicators are also better than the existing CNN model; Chouhan et al. [57] also combines transfer learning and deep learning. It mainly uses five pre-trained deep learning models to extract features and classify them to get the output, and then use an ensemble model to integrate the five pre-trained models. The resulting model combines the output of five pre-trained models, and its performance is better than that of a single model.
In addition to the traditional standalone model, a variety of fusion methods have also been derived. Saul et al. [58] designed a model combining convolutional neural network and residual network to classify images. Through the analysis of X-ray images, an accuracy rate of 78.73% was finally obtained, which is higher than the optimal previous independent model 76.8%; Wu et al. [59] creatively proposed a multi-view fusion model, as shown in Fig. 5. Its bottom layer uses the ResNet50 architecture. Based on the original model, the classic RGB channel image is changed to the three lungs view image as input. Compared with the single-view model, the use of medical images from a more comprehensive perspective reduces some one-sided problems caused by single factors, while avoiding overfitting and having better performance. The data set used is 495 high-resolution CT images to diagnose COVID-19, showing the great potential of deep learning to train models through CT images to achieve efficient screening of Convid-19. COVID-19 involves a large amount of patient information and involves many security and privacy issues. In terms of ensuring the security of medical data, Hossain et al. [60] proposed the B5G framework to detect some influencing factors of COVID-19 in daily life, and cleverly combine with blockchain technology to improve data security.
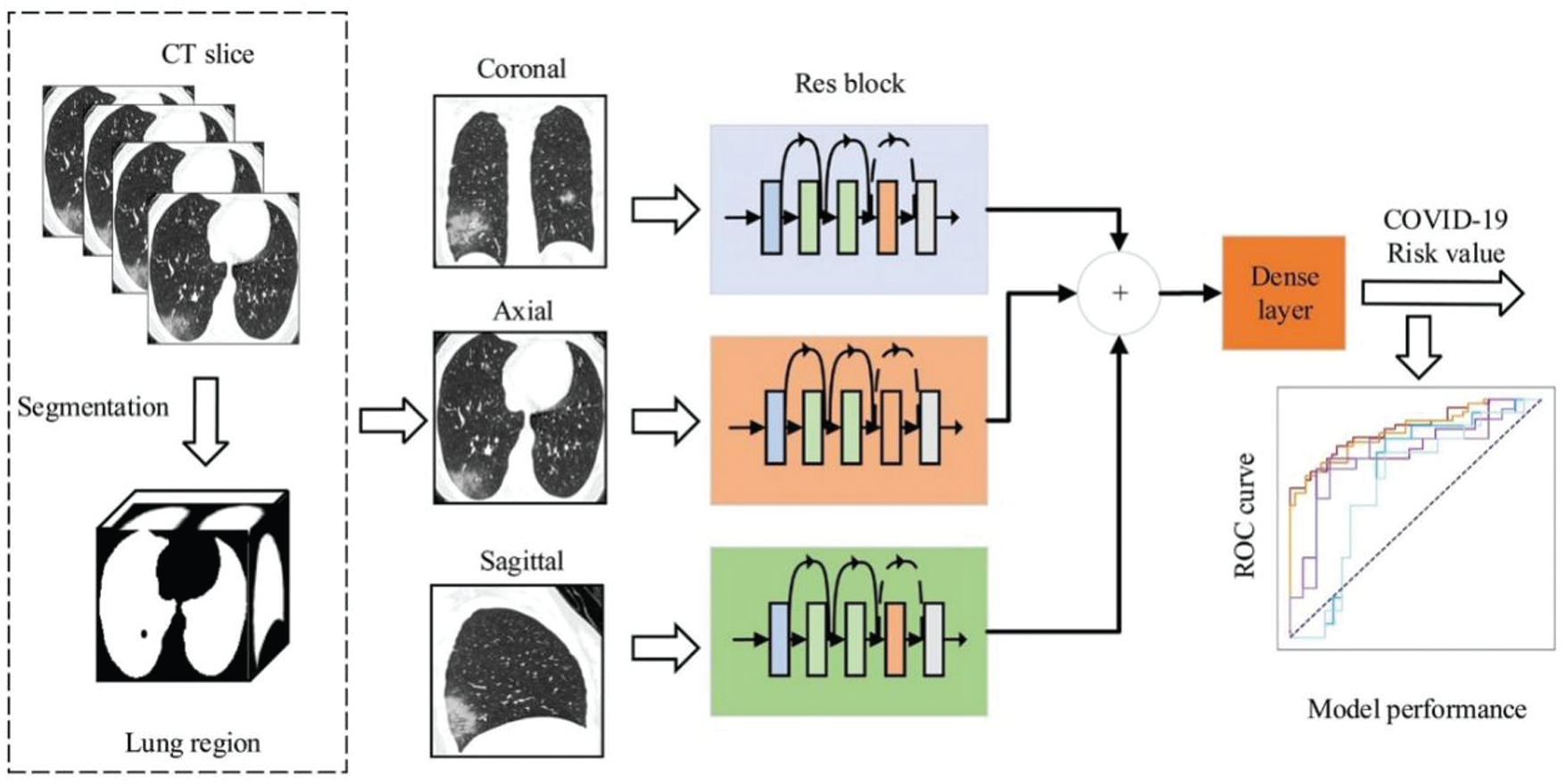
Figure 5: Multi-view deep learning fusion model [59]
Under the modern background of COVID-19, the epidemic has strongly impacted the global economy and occupies most of the medical resources in society. The relationship between economy and quality has become a problem that must be faced in reality. Reference [61] proposed an auction mechanism and decision-making model, which can assist product selection and effectively promote a win-win situation in economy and quality. The use of deep learning models can effectively alleviate the shortage of medical resources, reduce the pressure on doctors, and achieve rapid diagnosis as well accurate prediction of the individualized prognosis of COVID-19, which is essential for the management of COVID-19 [62]. In the research of the future, we should further combine DL with CT imaging, X-ray image, and other clinical information. More consideration can be given to comprehensive diagnosis, it means that predict at the same time through the patient's CT images, chest radiography and other information to avoid errors caused by single information.
3.2 Cardiovascular Disease (CVD)
Cardiovascular diseases have many different types, such as common coronary heart disease, hypertension, arrhythmia, and congenital heart disease. According to statistics, nearly 18 million people die from cardiovascular disease every year, accounting for 31% of the global deaths, and it is the world's rank first fatal disease [63]. Especially in the fast-paced society, excessive work pressure and irregular diets have made cardiovascular diseases more common. There are many factors that cause cardiovascular diseases, such as overt indicators like blood pressure and blood lipids, as well as recessive factors consist of genes and heredity. As the most common arrhythmia in clinical work, atrial fibrillation often causes the formation of brain thrombosis or embolization of other organs. The general detection is to analyze the changes in heart rhythm, but the actual clinical abnormality of heart rate is not obvious, and it is difficult to detect [64]. Deep learning has a unique processing method for high-dimensional complex features, and it can identify abnormal features among many influencing factors. In general, heart diseases are assisted in the analysis by Electrocardiograph (ECG), which can detect various abnormalities from arrhythmia to acute coronary syndrome [65]. Hannun et al. [65] used DNN to process ECG to classify the heart rhythm category, which exceeded the sensitivity of cardiologists, and could effectively reduce the misjudgment of abnormal heart rhythm diagnosis. DL can not only shorten the diagnosis time when helping CVD prediction, but also improve the accuracy, which provides many conveniences for the prevention and treatment of CVD.
During the past research, there are many practices using machine learning to predict CVD. With the development of deep learning, more and more DL methods have surpassed the traditional ML methods in many aspects. Ayon et al. [66] used seven models (including six comparative models) to predict coronary heart disease. The author utilized the Statlog data set and Cleveland data set to tests, and used five-fold and ten-fold cross-validation methods to make full use of the data. As shown in Fig. 6, only the indicators under the five-fold cross-validation of the Statlog data set are extracted here. Compared with the commonly used ML models, the accuracy, sensitivity and NPV of DNN reached 98.15%, 98.67% and 98.32%, all of which were better than the optimal ML model SVM. It is almost the same as SVM in Specificity and Precision and F1 score. In ten-fold cross-validation, DNN has the same excellent performance. And it can be seen from the figure that the performance of DNN on various evaluation indicators is very stable, and the classification effect in practical applications is also better. From this, it can be seen that when dealing with disease data with a wider dimension, the performance of DL is better than the general ML models.
Beyond that, clinical data of coronary heart disease often have unbalanced categories, and the quality of data collection is not reach the standard. Based on this problem, Dutta et al. [67] proposed an efficient neural network with convolutional layers model, which uses sub sampling methods to solve the problem of abnormal data categories, and compares it with a variety of latest machine learning models. Although it is equal to the optimal machine learning model in predicting the recall value (0.77), the accuracy rate reaches 82% when testing negative cases, which is higher than the optimal model SVM 76%, and the classification performance is also better than commonly used machine learning models. MIT-BIH arrhythmia database is the first set of more general evaluation materials for arrhythmia detection [68]. Shi et al. [69] used the improved XGBoost for the classification of abnormal heartbeat detection, combined with the MIT-BIH atrial fibrillation database (AFDB), and merged the threshold classifier on the hierarchical classifier to achieve multi-level classification of heartbeat conditions. Wang et al. [70] proposed a neural network only contains the full connection layer to detect abnormal heartbeat in accordance with AAMI standard (classifying heartbeat types into five categories: N, S, V, F and Q). The basic structure contains two layers of neural networks, and the results are further optimized through two classifications, and the effectiveness is verified by multiple data sets. Jin et al. [71] designed a residual network and combined domain adaptation methods in transfer learning. By learning and training from the source data domain, and then applying it to the target data domain, the average accuracy rate can be increased by 4.5%. Effectively solved the problem of unbalanced and unlabeled data sets.
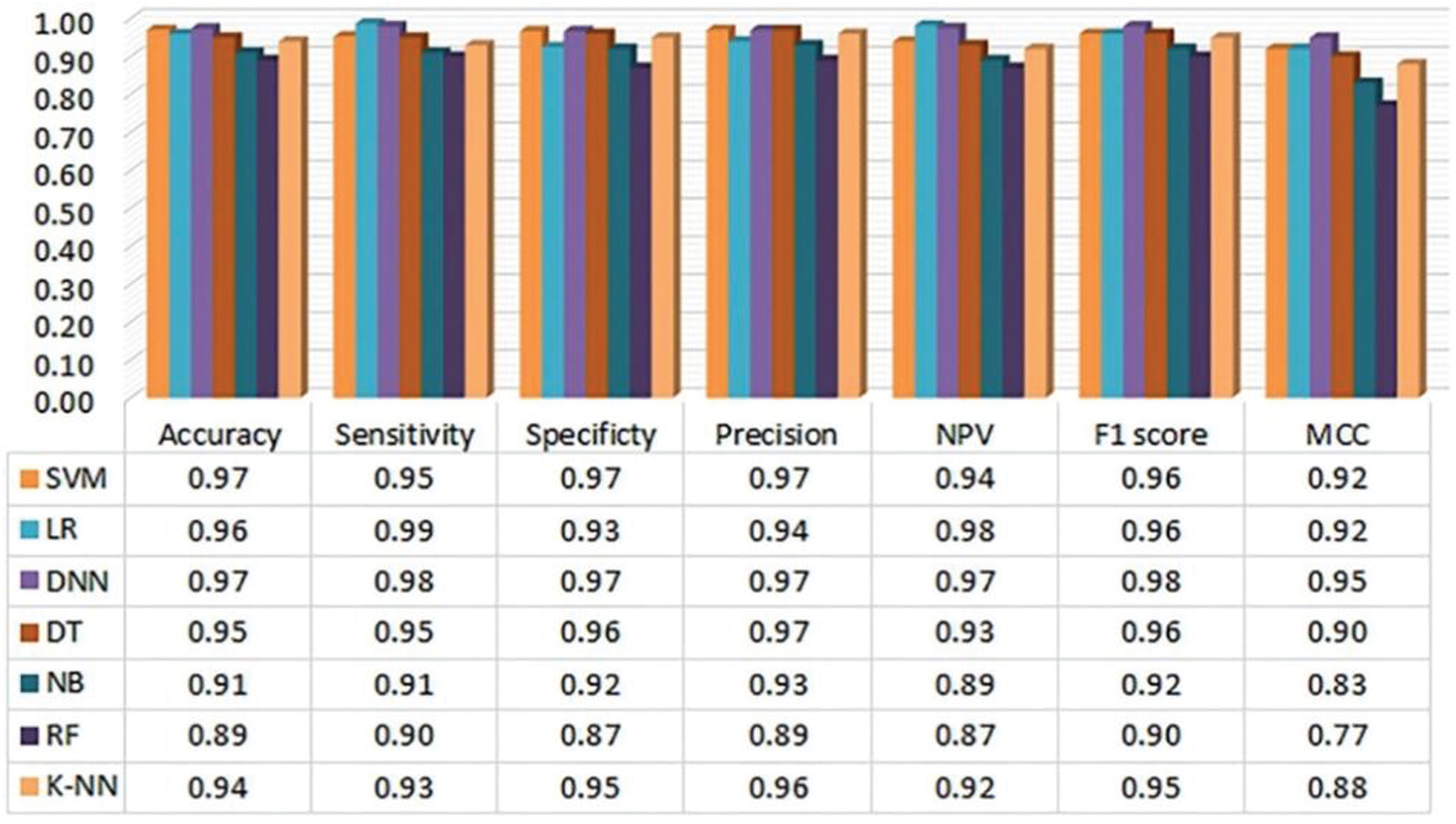
Figure 6: Evaluation metrics of the heart disease prediction (ten-fold) system using Statlog data set by Ayon et al. [66]
RNN also outperforms most ML algorithms when processing big data with delays and anomalous noise. The prediction of CVD generally needs to consider the patient's historical medical records to help decision-making. For data distributed along the timeline, RNN can be used to process. On the other hand, RNN's unique gate control structure can realize real-time monitoring of patients’ medical records and predict physical conditions at any time. Anandajayam et al. [72] Use the RNN model to predict heart disease based on the past medical records of patients with a history of heart disease, comparing the results with logistic regression, the accuracy of RNN is higher than 90%, which is better than logistic regression 85%. The result shows the advantages of RNN in processing large data set. Kuang et al. [73] created a dynamic LSTM model by improving the traditional LSTM. The unified processing of input data at different times is better than traditional LSTM in the final classification result. For the abnormal heartbeat phenomena that existed in the past, there are comparatively large individual differences. Jin et al. [74] proposed the hybrid LSTM model in his research, utilizes the advantages of active learning to automatically extract features and can study ECG fragments in a wider time span. It better solved the problems that arise due to individual differences.
What's more, using the combination of RNN and CNN to build a hybrid model can deal with more diverse disease prediction scenarios. And different model combination methods can further improve the performance of the hybrid model. Shi et al. [75] proposed a network structure that includes both CNN and LSTM, by combining several input layers for feature input. While realizing automatic feature extraction, it also makes full use of manual features to achieve a subject accuracy rate of 94.20%. Jin et al. [76] created a hybrid model called twin-attentional convolutional-LSTM, which uses CNN and LSTM as the bottom layer. It makes full use of LSTM's ability to capture time-domain information, and effectively makes up for the shortcomings of CNN in processing time-sensitive information. An accuracy rate of 98.51% is obtained on the AFDB data set. Ali et al. [77] constructed a hybrid model combining CNN and GRU to predict heart disease. Combining LDA and PCA feature algorithms for feature extraction, then improve accuracy through k-fold cross-validation, the final CNN-GRU accuracy rate is 94.5%, which is higher than 92% of LSTM and 93.7% of CNN-LSTM. Deep belief networks can also be applied to disease prediction, and it can be used to both supervised learning and unsupervised learning. Lu et al. [78] proposed an improved DBN model, combined with unsupervised training and supervised optimization, the model has excellent stability and prediction accuracy. In the final examination, the model achieved a classification accuracy of 91.26%. At the same time, the use of deep learning can help identify disease risk factors and find abnormal indicators of disease from multiple indicators. Poplin's et al. [79] used the monitoring of retinal characteristics to non-invasively detect the characteristics of blood vessels. Starting from the retinal image, combined with CNN to predict and verify the influencing factors of cardiovascular diseases, they successfully achieved the classification of multiple influencing factors.
Taking into account the characteristics of CVD, RNN can be used more to achieve early prediction, and its unique processing method for time series data can more comprehensively investigate the patient's medical history. At present, the popular use of Electronic health record (EHR) provides an opportunity for RNN to develop. In the future, EHR and RNN should be more combined to realize multi-faceted disease monitoring.
Alzheimer's disease is a syndrome, mainly manifested as a decline in cognitive and behavioral abilities, and is a degenerative disease of the nervous system. Approximately 50 million people worldwide suffer from AD. Although the initial mortality rate is not high, it has a serious impact on the burden of individuals and families [80]. The initial period of the patient's onset is not easy to detect, but will gradually get worse over time. Therefore, early AD is easily overlooked, and there is no cure for AD in the current medical field. Once suffering from AD, the medical community is still at a loss and can only be relieved by early diagnosis. The early prediction and prevention of AD is a global issue and a major challenge for global technology. Reference [81] pointed out the general development process of AD patients, from the initial normal state to mild cognitive impairment (MCI), and eventually may develop into AD. It is discovered in time that MCI can effectively alleviate and avoid deterioration to AD. The article also pointed out difficulties of AD prediction. Although there is still no comprehensive method to predict AD, deep learning technology has received more attention in the early detection and automatic classification of AD.
Magnetic resonance image (MRI) and Positron emission tomography (PET) are common medical image data used to diagnose AD. These two images have high-dimensional, multi-modal and other complex characteristics [82]. The previous processing methods are time-consuming and poor in performance. With the development of deep learning in image processing, more studies have begun to use DL models to process these brain medical images. Jo et al. [82] discussed the performance difference between the DL method and the hybrid method, as well as the impact of different AD image data, finally obtained the hybrid method with better performance, and the test indicators obtained when multiple images were mixed more acurrate. Ji's team [83] proposed a 3D-CNN model that can not only extract features from the two dimensions of time and space, but also capture more complete spatial features than the 2D model, which is more suitable for the requirements of medical imaging analysis. Cheng's team [84] based on the characteristics of MRI in the spatial and temporal dimensions, verify the effectiveness of 3D-CNN in diagnosing AD by MRI. The Alzheimer's Disease Neuroimaging Initiative (ADNI) database has extremely abundant sample types and enormous data size, including MRI, PET, Genetic Data, Biospecimen Data and other AD diagnostic data. Most of the influencing factors of AD are taken into consideration, and it is one of the most common databases for AD research. Wang et al. [85] improved on the basis of 3D-CNN, constructed the 3D-densely CNN model in Fig.7. The author uses the ANDI dataset and performed classification tests on AD/MCI/Normal, the accuracy was all in More than 93%. The 18F-FDG PET image has a unique distribution in the low-metabolic area of the cortex, which can help identify MCI and AD [86]. By comparing with the MRI of AD/MCI patients, 18F-FDG PET has higher sensitivity in helping to detect AD [81]. Using the CNN model of InceptionV3, combined with 18F-FDG PET images, proved that the DL model can correctly classify lesions.
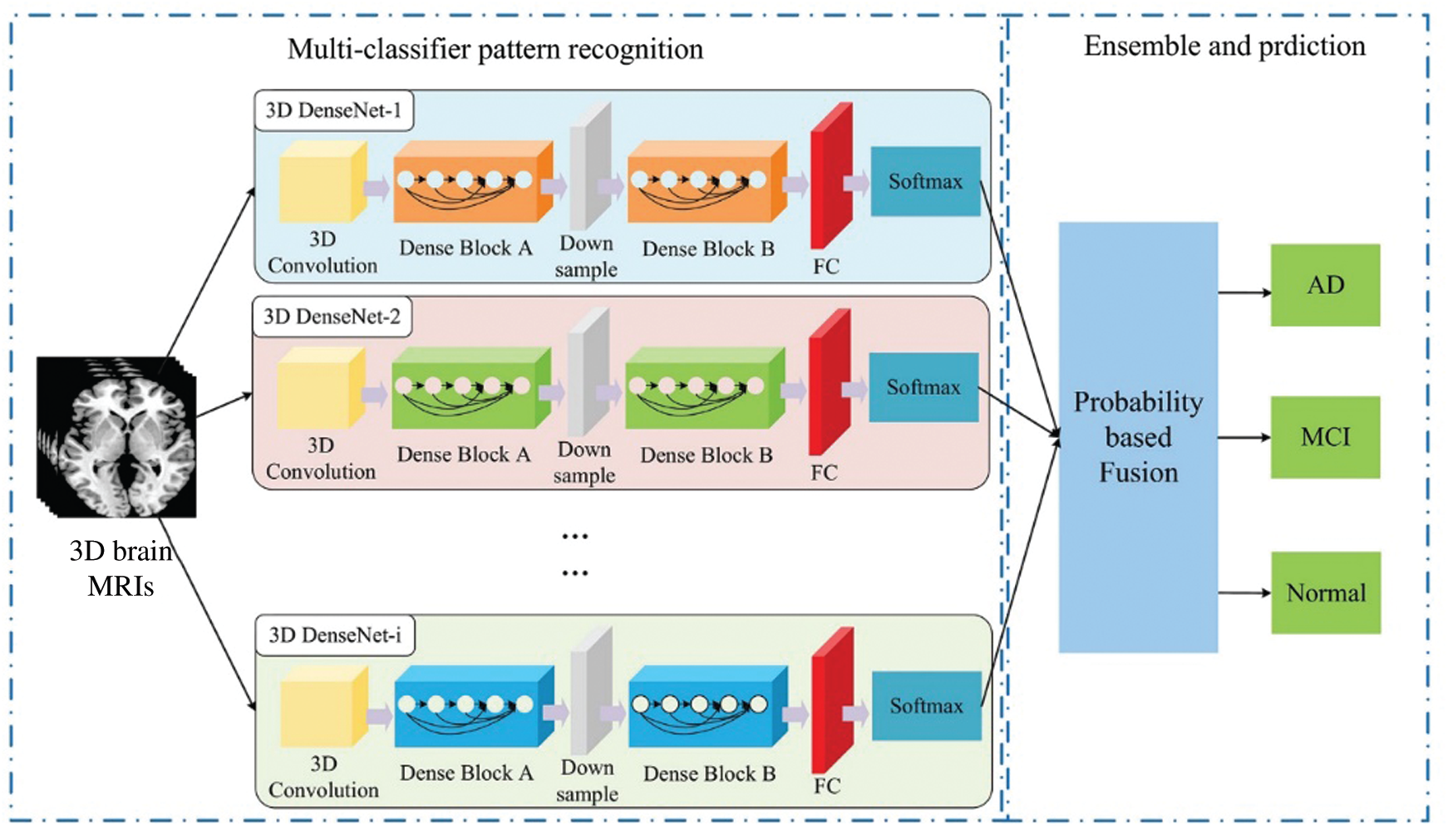
Figure 7: Wang proposed ensemble 3D-DenseNet framework for AD and MCI diagnosis [85]
In order to get rid of some limitations of identifying and classifying brain abnormalities, Cui et al. [87] proposed a framework that combines RNN and CNN. CNN learns spatial features, RNN extracts longitudinal features, and the results are tested on the ANDI dataset. The result is better than the CNN average method. Reference [88] pointed out that the bilateral hippocampus of AD/MCI patients will show lesion features, which can be identified by MRI images. Li et al. [89] used a hybrid convolutional and recurrent neural network to diagnose AD by identifying the lesions in the hippocampus in MRI, the consequence can get 91.0% AUC when classifying AD with normal controls.
Synthesize these papers, the deep learning model considers a more comprehensive perspective when processing brain medical images. Compared with traditional medical methods and medical experts, it can dig deeper brain abnormalities. However, the test results obtained through the model cannot provide interpretable reasons for decision-making. In practical applications, the image quality requirements are also tough to reach, and the performance in different image sets is not stable enough. There are many brain pathological factors, and the changes are extremely rapid. There is still a lot of room for the prediction of AD. The medical field should also further strengthen research on the prevention and treatment of AD, which can better provide basic knowledge for AI to provide medical services. It can also better to tap and develop DL technical capabilities.
Cancer is a general term for a large group of diseases. Mainly refers to the irregular growth of cells under the influence of disease-inducing factors, which become tissue masses. Cancer has a high mortality rate and ranks second in the global cause of death [90]. Cancer, also known as tumor, is generally divided into benign and malignant. Benign tumors are easy to clear and have a low recurrence rate; malignant tumors are more stubborn, easy to recur, and easy to metastasize, which can damage other tissues and organs. Although malignant tumors are difficult to cure, if timely prevention and detection and treatment can prevent the tumor from developing in the direction of deterioration [91,92]. According to statistics, 30%‒50% of cancer deaths can be prevented, early diagnosis and screening are essential. Because of there are many types of cancerous cells and large differences between cells, the comprehensiveness of prediction technology is required. In recent years, deep learning has shown its unique advantages in the classification of cancer lesions by virtue of the ability to automatically extract key data from the original data [93].
The traditional single-omics method can no longer meet the needs when studying complex diseases [94]. Cancer is a disease with a long development cycle and the possibility of multiple gene mutations. More data and more related genes must be considered to obtain more effective information. The multi-omics joint method can obtain a more comprehensive flow of disease information, and fundamentally promote the accurate diagnosis of cancer diseases, it will become a new trend in cancer research [95]. The UCSC Xena database has a wealth of tumor data sets. The Cancer Genome Atlas (TCGA) contains rich multi-omics data resources. It is conducive to multi-omics research on cancer diseases. Tan et al. [96] proposed a MOSAE (Multi-omics Supervised Autoencoder) model for genomics research, through specific AE to achieve the processing and fusion of multiple omics structures, use TCGA Pan-Cancer data to compare with six commonly used machine learning models, the ROC and AUC obtained are better than other models. Tong et al. [97] developed two different AEs based on the original AE to achieve different functions. ConcatAE was used to fuse modal information, and CrossAE was used to perform multi-modal denoising. These two models are applied to breast cancer prediction, and combined with multi-omics data to test the performance of these models, both are better than the single model. Asada et al. [98] combined RNA, miRNA expression and clinical information as assembled dataset, tested the types of genes related to the prognosis of lung cancer through ML and DL, and successfully discovered new genes related to patient survival. Takahashi's team [99] used AE to process six multi-omics data sets, reduce dimensionality and unify features, then utilize ML to establish a lung cancer prognosis prediction model. The model shows advantages in multiple indicators. After searching for keywords in multiple omics, the current research on the combination of deep learning and multiple omics for cancer diagnosis prediction is still relatively scarce. The development of existing genomics technology is more conducive to the acquisition of cancer genomics data. In the future, we should make full use of the advantages of DL technology for multi-data processing, and focus on multi-omics integration research. For the dynamic and unstable development of cancer, the condition of cancerous cells can be detected from more angles, and at the same time, it can help to discover new factors for cancer monitoring or evaluation, achieving accurate cancer prediction.
DL model can better extract potential features when processing diseased cell data, and then enhance the data through AE or GAN, solve unlabeled and irregular data, optimize the noise data, which further benefit detect the disease of cancer cells. Khamparia et al. [100] uses CNN and variational autoencoder for the diagnosis and classification of cervical cancer. The main structure is CNN to extract high-dimensional features, and then through AE to reduce the dimensionality, while enhancing the data, finally handed over to the dense layer and Softmax layer for classification. After setting different filters and comparing with the existing method, the accuracy of 99.4% when the size is 3 × 3 is obtained and it is better than the comparison method. Iizuka et al. [101] uses InceptionV3 network combined with RNN to help classify and diagnose adenocarcinoma, adenoma, and non-tumor. Through the classification test in the WSI image dataset, the AUC is above 96.0%. Khan et al. [102] uses the ResNet101 model at the bottom layer, which can extract deeper features than the traditional CNN model, and at the same time, with the help of transfer learning, it reduces the data demand of the model. After applying the improved model to the WCE image database (including four different types of stomach abnormalities), through the optimization algorithm and the fusion method of maximum correlation, the accuracy performance increased from the initial 87.45% to 99.46%. By comparing with other neural networks, it has obvious advantages in accuracy, which is better than all previous methods.
Apart from predicting cancer lesions, DL can also be used for the detection of anti-cancer drugs. Baptista et al. [103] used the DL method to help predict the sensitivity of anti-cancer drugs, which greatly reduced the limitations of drug testing in clinical use. Mencattini et al. [104] studied cancer drug response at the cellular level, it can be evaluated in vitro, combined with the Alex-NET framework for drug response classification, using the basic circuit shown in Fig. 8, and the final classification effect is obvious. Kuenzi et al. [105] also study the reaction of drugs in the process of carcinogenesis from the cellular level. Realize the clinical research of anti-cancer drugs from a new perspective.
Advances in the medical field and the rise of a new generation of computers will promote the emergence of new methods of cancer treatment. Traditional cancer diagnosis methods are done manually by radiologists. A medical image has several million pixels, but the cancerous part may only occupy more than a dozen pixels. Manual diagnosis is time-consuming, and there are certain misjudgments. Moreover, the requirements for radiologists are higher. The use of deep learning can identify deeper cancer cell lesions, while considering horizontal and vertical characteristics to provide a more comprehensive diagnosis basis. To a greater extent, the pressure on medical diagnosis can be reduced, and while improving the defects of artificial care, it can achieve earlier and more accurate cancer cell diagnosis [106].
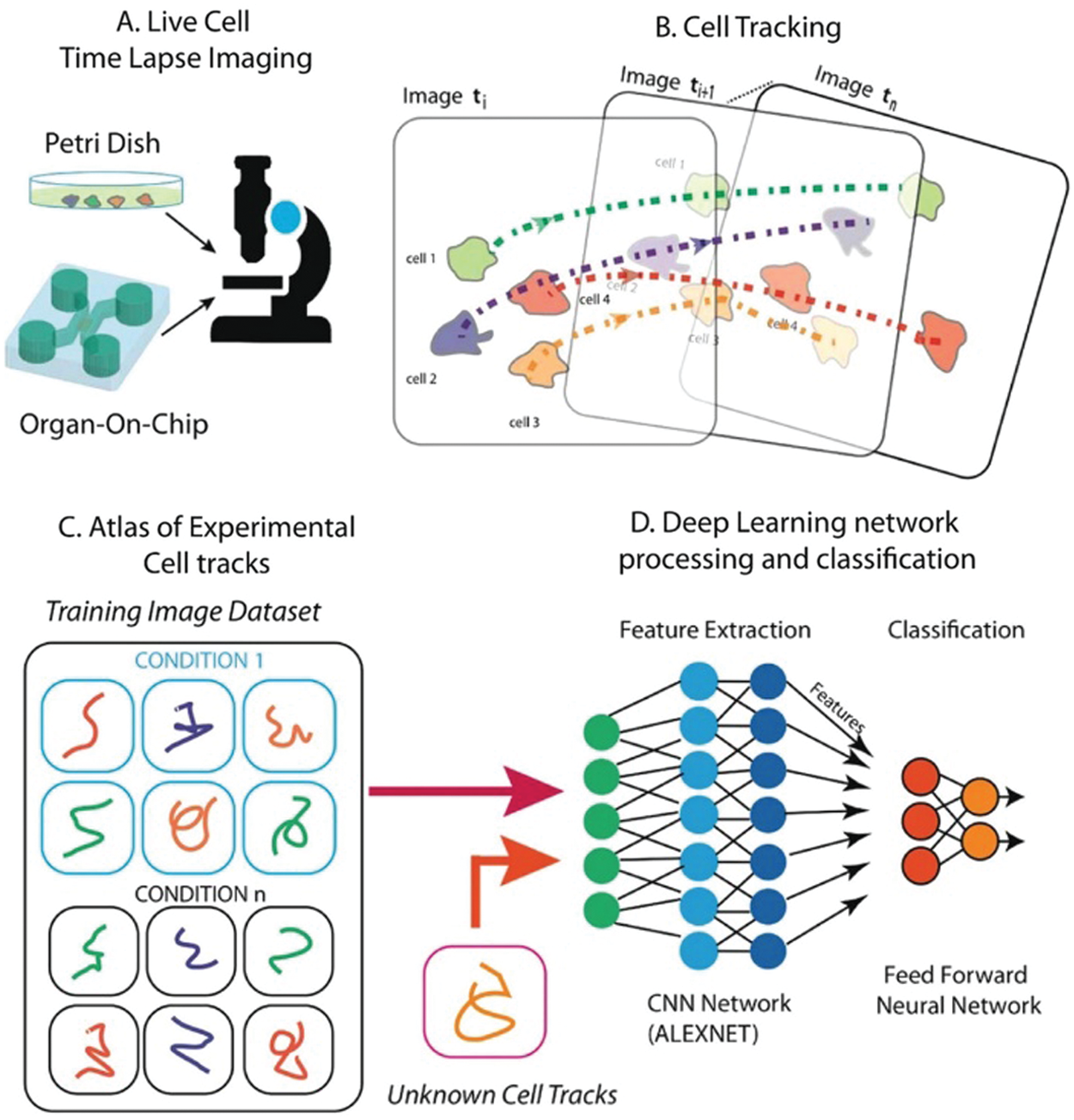
Figure 8: Schematic diagram of the method proposed by Mencattini et al. [104]
Diabetes mellitus is mainly manifested as an abnormal increase in blood glucose levels, exceptional insulin secretion or unusual functional absorption, which is a metabolic disease. If it cannot be diagnosed and treated in time, diabetes can affect multiple organs, and complications can seriously threaten human health. Diabetes is divided into Type 1 and Type 2. Both types show a certain genetic correlation, so it is essential for genetic research to prevent the development of diabetes from the root cause. Deep learning's screening of diabetes-related features can not only help diagnose diabetes, but also achieve accurate disease classification.
In medical diagnosis, blood glucose levels are often used to test for diabetes, and ECG signals can also help in diabetes monitoring. Besides, the abnormality of Heart Rate Variability (HRV) is used as a voucher to identify diabetic lesions [107]. Ayon et al. [108] used DNN for diabetes feature monitoring, combined with two cross-validation method tests, and compared with a variety of machine learning methods. The accuracy of 98.35% in five-fold cross-validation is obtained, which is higher than 97.47% of the optimal SVM. Padmapritha [109] uses LSTM to help test blood glucose (BG) levels. Li et al. [110] proposed a CRNN model. The bottom layer includes CNN and an improved LSTM structure, which can quickly identify blood glucose abnormalities and has high sensitivity in predicting BG levels. Swapna et al. [107] also used the method of combining CNN and LSTM to diagnose with the help of HRV data, which can achieve an accuracy of 95.7%. Pavithra's team [111] used Deep GRU and LSTM to test on the diabetes dataset and found that the accuracy of GRU is higher than that of LSTM. Perdomo et al. [112] creatively proposed a combination of obstructive sleep apnea and Type 2 diabetes to diagnose, combining DBNN and CNN to build a training network, and the fusion model is better than independent DBN and CNN with 98.80% accuracy.
Diabetes data monitoring in daily medical treatment relies on diabetes monitoring equipment. In China, the treatment rate and control rate of diabetes have not reached 50%. Whether it is a disease analysis platform or basic equipment, there are some gaps in the real-time monitoring of diabetes. The huge demand provides basic conditions for the practical application of the Internet of Things (IoT) and promotes the development of the global IoT. To a certain extent, it has also promoted the advancement of medical treatment, combined with the advanced processing methods of deep learning, to provide more intelligent measures for future disease diagnosis. Diabetes monitoring can be found from various angles, tiny retinal cracks [112], and facial features to identify the disease. In actual applications, a richer diagnosis model should be established and applied to more scenarios. AI help is the only way to diagnose diabetes in the future.
Over the above categories of diseases, deep learning technology is also widely used in other diseases, the following is a brief summary.
Hepatitis belongs to a type of acute and chronic inflammation. There are five classifications in medicine. Different types of hepatitis have different manifestations and alternate phenomena. Most people have no obvious symptoms at the initial stage, and more methods are needed to help predict. Failure to diagnose hepatitis in time may lead to liver cirrhosis and even worsen liver cancer. Timely discovery and vaccination can achieve a greater degree of prevention and treatment. Jyoti et al. [113] used DNN to predict hepatitis and tested different activation functions, through the result obtained an accuracy of 92.3% when using the swish function, which is higher than other methods. Yang's team [114] proposed a 7-layer AlexNet CNN structure, combined with transfer learning to apply the pre-trained model to the detection of liver lesions. While realizing the algorithm automation, the accuracy is no less than the previous ML method and traditional neural network. Xiao et al. [115] creatively studied a variety of hepatobiliary diseases from eye feature images, combined with the ResNet-101 network to help identify hepatitis, and proved the correlation between abnormal eye function and hepatobiliary diseases. Frid-Adar et al. [116] used GAN to enhance medical data, synthesized high-quality liver lesion images, combined with CNN to complete lesion classification, obtained 85.7% sensitivity and 92.4% specificity, which are better than the comparison method 78.6% and 88.4%. Lu et al. [117] used Raman spectroscopy to build an MSCIR model. The standard architecture was combined with CNN and RNN, and the data processing was strengthened from multiple levels. The accuracy rate was 96.15%, higher than the 91.53% of traditional NN. Wang et al. [118] also identified patients with type B pneumonia from Raman spectroscopy, used PCA to reduce dimensionality, and then used LSTM to process long-span data, finally reaching an accuracy of 97.32%. Both papers use PCA for dimensionality reduction. If you change to AE or Gan, you may get better results.
Generally speaking, it is infection with multiple pathogens such as bacteria and viruses, causing serious damage to multiple organs of the body. There are many ways of transmission and there is no specific trend. Infectious shows high fulminant, once spread, it is difficult to control. The source of infectious diseases must be cut off early and the characteristics of the pathogen must be explored. COVID-19, which still plagues the world, is a viral infectious disease. As a thorny global medical problem, many teams currently use DL to help predict and diagnose. Zhu et al. [119] used BiPathCNN combined with virus sequences to predict the possible source of the virus and proved that COVID-19 has the same infectivity as other coronaviruses. Fu et al. [120] used ResNet-50, and combined with transfer learning to identify viral pneumonia from a variety of lung diseases through CT images, with an accuracy rate of 98.8%.
AI contributes to the discovery and high-risk factors timely control. It also helps effectively improve the prevention and control of infectious diseases [121]. Using DNN and LSTM to predict infectious diseases, and testing with optimal control parameters, the average performance of the ARIMA model is improved by 24% and 19%, respectively. The DNN is still stable when the infectious disease spreads, and the average performance very well, LSTM is more accurate. In recent years, the development of big data has played a significant role in the prevention of infectious diseases. Regarding the variability and unpredictability of infectious diseases, big data has realized deeper mining to solve difficult problems. The existence of big data must have corresponding processing technology, and deep learning can effectively solve this demand [122].
Kidney diseases include renal functional damage and tissue inflammation. The typical one is chronic kidney disease (CKD). If early diagnosis and treatment are ignored, it may lead to kidney cancer. In the latest study, Alnazer et al. [123] evaluated the medical imaging methods used for CKD detection, including MRI, CT, PET, etc. These complex medical images can more comprehensively analyze kidney disease conditions and provide a data basis for DL's disease research. Wu et al. [124] created an automated architecture, a multifunctional network (MF-Net) mainly composed of CNN, for kidney abnormality detection, and obtained the best average classification accuracy of 94.67% compared with multiple fusion models rate. Navaneeth et al. [125] improved the traditional convolutional neural network and introduced dynamic processing in the pooling layer. And proposed to identify chronic kidney disease by the level of urea in saliva, then use SVM as a classifier to obtain 97.67% classification accuracy, which is better than the 96.12% of traditional CNN. Bhaskar et al. [126] also used saliva urea abnormalities to detect diseases, developed an ammonia sensor, and obtained data from it, and then used the CNN-SVM model for classification, which can achieve an accuracy of 98.04%. Khamparia et al. [127] use stacked AutoEncoder for feature enhancement, then use the Softmax function as the classifier, and even reached 100% accuracy when performing CKD classification. Ma's team [128] proposed a Heterogeneous Modified ANN model. This model combines multiple classifiers to achieve the prediction of chronic kidney disease. During the experiment, the data integration is carried out through the medical Internet of Things, which ensures the reliability of the data and reduces the calculation time. Chen et al. [129] used an Adaptive hybridized deep CNN structure to detect kidney disease, combined with the medical Internet of Things, and conveniently realized model optimization and prediction, which was significantly better than other NN models in AUC.
4 Existing Problems and Technical Obstacles
4.1 Insufficient Interpretability
Although the deep learning model has excellent performance in feature extraction and classification capabilities, its poor model interpretability has become a criticism that is hard to ignore. As two independent branches of science, deep learning and medicine will inevitably be heterogeneous in all aspects in research. Some existing disease prediction methods separate medical domain knowledge from deep learning methods. How to properly integrate these two kinds of subjects is very important, which requires attention to the interpretability of the model. The interpretability of the model indicates the extent to which humans can understand the reasons for the decision. In general, the model is required to answer the “why” question, that is, how the model predicts. Some traditional statistical methods are manually selected features based on medical knowledge, this approach is closely integrated with medical knowledge. The deep learning method is completely based on data, and rarely considered the previous domain knowledge or experience and some known risk factors.
Deep neural network is like a black box model. After the model is fully trained, new data is input to the model and the prediction results are output. The model only lets people get the prediction result by the corresponding input data, but does not show people how to predict, which inevitably leads to some lack of trust. In order to apply the deep learning method to the real disease prediction and healthcare system, realize the intelligence of disease prediction, it is necessary to solve the basic credibility problem and make the model convincing. The credibility of deep learning models needs to be based on certain model interpretability. Only when the problem of model interpretability is solved, deep learning can be more widely applied to the actual scenes in the medical field and achieve the purpose of combining with the real application. In conclusion, more attention should be paid to the study of model interpretability in future research. Whether it is the criteria, scope or role of interpretability, appropriate methods should be selected and combined with realistic requirements to make the model “humanized”. Only in this way, it can help elevate the believability while using the deep learning ideas. In this way not only provide rapid and accurate diagnosis for patients, but also provide explainable diagnosis basis.
4.2 Clinical Implementation Issues
Most of the current deep learning methods combined with disease prediction are still in the theoretical stage and have not been applied to the clinic. There are several reasons for this situation.
The poor generalization ability of the model has led to doubts about the practicality and accuracy of the model in actual use. For example, in some areas with poor medical equipment, high-quality medical images cannot be extracted, and it is difficult to meet the basic requirements of the model for input data. Reference [72] pointed out that although DNN has obvious advantages in various indicators, it will show instability when applying different test sets. The main reason is that DL has higher data requirements. For this situation, on the one hand, CNN or other deep learning models with denoising function can be designed [130]. On the other hand, image enhancement technology can also be applied, using appropriate filters, to enhance the overall or local features of the image, amplify key features, and improve the applicability of the image. At the same time, make full use of the existing excellent image fusion technology, and integrate various image data to enrich the diagnosis basis [131]. For medical data, the dataset is small, and there are still many problems of class imbalance. In the future, more attention should be paid to the research of data availability to improve the generalization ability of the model.
Whether the deep learning model is stable in practical applications is still unknown. To truly apply deep learning to clinical applications, a high degree of stability is one of the necessary conditions. In reality, medical data are spotty and vary widely. When the neural network model is applied to the actual medical situation, it may face the situation that the training sample does not match the real sample. When the model refined by training samples is processing real samples, there may be some abnormal features, insufficient classification accuracy or underfitting, and the model is not robust enough. If the stability of the model cannot be guaranteed, it will not only lead to a decrease in the performance and efficiency of the algorithm, inaccurate disease prediction, but also some clinical safety problems may occur, and even the life safety of medical patients may be threatened. Therefore, the transition to clinical application should be realized under the premise of ensuring the stability of the model.
When using deep learning methods to predict diseases, the medical records and personal data of disease patients are generally involved, so the protection of data and the privacy of patients should naturally be fully considered. How to protect the privacy of patients and prevent the leakage of personal information is particularly important. Lv et al. [132] proposed the use of differential privacy technology to process medical data, which can effectively ensure data security. Reference [60] used surveillance systems combined with blockchain technology enables reliable protection of data in telemedicine. The so-called technology-generated problems should be solved by technology, and the problems generated by the application of deep learning technology should also be supported by corresponding technologies. Although distributed technology has made some achievements in data protection in recent years, and the goal of storing patient data in the original medical institution or original storage device has been achieved [133]. However, distributed technology is still relatively independent, and the research on integration with deep learning technology is not enough. In future research, more attention should be paid to the protection of doctors and patients’ privacy [88]. There are many types of medical data, with large differences, and the quality of the data is uneven due to the influence of medical conditions. Therefore, while using existing technologies, we should develop application technologies that are more suitable for medical data and can ensure privacy and security. Let patients no longer worry about the exposure of personal information, so as to promote the realization of clinical applications more quickly.
In addition to the three points mentioned above, there are also some influencing factors. For example, in disease prediction, the input data of the model is manually labeled data. It is inevitable that there will be some personal biases, which may cause ethical problems. This requires the establishment of a realistic ethical framework to avoid the influence of personal emotions, and it is obviously not easy to establish a global consensus on ethics. In addition, the application threshold of deep learning technology is relatively high, most areas still do not have the basic conditions, and there is a shortage of talents in related fields. Therefore, there is still a long way to go if really want to apply deep learning technology to actual clinical practice.
4.3 The Challenge of Model Training
The deep learning models mentioned above still have some shortcomings, such as AE or GAN. The problems include the credibility of synthetic data. Because of the lack of some necessary associations and mappings, it is not enough to be convincing. Instability and the uncertainty of evaluation indicators are both problems that need to be solved urgently. Only when these problems and deficiencies are resolved can it become a more trustworthy technology in the field of healthcare. In addition, under the characteristics of medical data, there are more over-fitting and under-fitting phenomena during model training [134]. The lack or redundancy of the training data set will cause the above problems. It can be solved by appropriately increasing or decreasing the size of the dataset, starting from the model itself, and changing the model structure [135].
What's more, data quality is still the biggest challenge in model training. The excellent performance of DL models in disease prediction relies on high-quality medical data. Although it is easy to obtain medical data under existing conditions, the data quality is still low. Many medical data need to have a wealth of experience in medical experts to give a proprietary label. The processing of image features is particularly important [136], and medical data sets involve many privacy issues, and they are stored in independent institutions. The data sets are closed, not open, and a large number of datasets cannot be used in actual research. The inability to get adequate training [137] also hinders the development of many innovative models.
5.1 Deep Learning Drives Precision Medicine (PM)
Precision medicine is extremely significant in future research. No matter in patient diagnosis, treatment, or prognosis management, precision medicine has achieved many breakthroughs under the support of the development of big data. Compared with traditional medical treatment, it can provide patients with more effective and timely medical services. Since the introduction of precision medicine in 2015 to the present, it has always been the key to the future global healthcare and is a golden target for development plans in many countries around the world [138]. Its unique diagnostic paradigm and basic concepts have opened up a new idea for human health and medical care [139–141]. Afzal et al. [138] discussed the development process and future direction of PM from the perspective of informatics, and proposed the PM framework shown in Fig. 9, which fully displays the source of data, related processing, practical applications and services object. It can be seen that precision medicine will be combined with multi-domain applications in the next era development. A set of relatively complete operation methods can realize the systematic operation of medical diagnosis and promote the development of medical treatment to more intelligent.
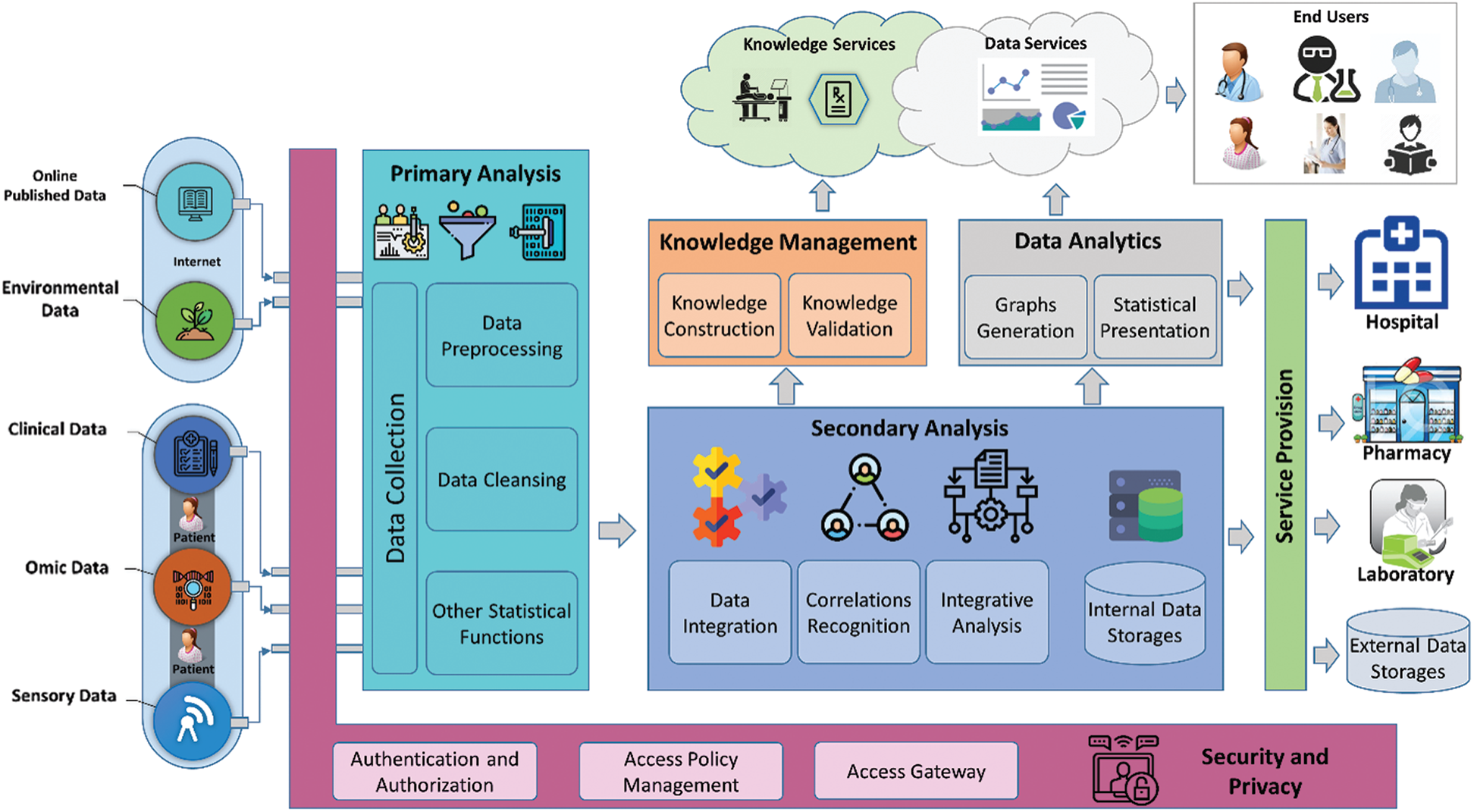
Figure 9: A holistic integrated precision medicine framework [138]
Just as tailored, precision medicine pays more attention to the study of individual differences [142], and refines the impact of each individual's personal factors on disease [143]. Evaluation of personal health from genomics, living environment and other levels, coupled with clinical data analysis, has a higher resolution. For example, Panayides et al. [144] proposed that starting from the method of radio genomics, combined with precision medicine, abnormal diseases can be found faster when dealing with complex diseases. PM can be summarized in three correct terms, namely, the right time, the right patient, and the right medicine to achieve timely diagnosis of the patient. And precision medicine also has a relatively good performance in the prevention of malignant diseases, especially when dealing with cancer [145,146], tumor [147] and other related diseases. Its unique diagnosis and treatment methods have promoted disease prediction. And treatment is developing towards the era of precision medicine [148].
At present, precision medicine in Western countries has made many achievements, while the development of the Asia-Pacific region is still in its infancy. On the one hand, the urgent problem to be solved at the moment is to ensure the diversity and high quality of gene collection. On the other hand, the crux needs a firm promise that the extraction is consistent with the genetic characteristics of the population in the Asia-Pacific region. Reference [138] shows the close relationship between precision medicine and artificial intelligence. The rapid development of AI and the emergence of related platforms have provided many conveniences for precision medicine [149]. With the advent of the era of big data, combined with deep learning as the engine of the era, it fundamentally promotes the development of precision medicine [150].
Regarding the remaining constraints between precision medicine and big data [151], it is necessary to fundamentally handle the relationship between PM, big data and deep learning. Deep learning not only provides advanced technical means, but also arouses people's understanding of the future medical multi-angle thinking. As a part of precision medicine, accurate prediction of diseases reflects the advantages of future medical care and plays a key role in promoting the development of modern medical technology. The current precision medicine research is still in the exploratory development stage [152–154], the research status of different diseases is quite different, and the application of deep learning technology is relatively narrow. In the future, in the process of exploring the medical field, AI researchers should focus more on precision medicine. Combining the research on pathological changes caused by gene mutations in the medical field, construct a deep learning model that is more in line with the requirements of precision medicine. Applying deeper and more changeable models to it. While promoting the development of precision medicine, it also drives the multi-faceted extension of deep learning, making medical care conform to the needs of the society in the future.
5.2 Combine with the Internet of Things
The maturity of the Internet of Things technology has strengthened the development of intelligent society. Especially the arrival of the current 5G era has brought qualitative changes to the application of IoT technology. Broader coverage of the Internet of Things, low-latency and high-throughput performance accelerate the spread of data, and the high demand for health and medical modernization, all of which promote the integration of the Internet of Things, medical and DL fields. E-healthcare is an important application combining medical care and the Internet of Things in recent years. It can not only provide “interaction” of medical information, but also provide a system and platform for telemedicine services. Reference [155] shows that in the current raging COVID-19, the healthcare system has played a great role, providing important help for better tracking patients and monitoring disease symptoms. The combination of the Internet of Things and AI has further improved the previous single disease prediction method. The cloud-based electronic physical localization system proposed by Hossain [156] can effectively process speech and EEG signals. Bisio et al. [157] proposed an eHealth system based on the Internet of Things (IoT) to help remotely monitor the rehabilitation of stroke patients. Compared with traditional medical systems, it facilitates remote monitoring. Once promoted, the DL method can be used to efficiently process data, which can provide a wider range of remote end-to-end services for patient rehabilitation.
The Internet of Medical Things (IoMT) has developed rapidly in the past few years [158]. The infrastructure such as common life wearable devices (bracelet, belt, diagnosis and treatment glasses, intelligent underwear, etc.) [159], intelligent home devices [160], has become an important source of medical data acquisition. Reference [161] created a medical system that can detect and perceive emotions, and obtain patient status by monitoring emotions. The daily monitoring of personal activities can be realized, the operation is simple, and the system provides dynamic data. On this basis, through the processing of dynamic data with deep learning, real-time monitoring of patient health can be achieved [162]. Reference [163] proposes a new type of intelligent medical equipment, and integrates deep learning methods in disease monitoring, realizes physical condition detection by acquiring all-round information of patients, which greatly improves the performance of the detection system. In addition, combined with the use of sensors, it is possible to track key physical indicators during the postoperative recovery phase of the patient. Another combined with the DL method, the longitudinal timeline data stream is processed to remotely monitor the patient's recovery status. Realize the symbiotic development with Deep Learning through these medical IoT devices. As shown in Fig. 10, personal medical data is first obtained at the terminal, and then transmitted through various networks. The data and personal information are handed over to the IoMT platform. IoMT uses DL to process massive amounts of data information, thereby realizing all the data on the IoMT platform. Required medical operations, including interactions with medical service systems and hospitals. This describes the complete framework of the combination of the medical Internet of Things and DL.
In prospective work, more intelligent processing of data should be realized through the Internet of Things to provide higher-quality basic data for deep learning models. At the same time, it is necessary to develop more IoT medical system platforms, produce more smart hardware, and build a truly smart city. Smart medical care is an important link in a smart city, and a highly interactive medical platform is even more critical. The medical monitoring framework proposed by [164] can simultaneously receive voice signals and EGG signals, and achieves 93% accuracy in voice case monitoring. It greatly improves the shortcomings of difficulty in interaction. On the basis of ensuring the security of the Internet of Things platform, it further broadens the application coverage and more comprehensively serves the user population [165]. Intelligence is the key word for future medical health and urban development. To truly realize comprehensive medical intelligence, better integration of medical Internet of Things and deep learning technology is essential. A new technological model is used to help disease diagnosis and reduce pressure on medical resources.
5.3 Specialized Model Improvement
A very important feature of the deep learning model is that the architecture is very changeable. Different models have many features that can be changed. They can be changed at will according to the needs. Regardless of the data or the scene, the general actual needs can be based on the general model to make a slight change to help implement it. This feature is very compatible with the medical field, and can meet the needs of different diseases, individual differences in different patients, and complex medical conditions.
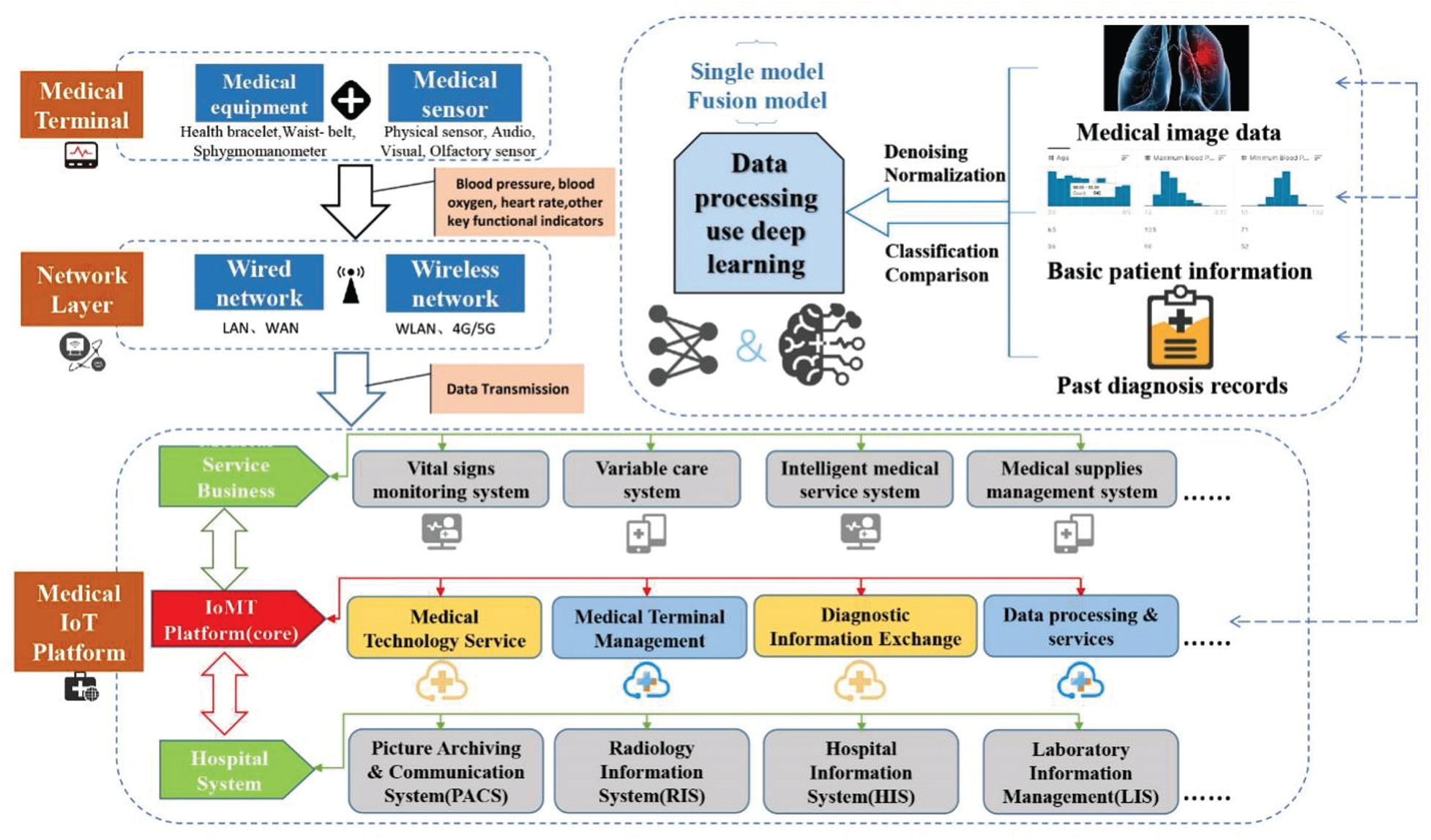
Figure 10: Integrated architecture diagram of medical IoT platform
Specifically, for example, when dealing with complex structures and high-dimensional features, DL models tend to have deeper levels, which leads to the possibility of gradient disappearance during the gradient propagation process. Based on the characteristics of medical data, a DL model that is more conform to the characteristics of medical data should be designed [166–168]. Comprehensive consideration of various influencing factors, including different training sets, optimization methods, training times, parameter effects, etc. Improve the utilization of important data features in medical data, reduce unnecessary parameters, avoid over-fitting, and fundamentally optimize the efficiency of the classifier. In addition, according to different types of data used in medical diagnosis, such as electronic medical records arranged in a time series, the advantages of transfer learning can be fully utilized here to design a framework that combines multiple models [169]. Combined with the existing results, as much as possible to reduce unnecessary parameter training process, maximize the expansion of the functions of existing models. While making good use of transfer learning to achieve specific model optimization, it can also be combined with multi-domain applications, such as Internet of Things related equipment [170], and a big data foundation to form a more complete integrated framework.
For different diseases, it will be affected by many factors when predicting. Moreover, the actual situation in medical diagnosis varies greatly, and there is no fixed objective law. In addition, sometimes, there is a certain correlation between different diseases, and the complications of one disease may cause the production of other diseases. This adds many limitations to practical applications. According to the characteristics of different diseases, the corresponding DL model should be designed according to the unique usage data and prediction methods, so as to facilitate the rapid expansion of the model according to different situations in practical applications. To ensure that model research develops towards diversification, while establishing standard treatment frameworks corresponding to different diseases.
The development of medical healthcare in the future has more modern prospects under the background of the era of big data and the coverage of the Internet of Things. In the face of the high dimension and instability of medical data, deep learning has become the main driving force for the future development with its unique feature processing method and variable model structure. This article introduces a series of popular deep learning prediction methods and frameworks for different disease, and introduces the optimal selection method based on commonly used medical diagnosis data. Pointed out some existing medical practical problems, as well as the limitations of deep learning itself. In the outlook, it is proposed to combine with precision medicine to conduct individual disease research on the genetic level; combine with the Internet of Things to develop intelligent medical platforms and equipment; at the same time, it proposes model improvement ideas for the particularity of medical data. In the future, we believe that disease diagnosis and deep learning will develop in a more diversified direction, and will certainly extend from today's auxiliary diagnosis to decision-making diagnosis. For different diseases, more and more DL models have emerged. The models are intertwined and learn from each other, resulting in a richer DL system network. This corresponds to the complex medical system, which will facilitate the development of medical diagnosis and clinical application as well as promote the development of the medical field.
Funding Statement: This work was supported in part by the National Natural Science Foundation of China (Nos. 61902203, 61976242) and Key Research and Development Plan-Major Scientific and Technological Innovation Projects of Shandong Province (2019JZZY020101).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Cho, N., Shaw, J. E., Karuranga, S., Huang, Y., da Rocha Fernandes, J. D. et al. (2018). IDF diabetes atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Research and Clinical Practice, 138, 271–281. DOI 10.1016/j.diabres.2018.02.023. [Google Scholar] [CrossRef]
2. Steenman, M., Lande, G. (2017). Cardiac aging and heart disease in humans. Biophysical Reviews, 9(2), 131–137. DOI 10.1007/s12551-017-0255-9. [Google Scholar] [CrossRef]
3. Shickel, B., Tighe, P. J., Bihorac, A., Rashidi, P. (2017). Deep EHR: A survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE Journal of Biomedical and Health Informatics, 22(5), 1589–1604. DOI 10.1109/JBHI.2017.2767063. [Google Scholar] [CrossRef]
4. Sun, W., Cai, Z., Li, Y., Liu, F., Fang, S. et al. (2018). Data processing and text mining technologies on electronic medical records: A review. Journal of Healthcare Engineering, 2018, 9. DOI 10.1155/2018/4302425. [Google Scholar] [CrossRef]
5. Cao, C., Liu, F., Tan, H., Song, D., Shu, W. et al. (2018). Deep learning and its applications in biomedicine. Genomics, Proteomics & Bioinformatics, 16(1), 17–32. DOI 10.1016/j.gpb.2017.07.003. [Google Scholar] [CrossRef]
6. Shrestha, A., Mahmood, A. (2019). Review of deep learning algorithms and architectures. IEEE Access, 7, 53040–53065. DOI 10.1109/ACCESS.2019.2912200. [Google Scholar] [CrossRef]
7. Dumoulin, V., Visin, F. (2016). A guide to convolution arithmetic for deep learning. arXiv preprint arXiv:1603.07285v2. [Google Scholar]
8. Kaur, T., Gandhi, T. K. (2019). Automated brain image classification based on VGG-16 and transfer learning. 2019 International Conference on Information Technology, pp. 94–98. Bhubaneswar, India, IEEE. [Google Scholar]
9. Huang, G., Liu, Z., van der Maaten, L., Weinberger, K. Q. (2017). Densely connected convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708. Honolulu, HI, USA. [Google Scholar]
10. Kassani, S. H., Kassani, P. H., Khazaeinezhad, R., Wesolowski, M. J., Schneider, K. A. et al. (2019). Diabetic retinopathy classification using a modified xception architecture. 2019 IEEE International Symposium on Signal Processing and Information Technology (ISSPITpp. 1–6. Ajman, United Arab Emirates, IEEE. [Google Scholar]
11. Tang, S., Yuan, S., Zhu, Y. (2020). Convolutional neural network in intelligent fault diagnosis toward rotatory machinery. IEEE Access, 8, 86510–86519. DOI 10.1109/ACCESS.2020.2992692. [Google Scholar] [CrossRef]
12. Khare, S. K., Bajaj, V. (2020). Time-frequency representation and convolutional neural network-based emotion recognition. IEEE Transactions on Neural Networks and Learning Systems, 1–9. DOI 10.1109/TNNLS.2020.3008938. [Google Scholar] [CrossRef]
13. Siddiqi, R. (2019). Automated pneumonia diagnosis using a customized sequential convolutional neural network. Proceedings of the 2019 3rd International Conference on Deep Learning Technologies (ICDLT 2019pp. 64–70. New York, NY, USA. [Google Scholar]
14. Zhang, N., Cai, Y. X., Wang, Y. Y., Tian, Y. T., Wang, X. L. et al. (2020). Skin cancer diagnosis based on optimized convolutional neural network. Artificial Intelligence in Medicine, 102, 101756. DOI 10.1016/j.artmed.2019.101756. [Google Scholar] [CrossRef]
15. Hossain, M. S., Amin, S. U., Alsulaiman, M., Muhammad, G. (2019). Applying deep learning for epilepsy seizure detection and brain mapping visualization. ACM Transactions on Multimedia Computing, Communications, and Applications (TOMM), 15(1s), 1–17. DOI 10.1145/3241056. [Google Scholar] [CrossRef]
16. Zhang, Y., Liu, Y., Sun, P., Yan, H., Zhao, X., Zhang, L. (2020). IFCNN: A general image fusion framework based on convolutional neural network. Information Fusion, 54, 99–118. DOI 10.1016/j.inffus.2019.07.011. [Google Scholar] [CrossRef]
17. Sharma, H., Jain, J. S., Bansal, P., Gupta, S. (2020). Feature extraction and classification of chest X-ray images using cnn to detect pneumonia. 2020 10th International Conference on Cloud Computing, Data Science & Engineering (Confluencepp. 227–231. Noida, India, IEEE. [Google Scholar]
18. Sherstinsky, A. (2020). Fundamentals of recurrent neural network (RNN) and long short-term memory (LSTM) network. Physica D: Nonlinear Phenomena, 404, 132306. DOI 10.1016/j.physd.2019.132306. [Google Scholar] [CrossRef]
19. Long, D., Zhang, R., Mao, Y. (2019). Recurrent neural networks with finite memory length. IEEE Access, 7, 12511–12520. DOI 10.1109/ACCESS.2018.2890297. [Google Scholar] [CrossRef]
20. Lynn, H. M., Pan, S. B., Kim, P. (2019). A deep bidirectional GRU network model for biometric electrocardiogram classification based on recurrent neural networks. IEEE Access, 7, 145395–145405. DOI 10.1109/ACCESS.2019.2939947. [Google Scholar] [CrossRef]
21. Cui, Y., Zhao, S., Wang, H., Xie, L., Chen, Y. et al. (2018). Identifying brain networks at multiple time scales via deep recurrent neural network. IEEE Journal of Biomedical and Health Informatics, 23(6), 2515–2525. DOI 10.1109/JBHI.2018.2882885. [Google Scholar] [CrossRef]
22. Choi, E., Bahadori, M. T., Schuetz, A., Stewart, W. F., Sun, J. (2016). Doctor ai: Predicting clinical events via recurrent neural networks. Machine Learning for Healthcare Conference, pp. 301–318. Los Angeles, CA, PMLR 56. [Google Scholar]
23. Banerjee, I., Ling, Y., Chen, M. C., Hasan, S. A., Langlotz, C. P. et al. (2019). Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification. Artificial Intelligence in Medicine, 97, 79–88. DOI 10.1016/j.artmed.2018.11.004. [Google Scholar] [CrossRef]
24. Zhang, P., Wang, X., Zhang, W., Chen, J. (2018). Learning spatial–spectral–temporal EEG features with recurrent 3D convolutional neural networks for cross-task mental workload assessment. IEEE Transactions on Neural Systems and Rehabilitation Engineering, 27(1), 31–42. DOI 10.1109/TNSRE.2018.2884641. [Google Scholar] [CrossRef]
25. Qin, C., Schlemper, J., Caballero, J., Price, A. N., Hajnal, J. V. et al. (2018). Convolutional recurrent neural networks for dynamic MR image reconstruction. IEEE Transactions on Medical Imaging, 38(1), 280–290. DOI 10.1109/TMI.2018.2863670. [Google Scholar] [CrossRef]
26. Liang, G., Hong, H., Xie, W., Zheng, L. (2018). Combining convolutional neural network with recursive neural network for blood cell image classification. IEEE Access, 6, 36188–36197. DOI 10.1109/ACCESS.2018.2846685. [Google Scholar] [CrossRef]
27. Kim, J. C., Chung, K. (2020). Multi-modal stacked denoising autoencoder for handling missing data in healthcare big data. IEEE Access, 8, 104933–104943. DOI 10.1109/Access.6287639. [Google Scholar] [CrossRef]
28. Chen, M., Shi, X., Zhang, Y., Wu, D., Guizani, M. (2017). Deep features learning for medical image analysis with convolutional autoencoder neural network. IEEE Transactions on Big Data, 1, 1. DOI 10.1109/TBDATA.2017.2717439. [Google Scholar] [CrossRef]
29. Azarang, A., Manoochehri, H. E., Kehtarnavaz, N. (2019). Convolutional autoencoder-based multispectral image fusion. IEEE Access, 7, 35673–35683. DOI 10.1109/ACCESS.2019.2905511. [Google Scholar] [CrossRef]
30. Zhang, J., Lin, F., Xiong, P., Du, H., Zhang, H. et al. (2019). Automated detection and localization of myocardial infarction with staked sparse autoencoder and treebagger. IEEE Access, 7, 70634–70642. DOI 10.1109/ACCESS.2019.2919068. [Google Scholar] [CrossRef]
31. Wei, R., Mahmood, A. (2021). Recent advances in variational autoencoders with representation learning for biomedical informatics: A survey. IEEE Access, 9, 4939–4956. DOI 10.1109/ACCESS.2020.3048309. [Google Scholar] [CrossRef]
32. Yeh, R. A., Chen, C., Yian Lim, T., Schwing, A. G., Hasegawa-Johnson, M. et al. (2017). Semantic image inpainting with deep generative models. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 5485–5493. Honolulu, HI, USA. [Google Scholar]
33. Zhou, Z., Wang, Y., Guo, Y., Qi, Y., Yu, J. (2019). Image quality improvement of hand-held ultrasound devices with a two-stage generative adversarial network. IEEE Transactions on Biomedical Engineering, 67(1), 298–311. DOI 10.1109/TBME.2019.2912986. [Google Scholar] [CrossRef]
34. Bing, X., Zhang, W., Zheng, L., Zhang, Y. (2019). Medical image super resolution using improved generative adversarial networks. IEEE Access, 7, 145030–145038. DOI 10.1109/ACCESS.2019.2944862. [Google Scholar] [CrossRef]
35. Huang, J., Le, Z., Ma, Y., Fan, F., Zhang, H. et al. (2020). MGMDcGAN: Medical image fusion using multi-generator multi-discriminator conditional generative adversarial network. IEEE Access, 8, 55145–55157. DOI 10.1109/ACCESS.2020.2982016. [Google Scholar] [CrossRef]
36. Li, Y., Zhao, J., Lv, Z., Li, J. (2021). Medical image fusion method by deep learning. International Journal of Cognitive Computing in Engineering, 2, 21–29. DOI 10.1016/j.ijcce.2020.12.004. [Google Scholar] [CrossRef]
37. Yi, X., Walia, E., Babyn, P. (2019). Generative adversarial network in medical imaging: A review. Medical Image Analysis, 58, 101552. DOI 10.1016/j.media.2019.101552. [Google Scholar] [CrossRef]
38. Yang, Z., Chen, Y., Le, Z., Fan, F., Pan, E. (2019). Multi-source medical image fusion based on wasserstein generative adversarial networks. IEEE Access, 7, 175947–175958. DOI 10.1109/ACCESS.2019.2955382. [Google Scholar] [CrossRef]
39. Hong, H., Li, X., Wang, M. (2019). Gane: A generative adversarial network embedding. IEEE Transactions on Neural Networks and Learning Systems, 31(7), 2325–2335. DOI 10.1109/TNNLS.2019.2921841. [Google Scholar] [CrossRef]
40. Wang, C., Xu, C., Yao, X., Tao, D. (2019). Evolutionary generative adversarial networks. IEEE Transactions on Evolutionary Computation, 23(6), 921–934. DOI 10.1109/TEVC.2019.2895748. [Google Scholar] [CrossRef]
41. Kaur, M., Singh, D. (2020). Fusion of medical images using deep belief networks. Cluster Computing, 23(2), 1439–1453. DOI 10.1007/s10586-019-02999-x. [Google Scholar] [CrossRef]
42. Al-Fatlawi, A. H., Jabardi, M. H., Ling, S. H. (2016). Efficient diagnosis system for Parkinson's disease using deep belief network. 2016 IEEE Congress on Evolutionary Computation, pp. 1324–1330. IEEE. [Google Scholar]
43. Dua, M., Makhija, D., Manasa, P. Y. L., Mishra, P. (2020). A CNN–RNN–LSTM based amalgamation for Alzheimer's disease detection. Journal of Medical and Biological Engineering, 40(5), 688–706. DOI 10.1007/s40846-020-00556-1. [Google Scholar] [CrossRef]
44. Khan, S., Islam, N., Jan, Z., Din, I. U., Rodrigues, J. J. C. (2019). A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognition Letters, 125, 1–6. DOI 10.1016/j.patrec.2019.03.022. [Google Scholar] [CrossRef]
45. Talo, M., Baloglu, U. B., Yıldırım, Ö., Acharya, U. R. (2019). Application of deep transfer learning for automated brain abnormality classification using MR images. Cognitive Systems Research, 54, 176–188. DOI 10.1016/j.cogsys.2018.12.007. [Google Scholar] [CrossRef]
46. Hu, Z., Tang, J., Wang, Z., Zhang, K., Zhang, L. et al. (2018). Deep learning for image-based cancer detection and diagnosis−A survey. Pattern Recognition, 83, 134–149. DOI 10.1016/j.patcog.2018.05.014. [Google Scholar] [CrossRef]
47. Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B. et al. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine, 382, 727–733. DOI 10.1056/NEJMoa2001017. [Google Scholar] [CrossRef]
48. Bansal, M. (2020). Cardiovascular disease and COVID-19. Diabetes & Metabolic Syndrome: Clinical Research & Reviews, 14(3), 247–250. DOI 10.1016/j.dsx.2020.03.013. [Google Scholar] [CrossRef]
49. Nishiga, M., Wang, D. W., Han, Y., Lewis, D. B., Wu, J. C. (2020). COVID-19 and cardiovascular disease: From basic mechanisms to clinical perspectives. Nature Review Cardiology, 17, 543–558. DOI 10.1038/s41569-020-0413-9. [Google Scholar] [CrossRef]
50. Fan, D., Yang, J., Zhang, J., Lv, Z., Huang, H. et al. (2018). Effectively measuring respiratory flow with portable pressure data using back propagation neural network. IEEE Journal of Translational Engineering in Health and Medicine, 6, 1–12. DOI 10.1109/JTEHM.2017.2688458. [Google Scholar] [CrossRef]
51. Voroshnina, N., Shabley, A., Vaulin, S. (2020). Neural network approach for prediction of pneumonia. 2020 International Multi-Conference on Industrial Engineering and Modern Technologies (FarEastConpp. 1–5. Vladivostok, Russia, IEEE. [Google Scholar]
52. Rajpurkar, P., Irvin, J., Zhu, K., Yang, B., Mehta, H. et al. (2017). Chexnet: Radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv preprint. arXiv: 1711.05225. [Google Scholar]
53. Qjidaa, M., Ben-Fares, A., Mechbal, Y., Amakdouf, H., Maaroufi, M. et al. (2020). Development of a clinical decision support system for the early detection of COVID-19 using deep learning based on chest radiographic images. 2020 International Conference on Intelligent Systems and Computer Vision (ISCVpp. 1–6. Fez, Morocco, IEEE. [Google Scholar]
54. Phankokkruad, M. (2020). COVID-19 pneumonia detection in chest X-ray images using transfer learning of convolutional neural networks. Proceedings of the 3rd International Conference on Data Science and Information Technology, pp. 147–152. Xiamen, China. [Google Scholar]
55. Raghu, M., Zhang, C., Kleinberg, J., Bengio, S. (2019). Transfusion: Understanding transfer learning for medical imaging. arXiv preprint. arXiv: 1902.07208. [Google Scholar]
56. Liang, G., Zheng, L. (2020). A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Computer Methods and Programs in Biomedicine, 187, 104964. DOI 10.1016/j.cmpb.2019.06.023. [Google Scholar] [CrossRef]
57. Chouhan, V., Singh, S. K., Khamparia, A., Gupta, D., Tiwari, P. et al. (2020). A novel transfer learning based approach for pneumonia detection in chest X-ray images. Applied Sciences, 10(2), 559. DOI 10.3390/app10020559. [Google Scholar] [CrossRef]
58. Saul, C. J., Urey, D. Y., Taktakoglu, C. D. (2019). Early diagnosis of pneumonia with deep learning. arXiv preprint. arXiv: 1904.00937. [Google Scholar]
59. Wu, X., Hui, H., Niu, M., Li, L., Wang, L. et al. (2020). Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: A multicentre study. European Journal of Radiology, 128, 109041. DOI 10.1016/j.ejrad.2020.109041. [Google Scholar] [CrossRef]
60. Hossain, M. S., Muhammad, G., Guizani, N. (2020). Explainable AI and mass surveillance system-based healthcare framework to combat COVID-19 like pandemics. IEEE Network, 34(4), 126–132. DOI 10.1109/MNET.011.2000458. [Google Scholar] [CrossRef]
61. Abdulsalam, Y., Hossain, M. S. (2020). COVID-19 networking demand: An auction-based mechanism for automated selection of edge computing services. IEEE Transactions on Network Science and Engineering, 1–1. DOI 10.1109/TNSE.2020.3026637. [Google Scholar] [CrossRef]
62. Chowdhury, M. E., Rahman, T., Khandakar, A., Mazhar, R., Kadir, M. A. et al. (2020). Can AI help in screening viral and COVID-19 pneumonia? IEEE Access, 8, 132665–132676. DOI 10.1109/ACCESS.2020.3010287. [Google Scholar] [CrossRef]
63. Timmis, A., Townsend, N., Gale, C. P., Torbica, A., Lettino, M. et al. (2020). European society of cardiology: Cardiovascular disease statistics 2019. European Heart Journal, 41(1), 12–85. DOI 10.1093/eurheartj/ehz859. [Google Scholar] [CrossRef]
64. Shi, H., Wang, H., Qin, C., Zhao, L., Liu, C. (2020). An incremental learning system for atrial fibrillation detection based on transfer learning and active learning. Computer Methods and Programs in Biomedicine, 187, 105219. DOI 10.1016/j.cmpb.2019.105219. [Google Scholar] [CrossRef]
65. Hannun, A. Y., Rajpurkar, P., Haghpanahi, M., Tison, G. H., Bourn, C. et al. (2019). Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nature Medicine, 25(1), 65–69. DOI 10.1038/s41591-018-0268-3. [Google Scholar] [CrossRef]
66. Ayon, S. I., Islam, M. M., Hossain, M. R. (2020). Coronary artery heart disease prediction: A comparative study of computational intelligence techniques. IETE Journal of Research, 66, 1–20. DOI 10.1080/03772063.2020.1713916. [Google Scholar] [CrossRef]
67. Dutta, A., Batabyal, T., Basu, M., Acton, S. T. (2020). An efficient convolutional neural network for coronary heart disease prediction. Expert Systems with Applications, 159, 113408. DOI 10.1016/j.eswa.2020.113408. [Google Scholar] [CrossRef]
68. Moody, G. B., Mark, R. G. (2001). The impact of the MIT-bIH arrhythmia database. IEEE Engineering in Medicine and Biology Magazine, 20(3), 45–50. DOI 10.1109/51.932724. [Google Scholar] [CrossRef]
69. Shi, H., Wang, H., Huang, Y., Zhao, L., Qin, C. et al. (2019). A hierarchical method based on weighted extreme gradient boosting in ECG heartbeat classification. Computer Methods and Programs in Biomedicine, 171, 1–10. DOI 10.1016/j.cmpb.2019.02.005. [Google Scholar] [CrossRef]
70. Wang, H., Shi, H., Lin, K., Qin, C., Zhao, L. et al. (2020). A high-precision arrhythmia classification method based on dual fully connected neural network. Biomedical Signal Processing and Control, 58, 101874. DOI 10.1016/j.bspc.2020.101874. [Google Scholar] [CrossRef]
71. Jin, Y., Qin, C., Liu, J., Lin, K., Shi, H. et al. (2020). A novel domain adaptive residual network for automatic atrial fibrillation detection. Knowledge-Based Systems, 203, 106122. DOI 10.1016/j.knosys.2020.106122. [Google Scholar] [CrossRef]
72. Anandajayam, P., Krishnakoumar, C., Vikneshvaran, S., Suryanaraynan, B. (2019). Coronary heart disease predictive decision scheme using big data and RNN. 2019 IEEE International Conference on System, Computation, Automation and Networking (ICSCANpp. 1–6. Pondicherry, India, IEEE. [Google Scholar]
73. Kuang, J. W., Yang, H. Z., Liu, J. J., Yan, Z. J. (2019). Dynamic prediction of cardiovascular disease using improved LSTM. International Journal of Crowd Science, 3(1), 14–25. DOI 10.1108/IJCS-01-2019-0002. [Google Scholar] [CrossRef]
74. Jin, Y., Qin, C., Liu, J., Li, Z., Shi, H. et al. (2021). A novel incremental and interactive method for actual heartbeat classification with limited additional labeled samples. IEEE Transactions on Instrumentation and Measurement, 70, 1–12. DOI 10.1109/TIM.2021.3069021. [Google Scholar] [CrossRef]
75. Shi, H., Qin, C., Xiao, D., Zhao, L., Liu, C. (2020). Automated heartbeat classification based on deep neural network with multiple input layers. Knowledge-Based Systems, 188, 105036. DOI 10.1016/j.knosys.2019.105036. [Google Scholar] [CrossRef]
76. Jin, Y., Qin, C., Huang, Y., Zhao, W., Liu, C. (2020). Multi-domain modeling of atrial fibrillation detection with twin attentional convolutional long short-term memory neural networks. Knowledge-Based Systems, 193, 105460. DOI 10.1016/j.knosys.2019.105460. [Google Scholar] [CrossRef]
77. Ali, A. A., Hassan, H. S., Anwar, E. M. (2020). Heart diseases diagnosis based on a novel convolution neural network and gate recurrent unit technique. 2020 12th International Conference on Electrical Engineering, pp. 145–150. Cairo, Egypt, IEEE. [Google Scholar]
78. Lu, P., Guo, S., Zhang, H., Li, Q., Wang, Y. et al. (2018). Research on improved depth belief network-based prediction of cardiovascular diseases. Journal of Healthcare Engineering, 2018, 1–9. DOI 10.1155/2018/8954878. [Google Scholar] [CrossRef]
79. Poplin, R., Varadarajan, A. V., Blumer, K., Liu, Y., McConnell, M. V. et al. (2018). Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nature Biomedical Engineering, 2(3), 158–164. DOI 10.1038/s41551-018-0195-0. [Google Scholar] [CrossRef]
80. Lyu, M. J., Yuan, S. M. (2020). Cloud-based smart dog music therapy and pneumonia detection system for reducing the difficulty of caring for patients with dementia. IEEE Access, 8, 20977–20990. DOI 10.1109/ACCESS.2020.2969482. [Google Scholar] [CrossRef]
81. Ding, Y., Sohn, J. H., Kawczynski, M. G., Trivedi, H., Harnish, R. et al. (2019). A deep learning model to predict a diagnosis of Alzheimer disease by using 18F-fDG PET of the brain. Radiology, 290(2), 456–464. DOI 10.1148/radiol.2018180958. [Google Scholar] [CrossRef]
82. Jo, T., Nho, K., Saykin, A. J. (2019). Deep learning in Alzheimer's disease: Diagnostic classification and prognostic prediction using neuroimaging data. Frontiers in Aging Neuroscience, 11, 220. DOI 10.3389/fnagi.2019.00220. [Google Scholar] [CrossRef]
83. Ji, S., Xu, W., Yang, M., Yu, K. (2012). 3D convolutional neural networks for human action recognition. IEEE Transactions on Pattern Analysis and Machine Intelligence, 35(1), 221–231. DOI 10.1109/TPAMI.2012.59. [Google Scholar] [CrossRef]
84. Cheng, D., Liu, M., Fu, J., Wang, Y. (2017). Classification of MR brain images by combination of multi-cNNs for AD diagnosis. Ninth International Conference on Digital Image Processing (ICDIP 2017vol. 10420, pp. 1042042. Hong Kong, China, International Society for Optics and Photonics. [Google Scholar]
85. Wang, H., Shen, Y., Wang, S., Xiao, T., Deng, L. et al. (2019). Ensemble of 3D densely connected convolutional network for diagnosis of mild cognitive impairment and Alzheimer's disease. Neurocomputing, 333, 145–156. DOI 10.1016/j.neucom.2018.12.018. [Google Scholar] [CrossRef]
86. Riederer, I., Bohn, K. P., Preibisch, C., Wiedemann, E., Zimmer, C. et al. (2018). Alzheimer disease and mild cognitive impairment: Integrated pulsed arterial spin-labeling MRI and 18F-fDG PET. Radiology, 288(1), 198–206. DOI 10.1148/radiol.2018170575. [Google Scholar] [CrossRef]
87. Cui, R., Liu, M. (2019). RNN-Based longitudinal analysis for diagnosis of Alzheimer's disease. Computerized Medical Imaging and Graphics, 73, 1–10. DOI 10.1016/j.compmedimag.2019.01.005. [Google Scholar] [CrossRef]
88. Ahmed, O. B., Benois-Pineau, J., Allard, M., Amar, C. B., Catheline, G. (2015). Classification of Alzheimer's disease subjects from MRI using hippocampal visual features. Multimedia Tools and Applications, 74(4), 1249–1266. DOI 10.1007/s11042-014-2123-y. [Google Scholar] [CrossRef]
89. Li, F., Liu, M., Alzheimer's Disease Neuroimaging Initiative. (2019). A hybrid convolutional and recurrent neural network for hippocampus analysis in Alzheimer's disease. Journal of Neuroscience Methods, 323, 108–118. DOI 10.1016/j.jneumeth.2019.05.006. [Google Scholar] [CrossRef]
90. Siegel, R. L., Miller, K. D., Jemal, A. (2020). Cancer statistics. CA: A Cancer Journal for Clinicians, 70(1), 7–30. DOI 10.3322/caac.21590. [Google Scholar] [CrossRef]
91. Tiwari, A., Srivastava, S., Pant, M. (2020). Brain tumor segmentation and classification from magnetic resonance images: Review of selected methods from 2014 to 2019. Pattern Recognition Letters, 131, 244–260. DOI 10.1016/j.patrec.2019.11.020. [Google Scholar] [CrossRef]
92. Tsherniak, A., Vazquez, F., Montgomery, P. G., Weir, B. A., Kryukov, G. et al. (2017). Defining a cancer dependency map. Cell, 170(3), 564–576. DOI 10.1016/j.cell.2017.06.010. [Google Scholar] [CrossRef]
93. Sultan, H. H., Salem, N. M., Al-Atabany, W. (2019). Multi-classification of brain tumor images using deep neural network. IEEE Access, 7, 69215–69225. DOI 10.1109/ACCESS.2019.2919122. [Google Scholar] [CrossRef]
94. de Anda-Jáuregui, G., Hernández-Lemus, E. (2020). Computational oncology in the multi-omics era: State of the art. Frontiers in Oncology, 10, 423. DOI 10.3389/fonc.2020.00423. [Google Scholar] [CrossRef]
95. Hasin, Y., Seldin, M., Lusis, A. (2017). Multi-omics approaches to disease. Genome Biol, 18, 83. DOI 10.1186/s13059-017-1215-1. [Google Scholar] [CrossRef]
96. Tan, K., Huang, W., Hu, J., Dong, S. (2020). A multi-omics supervised autoencoder for pan-cancer clinical outcome endpoints prediction. BMC Medical Informatics and Decision Making, 20(3), 1–9. DOI 10.1186/s12911-020-1114-3. [Google Scholar] [CrossRef]
97. Tong, L., Mitchel, J., Chatlin, K., Wang, M. D. (2020). Deep learning based feature-level integration of multi-omics data for breast cancer patients survival analysis. BMC Medical Informatics and Decision Making, 20, 225. DOI 10.1186/s12911-020-01225-8. [Google Scholar] [CrossRef]
98. Asada, K., Kobayashi, K., Joutard, S., Tubaki, M., Takahashi, S. et al. (2020). Uncovering prognosis-related genes and pathways by multi-omics analysis in lung cancer. Biomolecules, 10(4), 524. DOI 10.3390/biom10040524. [Google Scholar] [CrossRef]
99. Takahashi, S., Asada, K., Takasawa, K., Shimoyama, R., Sakai, A. et al. (2020). Predicting deep learning based multi-omics parallel integration survival subtypes in lung cancer using reverse phase protein array data. Biomolecules, 10(10), 1460. DOI 10.3390/biom10101460. [Google Scholar] [CrossRef]
100. Khamparia, A., Gupta, D., Rodrigues, J. J., de Albuquerque, V. H. C. (2020). DCAVN: Cervical cancer prediction and classification using deep convolutional and variational autoencoder network. Multimedia Tools and Applications, 80, 1–17. DOI 10.1007/s11042-020-09607-w.
101. Iizuka, O., Kanavati, F., Kato, K., Rambeau, M., Arihiro, K. et al. (2020). Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Scientific Reports, 10(1), 1–11. DOI 10.1038/s41598-020-58467-9.
102. Khan, M. A., Sarfraz, M. S., Alhaisoni, M., Albesher, A. A., Wang, S. et al. (2020). Stomachnet: Optimal deep learning features fusion for stomach abnormalities classification. IEEE Access, 8, 197969–197981. DOI 10.1109/ACCESS.2020.3034217.
103. Baptista, D., Ferreira, P. G., Rocha, M. (2021). Deep learning for drug response prediction in cancer. Briefings in Bioinformatics, 22(1), 360–379. DOI 10.1093/bib/bbz171.
104. Mencattini, A., Di Giuseppe, D., Comes, M. C., Casti, P., Corsi, F. et al. (2020). Discovering the hidden messages within cell trajectories using a deep learning approach for in vitro evaluation of cancer drug treatments. Scientific Reports, 10, 7653. DOI 10.1038/s41598-020-64246-3.
105. Kuenzi, B. M., Park, J., Fong, S. H., Sanchez, K. S., Lee, J. et al. (2020). Predicting drug response and synergy using a deep learning model of human cancer cells. Cancer Cell, 38(5), 672–684. DOI 10.1016/j.ccell.2020.09.014.
106. Coccia, M. (2020). Deep learning technology for improving cancer care in society: New directions in cancer imaging driven by artificial intelligence. Technology in Society, 60, 101198. DOI 10.1016/j.techsoc.2019.101198.
107. Swapna, G., Vinayakumar, R., Soman, K. P. (2018). Diabetes detection using deep learning algorithms. ICT Express, 4(4), 243–246. DOI 10.1016/j.icte.2018.10.005.
108. Ayon, S. I., Islam, M. (2019). Diabetes prediction: A deep learning approach. International Journal of Information Engineering & Electronic Business, 11(2), 21–27. DOI 10.5815/ijieeb.2019.02.03.
109. Padmapritha, T. (2019). Prediction of blood glucose level by using an LSTM based recurrent neural networks. 2019 IEEE International Conference on Clean Energy and Energy Efficient Electronics Circuit for Sustainable Development (INCCESpp. 1–4. Krishnankoil, India.
110. Li, K., Daniels, J., Liu, C., Herrero, P., Georgiou, P. (2019). Convolutional recurrent neural networks for glucose prediction. IEEE Journal of Biomedical and Health Informatics, 24(2), 603–613. DOI 10.1109/JBHI.2019.2908488.
111. Pavithra, M., Saruladha, K., Sathyabama, K. (2019). GRU based deep learning model for prognosis prediction of disease progression. 2019 3rd International Conference on Computing Methodologies and Communication (ICCMCpp. 840–844. Erode, India, IEEE.
112. Perdomo, O., Rios, H., Rodríguez, F. J., Otálora, S., Meriaudeau, F. et al. (2019). Classification of diabetes-related retinal diseases using a deep learning approach in optical coherence tomography. Computer Methods and Programs in Biomedicine, 178, 181–189. DOI 10.1016/j.cmpb.2019.06.016.
113. Jyoti, O., Islam, N., Hasnain, F. M. S. (2020). Prediction of hepatitis disease using effective deep neural network. 2020 IEEE International Conference for Innovation in Technology (INOCONpp. 1–5. Bangluru, India, IEEE.
114. Yu, Y., Wang, J., Ng, C. W., Ma, Y., Mo, S. et al. (2018). Deep learning enables automated scoring of liver fibrosis stages. Scientific Reports, 8, 16016. DOI 10.1038/s41598-018-34300-2.
115. Xiao, W., Huang, X., Wang, J. H., Lin, D. R., Zhu, Y. et al. (2021). Screening and identifying hepatobiliary diseases through deep learning using ocular images: A prospective, multicentre study. The Lancet Digital Health, 3(2), e88–e97. DOI 10.1016/S2589-7500(20)30288-0.
116. Frid-Adar, M., Diamant, I., Klang, E., Amitai, M., Goldberger, J. et al. (2018). GAN-Based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing, 321, 321–331. DOI 10.1016/j.neucom.2018.09.013.
117. Lu, H., Tian, S., Yu, L., Lv, X., Chen, S. (2020). Diagnosis of hepatitis B based on Raman spectroscopy combined with a multiscale convolutional neural network. Vibrational Spectroscopy, 107, 103038. DOI 10.1016/j.vibspec.2020.103038.
118. Wang, X., Tian, S., Yu, L., Lv, X., Zhang, Z. (2020). Rapid screening of hepatitis B using Raman spectroscopy and long short-term memory neural network. Lasers in Medical Science, 35(8), 1791–1799. DOI 10.1007/s10103-020-03003-4.
119. Zhu, H., Guo, Q., Li, M., Wang, C., Fang, Z. et al. (2020). Host and infectivity prediction of wuhan 2019 novel coronavirus using deep learning algorithm. BioRxiv, DOI 10.1101/2020.01.21.914044.
120. Fu, M., Yi, S. L., Zeng, Y., Ye, F., Li, Y. et al. (2020). Deep learning-based recognizing covid-19 and other common infectious diseases of the lung by chest CT scan images. MedRxiv. DOI 10.1101/2020.03.28.20046045.
121. Chae, S., Kwon, S., Lee, D. (2018). Predicting infectious disease using deep learning and big data. International Journal of Environmental Research and Public Health, 15(8), 1596. DOI 10.3390/ijerph15081596.
122. Wong, Z. S., Zhou, J., Zhang, Q. (2019). Artificial intelligence for infectious disease big data analytics. Infection, Disease & Health, 24(1), 44–48. DOI 10.1016/j.idh.2018.10.002.
123. Alnazer, I., Bourdon, P., Urruty, T., Falou, O., Khalil, M. et al. (2021). Recent advances in medical image processing for the evaluation of chronic kidney disease. Medical Image Analysis, 69, 101960. DOI 10.1016/j.media.2021.101960.
124. Wu, Y., Yi, Z. (2020). Automated detection of kidney abnormalities using multi-feature fusion convolutional neural networks. Knowledge-Based Systems, 200, 105873. DOI 10.1016/j.knosys.2020.105873.
125. Navaneeth, B., Suchetha, M. (2020). A dynamic pooling based convolutional neural network approach to detect chronic kidney disease. Biomedical Signal Processing and Control, 62, 102068. DOI 10.1016/j.bspc.2020.102068.
126. Bhaskar, N., Manikandan, S. (2019). A deep-learning-based system for automated sensing of chronic kidney disease. IEEE Sensors Letters, 3(10), 1–4. DOI 10.1109/LSENS.2019.2942145.
127. Khamparia, A., Saini, G., Pandey, B., Tiwari, S., Gupta, D. et al. (2020). KDSAE: Chronic kidney disease classification with multimedia data learning using deep stacked autoencoder network. Multimed. Tools Appl., 79, 35425–35440. DOI 10.1007/s11042-019-07839-z.
128. Ma, F., Sun, T., Liu, L., Jing, H. (2020). Detection and diagnosis of chronic kidney disease using deep learning-based heterogeneous modified artificial neural network. Future Generation Computer Systems, 111, 17–26. DOI 10.1016/j.future.2020.04.036.
129. Chen, G., Ding, C., Li, Y., Hu, X., Li, X. et al. (2020). Prediction of chronic kidney disease using adaptive hybridized deep convolutional neural network on the internet of medical things platform. IEEE Access, 8, 100497–100508. DOI 10.1109/ACCESS.2020.2995310.
130. Collins, F. S., Varmus, H. (2015). A new initiative on precision medicine. New England Journal of Medicine, 372(9), 793–795. DOI 10.1056/NEJMp1500523.
131. Li, Y., Lv, Z., Zhao, J., Pan, Z. (2019). Improving performance of medical image fusion using histogram, dictionary learning and sparse representation. Multimedia Tools and Applications, 78(24), 34459–34482. DOI 10.1007/s11042-019-08027-9.
132. Lv, Z., Piccialli, F. (2020). The security of medical data on internet based on differential privacy technology. ACM Transactions on Internet Technology (TOIT), 1–10. DOI 10.1145/3382769.
133. Kumar, R., Khan, A. A., Kumar, J., Zakria, A., Golilarz, N. A. et al. (2021). Blockchain-federated-learning and deep learning models for COVID-19 detection using CT imaging. IEEE Sensors Journal, 1–1. DOI 10.1109/JSEN.2021.3076767.
134. Hesamian, M. H., Jia, W., He, X., Kennedy, P. (2019). Deep learning techniques for medical image segmentation: Achievements and challenges. Journal of Digital Imaging, 32(4), 582–596. DOI 10.1007/s10278-019-00227-x.
135. Shen, D., Wu, G., Suk, H. I. (2017). Deep learning in medical image analysis. Annual Review of Biomedical Engineering, 19, 221–248. DOI 10.1146/annurev-bioeng-071516-044442.
136. Yang, X., Zhang, T., Xu, C., Yan, S., Hossain, M. S. et al. (2016). Deep relative attributes. IEEE Transactions on Multimedia, 18(9), 1832–1842. DOI 10.1109/TMM.2016.2582379.
137. Levine, A. B., Schlosser, C., Grewal, J., Coope, R., Jones, S. J. et al. (2019). Rise of the machines: Advances in deep learning for cancer diagnosis. Trends in Cancer, 5(3), 157–169. DOI 10.1016/j.trecan.2019.02.002.
138. Afzal, M., Islam, S. R., Hussain, M., Lee, S. (2020). Precision medicine informatics: Principles, prospects, and challenges. IEEE Access, 8, 13593–13612. DOI 10.1109/ACCESS.2020.2965955.
139. Llovet, J. M., Montal, R., Sia, D., Finn, R. S. (2018). Molecular therapies and precision medicine for hepatocellular carcinoma. Nature Reviews Clinical Oncology, 15(10), 599–616. DOI 10.1038/s41571-018-0073-4.
140. Le Tourneau, C., Borcoman, E., Kamal, M. (2019). Molecular profiling in precision medicine oncology. Nature Medicine, 25, 711–712. DOI 10.1038/s41591-019-0442-2.
141. Wang, Y., Sun, S., Zhang, Z., Shi, D. (2018). Nanomaterials for cancer precision medicine. Advanced Materials, 30(17), 1705660. DOI 10.1002/adma.201705660.
142. Fujiwara, N., Friedman, S. L., Goossens, N., Hoshida, Y. (2018). Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. Journal of Hepatology, 68(3), 526–549. DOI 10.1016/j.jhep.2017.09.016.
143. Zhang, S., Bamakan, S. M. H., Qu, Q., Li, S. (2018). Learning for personalized medicine: A comprehensive review from a deep learning perspective. IEEE Reviews in Biomedical Engineering, 12, 194–208. DOI 10.1109/RBME.2018.2864254.
144. Panayides, A. S., Pattichis, M. S., Leandrou, S., Pitris, C., Constantinidou, A. et al. (2018). Radiogenomics for precision medicine with a big data analytics perspective. IEEE Journal of Biomedical and Health Informatics, 23(5), 2063–2079. DOI 10.1109/JBHI.2018.2879381.
145. Loomans-Kropp, H. A., Umar, A. (2019). Cancer prevention and screening: The next step in the era of precision medicine. NPJ Precision Oncology, 3(1), 1–8. DOI 10.1038/s41698-018-0075-9.
146. Regel, I., Mayerle, J., Mahajan, U. M. (2020). Current strategies and future perspectives for precision medicine in pancreatic cancer. Cancers, 12(4), 1024. DOI 10.3390/cancers12041024.
147. Steuer, C. E., Ramalingam, S. S. (2018). Tumor mutation burden: Leading immunotherapy to the era of precision medicine. J. Clin. Oncol., 36(7), 631–632. DOI 10.1200/JCO.2017.76.8770.
148. Hamamoto, R., Suvarna, K., Yamada, M., Kobayashi, K., Shinkai, N. et al. (2020). Application of artificial intelligence technology in oncology: Towards the establishment of precision medicine. Cancers, 12(12), 3532. DOI 10.3390/cancers12123532.
149. Ahmed, Z., Mohamed, K., Zeeshan, S., Dong, X. (2020). Artificial intelligence with multi-functional machine learning platform development for better healthcare and precision medicine. Database, 2020, baaa010. DOI 10.1093/database/baaa010.
150. Hulsen, T., Jamuar, S. S., Moody, A. R., Karnes, J. H., Varga, O. et al. (2019). From big data to precision medicine. Frontiers in Medicine, 6, 34. DOI 10.3389/fmed.2019.00034.
151. Prosperi, M., Min, J. S., Bian, J., Modave, F. (2018). Big data hurdles in precision medicine and precision public health. BMC Medical Informatics and Decision Making, 18, 139. DOI 10.1186/s12911-018-0719-2.
152. Hey, S. P., Gerlach, C. V., Dunlap, G., Prasad, V., Kesselheim, A. S. (2020). The evidence landscape in precision medicine. Science Translational Medicine, 12(540). DOI 10.1126/scitranslmed.aaw7745.
153. Dienstmann, R., Vermeulen, L., Guinney, J., Kopetz, S., Tejpar, S. et al. (2017). Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nature Reviews Cancer, 17(2), 79–92. DOI 10.1038/nrc.2016.126.
154. Dayem Ullah, A. Z., Oscanoa, J., Wang, J., Nagano, A., Lemoine, N. R. et al. (2018). SNPnexus: Assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Research, 46(W1), W109–W113. DOI 10.1093/nar/gky399.
155. Tebeje, T. H., Klein, J. (2020). Applications of e-health to support person-centered health care at the time of COVID-19 pandemic. Telemedicine and E-Health, 27, 150–158. DOI 10.1089/tmj.2020.0201.
156. Hossain, M. S. (2017). Cloud-supported cyber–Physical localization framework for patients monitoring. IEEE Systems Journal, 11(1), 118–127. DOI 10.1109/JSYST.2015.2470644.
157. Bisio, I., Garibotto, C., Lavagetto, F., Sciarrone, A. (2019). When eHealth meets IoT: A smart wireless system for post-stroke home rehabilitation. IEEE Wireless Communications, 26(6), 24–29. DOI 10.1109/MWC.001.1900125.
158. Dubey, H., Monteiro, A., Constant, N., Abtahi, M., Borthakur, D. et al. (2017). Fog computing in medical internet-of-things: Architecture, implementation, and applications. in Handbook of Large-Scale Distributed Computing in Smart Healthcare, pp. 281–321. Cham: Springer.
159. Moore, C. (2020). Medical internet of things-based healthcare systems: Wearable sensor-based devices, patient-generated Big data, and real-time clinical monitoring. American Journal of Medical Research, 7(1), 41–47. DOI 10.22381/AJMR7120206.
160. Harvey, P., Toutsop, O., Kornegay, K., Alale, E., Reaves, D. (2020). Security and privacy of medical internet of things devices for smart homes. 2020 7th International Conference on Internet of Things: Systems, Management and Security (IOTSMSpp. 1–6. Paris, France, IEEE.
161. Hossain, M. S., Muhammad, G. (2017). Emotion-aware connected healthcare big data towards 5G. IEEE Internet of Things Journal, 5(4), 2399–2406. DOI 10.1109/JIOT.2017.2772959.
162. Zhou, Z., Yu, H., Shi, H. (2020). Human activity recognition based on improved Bayesian convolution network to analyze health care data using wearable IoT device. IEEE Access, 8, 86411–86418. DOI 10.1109/ACCESS.2020.2992584.
163. Fouad, H., Hassanein, A. S., Soliman, A. M., Al-Feel, H. (2020). Analyzing patient health information based on IoT sensor with AI for improving patient assistance in the future direction. Measurement, 159, 107757. DOI 10.1016/j.measurement.2020.107757.
164. Hossain, M. S., Muhammad, G., Alamri, A. (2019). Smart healthcare monitoring: A voice pathology detection paradigm for smart cities. Multimedia Systems, 25, 565–575. DOI 10.1007/s00530-017-0561-x.
165. Al-Garadi, M. A., Mohamed, A., Al-Ali, A. K., Du, X., Ali, I. et al. (2020). A survey of machine and deep learning methods for internet of things (IoT) security. IEEE Communications Surveys & Tutorials, 22(3), 1646–1685. DOI 10.1109/COMST.2020.2988293.
166. Alloghani, M., Baker, T., Al-Jumeily, D., Hussain, A., Mustafina, J. et al. (2020). Prospects of machine and deep learning in analysis of vital signs for the improvement of healthcare services. Nature-Inspired Computation in Data Mining and Machine Learning, pp. 113–136. Cham: Springer.
167. Wang, T., Lei, Y., Tian, Z., Dong, X., Liu, Y. et al. (2019). Deep learning-based image quality improvement for low-dose computed tomography simulation in radiation therapy. Journal of Medical Imaging, 6(4), 043504. DOI 10.1117/1.JMI.6.4.043504.
168. Mohan, S., Thirumalai, C., Srivastava, G. (2019). Effective heart disease prediction using hybrid machine learning techniques. IEEE Access, 7, 81542–81554. DOI 10.1109/ACCESS.2019.2923707.
169. Liu, Y., Kang, Y., Xing, C., Chen, T., Yang, Q. (2020). A secure federated transfer learning framework. IEEE Intelligent Systems, 35(4), 70–82. DOI 10.1109/MIS.2020.2988525.
170. Chen, Y., Qin, X., Wang, J., Yu, C., Gao, W. (2020). Fedhealth: A federated transfer learning framework for wearable healthcare. IEEE Intelligent Systems, 35(4), 83–93. DOI 10.1109/MIS.2020.2988604.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |